Abstract
In recent years there has been an explosion of interest in how genes regulate alcohol drinking and contribute to alcoholism. This work has been stimulated by the completion of the human and mouse genome projects and the resulting availability of gene microarrays. Most of this work has been performed in drinking animals, and has utilized the extensive genetic variation among different mouse strains.
At the same time, a much smaller amount of effort has gone into the in vitro study of the mechanisms underlying the regulation of individual genes by alcohol. These studies at the cellular and sub-cellular level are beginning to reveal the ways in which alcohol can interact with the transcriptional, translational and post-translational events inside the cell. Detailed studies of the promoter regions within several individual alcohol-responsive genes (ARGs) have been performed and this work has uncovered intricate signaling pathways that may be generalized to larger groups of ARGs.
In the last few years several distinct ARGs have been identified from 35,000 mouse genes, by both the “top-down” approach (ex vivo gene arrays) and the “bottom-up” methods (in vitro promoter analysis). These divergent methodologies have converged on a surprisingly small number of genes encoding ion channels, receptors, transcription factors and proteins involved in synaptic function and remodeling. In this review we will describe some of the most interesting cellular and microarray work in the field, and will outline specific examples of genes for which the mechanisms of regulation by alcohol are now somewhat understood.
Keywords: Ethanol, Gene Expression, Microarray, Heat Shock Proteins, Transcription, Neurons
1. Introduction – biological adaptation to alcohol
Alcohol is an anxiolytic and sedative drug that has a variety of effects on human behavior. Drinking alcohol can have both beneficial and negative health effects in adults, with moderate drinking being associated with a lower incidence of cardiovascular problems such as stroke, while higher levels of alcohol use are associated with liver and kidney disease. In addition, chronic heavy drinking can lead to problems of physical dependence and tolerance that result from physiological adaptation to the drug and are broadly similar to those associated with addiction to other drugs of abuse.
Alcohol drinking by the human mother is also hazardous to the fetus. Even moderate alcohol drinking during pregnancy can result in a variety of birth defects described as fetal alcohol syndrome (FAS) or alcohol-related developmental syndrome (ARDS). A great deal of work has been done on the cellular and molecular mechanisms of FAS and ARDS, and it is not our intention to review this area here. The reader is referred to several excellent reviews on FAS (Goodlett et al., 2005; Sulik, 2005). In general, we will discuss the results of work done on adult animals and post mortem human brains, except for some studies carried out with cultured embryonic neurons.
Despite the negative social and health effects of alcoholism, people have consumed the drug for thousands of years (Li et al., 1977). Until very recently, however, the mechanisms by which alcohol affects the central nervous system (CNS) were poorly understood and its actions were considered to be “non-specific” in nature. Recent advances in neuroscience have led to significant progress in our understanding of the neural basis of alcohol intoxication and alcoholism (Chandler et al., 1998; Faingold et al., 1998). In particular, the earlier notion that alcohol exerts its effects purely on membrane lipids has been shown to be inconsistent with the experimental evidence accumulated during the last twenty years.
The remarkable progress of research in cellular and molecular neuroscience has resulted in some spectacular advances for alcohol research, most notably the identification of a number of candidate target molecules within the CNS that are sensitive to levels of alcohol relevant to acute human intoxication. Advances in our understanding of the mechanisms of neuronal excitability and synaptic transmission have shown that ethanol interacts with a variety of neurotransmitter systems, including glutamate (Lovinger et al., 1989; Valenzuela and Cardoso, 1999; Woodward, 2000; Crowder et al., 2002), dopamine (Gessa et al., 1985, Imperato and Di Chiara, 1986; Brodie et al., 1990, Budygin et al., 2001), adenosine (Dar et al., 1983; Dunwiddle, 1996), and γ-aminobutyric acid [GABA] (Wu et al., 1995; Charlton et al., 1997; Cagetti et al., 2003; Sanna et al., 2003; Follesa et al., 2004a; Follesa et al., 2004b; reviewed in: Kumar et al. 2009). Actions on all of these systems may converge to produce the varied behavioral effects of acute alcohol ingestion.
As far as the effects of chronic alcohol intake are concerned, a consensus has formed in the field that the development of alcohol tolerance and dependence result from alterations in brain structure and function over time. This most likely involves the remodeling of synaptic connections that are dependent upon changes in gene expression, initiated by the presence of alcohol (Wilke et al., 1994). In this respect, the adaptations to alcohol resemble to some extent the brain plasticity that is known to occur during long-term exposure to other drugs of abuse, such as cocaine and heroin (Koob, 2004). Brain areas of interest to researchers in the field are therefore those associated with the brain's reward system (ventral tegmental area, nucleus accumbens) as well as the circuits associated with impulse control (prefrontal cortex), levels of arousal (thalamus) and anxiety (amygdala).
In the case of alcohol dependence, it is presumed that a series of functional physiological changes are induced in the brain by alcohol intake that can profoundly alter the activity of the nervous system, and these changes are manifested both psychologically and physiologically in the alcoholic. The development of alcoholism and addiction to other drugs occurs in several stages. The initial behavioral changes are associated with a transition from social drug use to more compulsive drug-seeking, and are proposed to involve neuroplasticity in pathways involved in the brain's reward system (Kalivas and O'Brien, 2008). These changes in brain function often manifest as alterations in decision-making by the individual, which result in increased alcohol intake and frequency of drinking. These initial functional changes may then be consolidated over time so that the CNS becomes “hard-wired” for a drug-seeking state that is maintained over a long period (addiction), and there is evidence that cycles of alcohol withdrawal lead the individual to experience the negative reinforcement properties of alcohol that result in sensitization and enhance drug-seeking behavior and excessive drinking (Becker, 1998; 2000). As heavier drinking becomes less episodic and more routine, eventually more profound physiological changes occur that lead to a state of enhanced CNS excitability (physical dependence), perhaps in order to counteract the depressant actions of alcohol on the brain. The extensive nature of this rewiring is clearly revealed in the hyperirritability, tremor and sympathetic activation seen in alcoholics during withdrawal from the drug (Becker, 1998; 2000). The intense withdrawal symptoms seen in long-term alcoholics provide strong evidence for such adaptations; normal CNS activity simply becomes impossible when alcohol is withdrawn from the chronic drinker, a state known as physical dependence. In addition, the physical manifestations of alcohol withdrawal syndrome (delirium tremens, extreme anxiety, hyperalgesia, autonomic activation, physical irritability, and incidence of seizures) strongly suggest a shift towards increased neuronal excitability in the alcoholic brain (Veatch and Gonzales, 1996). Extreme examples of this process of physiological adaptation to alcohol are observed in the biology of alcoholic individuals. Some long-term alcoholics routinely tolerate extremely high blood alcohol concentrations (>100mM), which would cause severe intoxication, profound sedation or death in naïve individuals (Urso et al., 1981). The development of this extreme behavioral tolerance to alcohol shows that the CNS of the alcoholic undergoes significant functional adaptations in order to cope with the continued presence of the drug.
These profound adaptations, however, presumably result from a complex chain of events that is initiated in the brain of the drinker long before a state of dependence is reached. Several studies have shown that even brief exposure to alcohol in animal models can modify the expression of a variety of genes (Bachtell et al., 1999). For example, it has been demonstrated in several strains of mice that a single exposure to low doses of ethanol (1 to 3 g/kg) can induce the expression of immediate-early genes (IEGs) such as c-fos (Zoeller and Fletcher, 1994; Hitzemann and Hitzemann, 1997; Demarest et al., 1999; Eiseman et al., 2002). The IEGs encode nuclear proteins with transcriptional activities that in turn regulate the expression of many other genes. The activation of these transcription factors may be the earliest events in a sequence of changes in gene expression that ultimately result in functional alterations of critical brain circuitry. These functional changes manifest themselves in the development of behavioral tolerance and adaptation seen in the advanced stages of chronic alcoholism.
The field has greatly benefited from recent developments in technology that have enabled many investigators to adopt a “global” approach to study the alterations of gene expression induced by alcohol, using gene microarrays to screen an entire genome and to identify large numbers of alcohol-responsive genes (ARGs) (e.g. Lewohl et al., 2000; Mulligan et al., 2006). Microarray studies performed on post mortem human brain of subjects with a long history of alcohol abuse and dependence showed profound changes in the expression of many genes, including those encoding cell adhesion proteins, myelin-related proteins, components of the protein synthesis and the ubiquitinproteasome protein degradation systems, mitochondrial and energy metabolism proteins, and proteins involved in signal transduction and gene transcription. All of these changes are consistent with the concept of extensive alcohol-induced adaptation of the CNS (Lewohl et al., 2000; Sokolov et al., 2003; Liu et al., 2004).
2. Regulation of neuronal gene expression
Neuronal gene expression is constantly modulated in a precise, coordinated manner throughout development, and in response to external stimuli. This regulation can occur on two levels: transcriptional control of mRNA synthesis, and by post-transcriptional modifications that include mRNA splicing and editing, as well as control of mRNA stability, translation and translocation. Functional dissection of neuronal gene promoters has elucidated some of the mechanisms underlying transcriptional control, including critical mechanisms that enable gene silencing in non-neuronal cells and gene activation in specific neuronal sub-populations (Quinn, 1996).
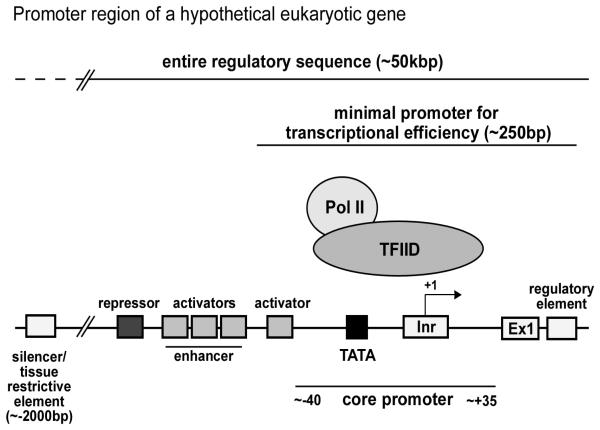
The typical eukaryotic gene structure contains the promoter core, a short region of 70-80 bp immediately spanning the transcription start site, and a subset of canonical DNA motifs (6-12 bp) that recruit the basal transcription factors necessary for initiation. In general, the multi-subunit complex transcription factor for RNA polymerase IID (TFIID) binds in a sequence-specific manner to one or several of these promoter elements and actively recruits the Pol II holoenzyme (containing RNA polymerase II and other associated proteins) to initiate transcription. It is important to note that while there is no universal core promoter sequence, there are several known DNA motifs and, undoubtedly, more remain to be discovered (reviewed in: Smale and Kadonaga, 2003).
The TATA box is usually located −31 to −24 bp upstream of the transcription start site and interacts specifically with the TATA-box binding protein (TBP) subunit of TFIID (Fig. 1). Another common motif, the initiator element (Inr), spans the initiation site (−2 to +5 bp) and is recognized by the TATA box binding protein (TBP)-associated factor 2 (TAF2) subunit of TFIID. While TATA box and Inr motifs are able to initiate transcription individually, both operate in conjunction with other core promoter elements (reviewed in: Smale and Kadonaga, 2003; Thomas and Chiang, 2006). These regions can either stimulate or repress transcriptional activity, depending on the context of the interactions. This core promoter region, primarily responsible for recruiting Pol II, is often transcriptionally silent and requires additional activating elements to elicit basal levels of transcription. Defined as the minimal promoter for transcriptional efficiency, this region spans ~250 bp across the transcription start site. In particular, the GC-box and/or CCAAT-box motifs located in the region −100 to −60 bp upstream of initiation are common activators that can cluster to form an enhancer (Thomas and Chiang, 2006). Additional regulatory elements are located more distally and can function as binding sites for activator and repressor proteins; these operate synergistically with the minimal promoter to modulate the rate of transcription. In general, these transcription factors fall into two classes: those that enhance the transcription rate above basal levels and those that either repress or silence transcription.
Figure 1.
Schematic diagram of a hypothetical eukaryotic gene promoter region with the canonical elements and binding proteins that regulate gene expression. In general, the TFIID complex binds to one or several of these core promoter elements to actively recruit the Pol II holoenzyme and initiate transcription. The TATA box and Inr elements are able to initiate transcription individually, but often operate synergistically with other motifs to recruit TFIID. This core promoter region is often transcriptionally silent and with additional activating elements forms the minimal promoter for transcriptional efficiency. [Adapted from Smale and Kadonaga, 2003; Deng and Roberts 2006; Thomas and Chiang, 2006; and Sadhale et al., 2007.]
In the simplest level of regulation, activators and repressors bind to their corresponding elements relatively near the transcription start site and either physically promote or inhibit local transcription. As such, these elements are classical, position-independent and direct active regulation. In particular, repressors can compete for activator binding sites, directly interact with an activator's activating region or indirectly interfere with subunits of the transcriptional machinery. In contrast, silencing is a positional effect, in which a gene is repressed because of its location, not in response to a specific environmental factor. This passive regulatory mechanism often spreads over large stretches of DNA, switching off several genes that may be distant from the actual silencer element. These elements have been shown to restrict promoter activity to the appropriate physiological cell type (such as neurons), in both transient expression systems and in transgenic mouse models (Quinn, 1996). Since the promoter sequences of eukaryotic genes contain multiple binding sites for transcription factors, each gene responds to multiple signaling pathways in a coordinated manner to facilitate the fine-tuning of transcript levels. Therefore, the complement and relative positioning of all of these diverse elements define the promoter architecture, and presumably determine how a given promoter responds to the many transcription factors in the cell.
3. Molecular mechanisms of gene regulation by ethanol
Despite numerous studies on the regulation of gene expression by alcohol, there are very few examples of genes for which the mechanisms by which ethanol alters transcription are known. In this section, we will summarize the most detailed studies of ARGs, in which detailed analysis of the promoter regions for these genes have revealed that ethanol exposure activates signaling pathways that result in changes in gene expression, and that in turn may be generalized to other ARGs.
3.1. Neuron-restrictive silencer element (NRSE) or repressor element-1 (RE-1)
The excitatory neurotransmitter L-glutamate acts on a variety of ionotropic and metabotropic receptors, notably the α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate (AMPA)-preferring and N-methyl-D-aspartate (NMDA)-preferring receptors. The NMDA receptor forms a ligand-gated ion channel; channel activation requires simultaneous binding of two co-agonists, glutamate and glycine, and the receptor is also “gated” by virtue of a voltage-dependent block by Mg2+ (Mayer and Westbrook, 1984; Nowak et al., 1984). Removal of the Mg2+ block by membrane depolarization allows influx of Na+ and Ca2+, leading to additional depolarization and generating a slow excitatory postsynaptic potential (EPSP) in the neuron (Forsythe and Westbrook, 1988).
Acute ethanol exposure has long been known to inhibit the excitatory action of glutamate at NMDA receptors (Lovinger et al., 1989; see also: Hoffman et al., 1989; Nie et al., 1994; Martin et al., 1995; Tabakoff and Hoffman, 1996; Tsai and Coyle, 1998; Calton et al., 1998; Woodward, 1999; Criswell et al., 2003; Loftis and Janowsky, 2003; Roberto et al., 2004), while chronic treatment with ethanol appears to up-regulate NMDA receptor expression in the brain (Gulya et al., 1991; Hu and Ticku, 1995; Kumari and Ticku, 2000; Nagy et al., 2004). This increase in receptor expression presumably offsets the effects of acute ethanol and may be one of the mechanisms that contributes to hyperexcitability during alcohol withdrawal (Nelson et al., 2005; Hendricson et al., 2007).
The NMDA receptor occurs in the brain as a heteromeric complex composed of the essential NMDAR1 (NR1) subunit in combination with NMDAR2 (NR2A-D) subunits, although additional NMDAR3 (NR3A-B) subunits have been reported (reviewed in: Cull-Candy et al., 2001). The stoichiometry of the receptor is a tetramer of two NR1 and two NR2 subunits. Selective splicing of NR1 transcripts and the differential expression of NR2 subunits leads to the generation of multiple receptor isoforms with distinct regional distributions and biophysical and pharmacological properties.
Detailed investigation into the effects of ethanol on NMDA subunit expression has revealed that the NR2B subunit, in particular, is highly regulated by ethanol (Follesa and Ticku, 1996; Hu et al., 1996; Kumari and Ticku, 1998; 2000). Analysis of the regulatory sequence of the NR2B gene in the mouse (Grin2b) has uncovered an upstream 800 bp region that is sufficient to direct neural-specific gene expression in a transgenic mouse. This region extends from 550 bp upstream of the first transcription site into the first 255 bp of the non-coding region (5′-UTR) of exon 1 (Sasner and Buonanno, 1996). The minimal promoter for activity is found in the region approximately 170 bp upstream of the first exon, and drives reporter gene expression in both neuronal and non-neuronal cell lines (Klein et al., 1998). Further upstream are several gene expression regulatory elements, including five highly conserved putative neuron-restrictive silencer element (NRSE) binding sites between −1407 to −2741 bp (Qiang et al., 2005; Fig. 2A).
Figure 2.
Location of the 5 putative NRSE, AP-1 and CRE cis-element in the 5′-flanking region of the NR2B (Grin2b) gene. A, Schematic diagram and relative location of the five putative NRSE sites present in the mouse Grin2b gene. The boxes with the numbers 1-5 represent the location of these elements. Below are the sequence and the specific location of each of these elements with the consensus nucleotides indicated in bold letters. Grin2b also poses an AP-1 and a CRE site proximal to the transcription initiation site as indicated in the diagram. The specific location and consensus sequence is indicated in the box below the promoter diagram. B, Determination of cis-acting elements involved in ethanol (E) response in Grin2b promoter. A set of Grin2b promoter regions differing in location and length were cloned in a promoterless plasmid containing the firefly luciferase reporter gene and several deletions and mutations were generated. Constructs were transfected into cortical neurons and firefly luciferase activity assayed and normalized to the activity of the co-transfected control plasmid containing Renilla luciferase. Two-way ANOVA revealed a significant sequence-dependent repression and ethanol-response effect compared to wild type for the NRSE sites. Mutation of the AP-1 site significantly decreased the expression of luciferase while mutation of CRE site reduce the stimulatory effects of ethanol (P < 0.05 by two-way ANOVA with post hoc test). Data are displayed as the mean ± s.e.m. [Reprinted with permission from Rani et al., 2005; Qiang et al., 2005; Qiang and Ticku, 2005].
The NRSE (repressor element-1, RE-1), has been identified as a key regulator in eukaryotic gene regulation (Ogbourne and Antalis, 1998). The NRSE blocks the expression of numerous neural-specific genes in non-neural cells (Mori et al., 1990, 1992; Kraner et al., 1992; Chong et al., 1995; Schoenherr and Anderson, 1995; Paquette et al., 2000). Examples of this type of non-neuronal repression include the silencing of genes that encode the voltage-dependent sodium channel (Chong et al., 1995), synapsin I (Li et al., 1993), the m4 muscarinic receptor (Wood et al., 1996), and the glutamate receptor subunits GluR2 (Huang et al., 1999) and NR2B (Qiang et al., 2005). Several studies have shown that this NRSE-mediated transcriptional repression of neuron-specific genes acts via interaction with the neuron-restrictive silencer factor (NRSF) or repressor element-1 silencing transcription factor (REST) (Chong et al., 1995; Schoenherr and Anderson, 1995; Wood et al., 1996; Bessis et al., 1997; Palm et al., 1998; Huang et al., 1999; Palm et al., 1999).
An analysis of the NR2 gene using electrophoretic mobility shift assay (EMSA) and reporter gene assays identified NRSF interaction with the NRSE2 and 3 binding sites as responsible for repression of the NR2B gene. The same study found that 100 mM ethanol treatment of neurons for 5 days reduces NRSF expression, causing decreases in NRSF/NRSE binding activity and an increase in NR2B promoter activation (Qiang et al., 2005). The expression of NRSF itself is developmentally controlled, with the highest levels seen during embryogenesis, and then declining with age, but little else is known about NRSF regulation (Palm et al., 1998). It is not clear at present how ethanol reduces NRSF expression to release the inhibition of the NR2B gene.
3.2. Regulation via cAMP-response element (CRE) and its complementary binding protein (CREB)
Two prominent genes were identified in neuroblastoma cell lines whose ethanol-induced regulation occurs via cyclic AMP (cAMP)-response element (CRE) sites; these genes are tyrosine hydroxylase (TH) (Gayer et al., 1991) and dopamine β-hydroxylase (DBH) (Hassan et al., 2003). The enzymes encoded by these two genes are critical for neurotransmitter synthesis and are therefore, of obvious importance to the neuropharmacology of alcohol. Deletion analysis of the 5′-proximal region (−262 to −142 bp) of the human DBH gene revealed that the induction of gene expression by alcohol is controlled by a pathway involving the interaction of the complementary binding protein (CREB) with CRE sites. These findings have been extended to a wider set of alcohol-sensitive genes that contain CRE sequences within their promoter regions, including TH (Hassan et al., 2003). The Grin2b promoter also contains a CRE site (Fig. 2A), located in the upstream region −410 to −403 bp, which is involved in the regulation of gene expression by ethanol (Klein et al., 1998). Mutation of this site prevents interaction with CREB, and abolishes the stimulatory effect of chronic ethanol treatment on Grin2b expression (Rani et al., 2005; Fig. 2B). In addition, ethanol treatment is known to increase levels of activated CREB by mediating its Ser133 phosphorylation via environmental-regulated kinase (ERK) (Rani et al., 2005), thereby regulating the binding of CREB to the CRE and hence modulating gene expression.
3.3. Specificity protein-1 (Sp1) and activator protein-1 (AP-1) elements
Ethanol treatment also regulates the expression of genes encoding several classes of molecular chaperones and proteins that bind to nascent polypeptides to facilitate correct folding (Miles et al., 1991; 1994; Hsieh et al., 1996; Wilke et al., 2000). Miles and colleagues investigated ethanol-induced changes in heat shock cognate protein 70 (Hsc70, also known as Hsp73) gene expression in neuroblastoma-glioma hybrid cells (NG108-15 cells). Hsc70 is a member of the heat shock protein (Hsp) family, and has 85% identity with human Hsp70. It shows both constitutive and heat-inducible expression.
The promoter of the Hsc70 gene has three putative specificity protein-1 (Sp1) transcription factor-binding sites localized at −263, −173 and −67 bp upstream from the transcription initiation site, two sets of overlapping heat-shock regulatory elements (HSE) located between −271 and −193 bp, and −63 and −39 bp, two inverted CCAAT boxes at −420 and −79 bp and a TATA box at −32 bp (Fig. 3A). The minimal promoter required for constitutive transcriptional efficiency and for heat shock-induced expression in HeLa cells is contained within a region 343 bp upstream of the transcription initiation site (Sorger and Pelham, 1987). Deletion analysis of a Hsc70 promoter-reporter construct transiently expressed in NG108-15 cells also showed that the upstream proximal 74 bp promoter region was strongly regulated by ethanol, suggesting that ethanol-responsive sequences should be contained within that region (Fig. 3B).
Figure 3.
Organization of the Hsc70 promoter region and identification of conserved regulatory elements. A, The diagram presents the relative position of regulatory elements in the rat Hsc70 gene. This gene contains three putative Sp1 sites, four overlapping HSE sites, two inverted CCAAT boxes and a TATA box. The specific sequences and location of all these sites is indicated in the boxes below the graph with the consensus sites shown bold letters. B, Identification of cis-acting ethanol responsive sequence in Hsc70 promoter. A plasmid containing −2500 to +60 bp of the Hsc70 region was coupled to a chloramphenicol acetyl-transferase (CAT) reporter gene and several deletions were generated. Constructs were transfected into NG108-15 cells and CAT activity assayed and presented as percentage vs. control (cells without ethanol). Data are displayed as mean ± s.e.m. (n > 6) compared by single-group t test with Bonferroni correction for multiple group (for differences from control cells) or ANOVA with post hoc analysis to determine difference between constructs. *P < 0.05 vs. control (t test); ** P < 0.05 vs. −74 bp construct (ANOVA). [Reprinted with permission from Sorger and Pelham, 1987; Wilke et al., 2000].
In fact, the Sp1 site located at −67 bp is crucial for alcohol regulation. By co-transfection of the Hsc70 −74 bp promoter-reporter construct and a construct expressing Sp1 transcription factor into cells naturally devoid of Sp1, it was demonstrated that Sp1 protein can enhance the increased expression of this gene in response to alcohol (Wilke et al., 2000). Studies with promoter-reporter constructs containing one or several Sp1 sites without the surrounding promoter structure reveal that the Sp1 site is necessary, but not sufficient, for ethanol regulation. Instead, the sequence context of the promoter where the Sp1 is localized is important for ethanol regulation, suggesting that the Sp1 factor may need to interact with other transcription factors or co-factors to mediate alcohol sensitivity (Wilke et al., 2000). The overlap of two HSE sites with the Sp1 site at −67 bp could mean that an interaction of Sp1 with the transcription factor, heat shock factor 1 (Hsf1) is necessary to confer the observed ethanol sensitivity.
The Grin2b promoter also includes four Sp1 sites located at −504, −346, −131 and −30 bp (Klein et al., 1998) and an activator protein 1 (AP-1) consensus sequence from −1107 to −1084bp (Qiang and Ticku, 2005; Fig. 2A). Although these Sp1 sites have now been implicated in alcohol regulation of this gene, chronic treatment of neurons with ethanol (75 mM; 5 days) also increased the binding activity of the AP-1 complex to its cis-acting element and enhanced the activity of the promoter construct (Fig. 2B). Since a variety of dimerized c-Fos, FosB, c-Jun, JunD and pCREB proteins comprise the AP-1 family (Qiang and Ticku, 2005), the mechanism by which alcohol stimulates AP-1 binding to its promoter element is presently unknown. It is likely that ethanol changes the expression and/or phosphorylation of the proteins involved, thereby affecting the ratios of the different dimers in the AP-1 complex.
3.4. Alcohol response element (ARE) and heat shock factor 1 (Hsf1)
Although the behavioral effects of alcohol present a complex picture, many of its actions are shared by drugs that modulate the actions of GABA; for example, the benzodiazepines produce a similar spectrum of anxiolytic, hypnotic and sedative effects to alcohol (Koob, 2003). For this reason, it has long been hypothesized that ethanol achieves some of its behavioral effects via regulation of the GABAA receptor (GABAA-R; reviewed in: Kumar et al. 2009). The GABAA-R is a member of the ligand-gated ion channel superfamily that mediates fast inhibitory synaptic transmission and also a form of tonic extrasynaptic inhibition in the CNS (Farrant and Nusser, 2005). The GABAA-R is a heteromeric assembly of five subunits that is encoded by nineteen genes (Barnard et al., 1998). Most of these genes contain a minimum of nine exons, and each typically has at least one very large intron of ~10-50 kb towards the 3′-end of the gene (e.g. Gabrb3; Kirkness and Fraser, 1993). Studies on ethanol-induced GABAA-R subunit gene expression changes have found contradictory results, with two exceptions: the gene encoding the α1 subunit (Gabra1) is modestly down-regulated by alcohol, and the α4 subunit gene (Gabra4) shows a remarkable degree of up-regulation. (Devaud et al., 1995; 1997; Petrie et al., 2001; Sanna et al., 2003; Cagetti et al., 2003; Liang et al., 2006; 2007; 2008).
Receptors that contain the α4 subunit are expressed at high levels in neurons of the thalamus, striatum and dentate gyrus and at more modest levels in the cerebral cortex (Pirker et al., 2000; Roberts et al., 2005; 2006). α4 subunits generally combine with δ subunits to form extra-synaptic GABAA-Rs, which are sensitive to very low concentrations of ambient GABA (Farrant and Nusser, 2005; Jia et al., 2005; Chandra et al., 2006). Levels of Gabra4 mRNA and its corresponding protein are up-regulated by chronic ethanol administration and/or subsequent withdrawal, in both in vivo and in vitro studies (Devaud et al., 1995; 1997; Cagetti et al., 2003; Sanna et al., 2003, reviewed in: Worst and Vrana, 2005; and see also: Liang et al., 2007; Kumar et al. 2009). This indicates an important potential role for Gabra4 in mediating the effects of ethanol and in homeostatic adaptation to the continued presence of the drug.
The minimal promoter required for the efficient transcription of Gabra4 is a 450 bp sequence that spans −444 to 19 bp across the transcription start site and lacks a TATA-box. While two Inr sites are found at −488 and −86 bp, neither is likely to be involved with initiation since they are located far from the transcription start site (Ma et al., 2004; (Fig. 4A). Although this Gabra4 promoter shows high activity in cultured mouse cortical neurons, it does not appear to possess a NRSE site for the direction of neural-specific expression, and is not sensitive to alcohol. This promoter has a binding site for the transcription factor Egr3 (located at −50 bp in the rat) that appears to be critical for increased Gabra4 expression after seizure activity (Roberts et al., 2005). Egr3 expression is activated by brain-derived neurotrophic factor (BDNF) via protein kinase C and MAP kinase-dependent pathways, causing an increase in Gabra4 expression (Roberts et al., 2006).
Figure 4.
The ARE is essential for ethanol (ETOH) and heat sensitivity of the Gabra4 promoter in cultured cortical neurons. A, Schematic of the basal Gabra4 promoter-reporter construct (pLuc-P7), shown together with the extended construct (pLuc-P7-EX2) and a third construct (pLuc-P7-EX2M), containing a mutated ARE sequence. The ARE sequence, shown in bold and underlined, is aligned to the consensus sequence found in ethanol sensitive genes in C. elegans. B, The extended Gabra4 promoter is sensitive to EtOH. 60 mM EtOH increased relative luc activity in neurons transfected with pLuc-P7-EX2 but not pLuc-P7 or pLuc-EX2M ((*P < 0.05, ***P < 0.001, significantly different from pLuc-P7/pLuc-P7-EX2M by one-way ANOVA with Tukey's Multiple Comparison post hoc test, all pair of columns compared, n ≥ 8). The values are mean ± s.e.m. expressed as percentage of increase vs. control (cells treated with vehicle). C, EtOH induces the formation of Hsf1 “stress granules” in the nucleus of cortical neurons in culture. Immunocytochemistry of cortical neurons stained with anti-Hsf1 antibody (red) and DAPI (blue) reveals the translocation of Hsf1 to the nucleus and the formation of aggregates or “stress granules”. The scale bar represents 5 μm. The graph shows the quantification of the number of Hsf1 granules per cell nucleus (by one-way ANOVA with Dunnett's Multiple Comparison post hoc test vs. control cells, n ≥ 20 cells from two independent cultures). Data are the mean ± s.e.m. (**P <0.01). [Adapted from Pignataro et al., 2007].
The minimal Gabra4 promoter described above, drives expression of the luciferase reporter gene in mouse cortical neurons (pLuc-P7) but is insensitive to ethanol. An extended construct, containing the minimal promoter and an additional downstream sequence spanning the first and second exons, is needed to generate a response to acute ethanol treatment (pLuc-P7-EX2, Fig. 4B; Pignataro, et al., 2007). At the end of exon 2 there is an 11-base sequence that is extremely similar to a consensus sequence found in a subset of C. elegans genes that respond to ethanol treatment (Kwon et al., 2004) [tCTGcGTCtCt, where uppercase letters indicate absolute conservation and lower case letters denote a degree of degeneracy at the position]. We have termed this sequence the alcohol response element (ARE) (Fig. 4A).
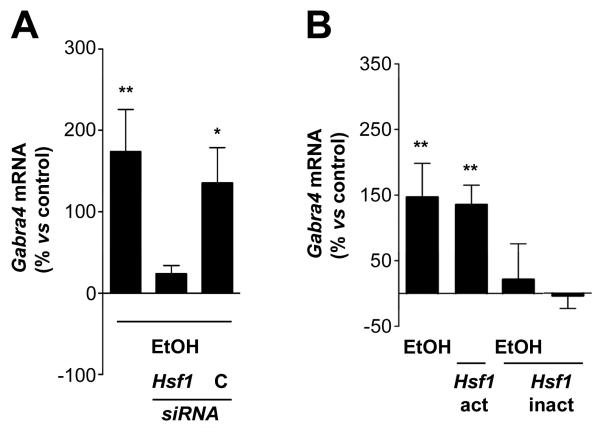
A genome-wide search in C. elegans for this element revealed that many of the genes that contain this sequence in their promoter region are members of the broad family of heat shock proteins, which are known to be activated by stress in many organisms. Mutation of the ARE in the extended Gabra4 promoter-reporter construct completely abolished the sensitivity of the gene to ethanol. Further analysis revealed that the ARE sequence is a consensus binding site for the transcription factor heat shock factor 1 (Hsf1). EMSA and “super-shift” experiments confirmed the binding of Hsf1 to the ARE sequence present in Gabra4 and demonstrated that this binding activity was enhanced by acute treatment with ethanol. Alcohol induced the translocation of Hsf1 from the cytoplasm to the nucleus of neurons, a known pre-requisite for the activation of Hsf1-dependent genes (Morimoto et al., 1998), and also caused nuclear aggregation of Hsf1 into stress granules (Cotto et al., 1997; Fig. 4C). Inhibition of Hsf1 protein expression with small interfering RNA (siRNA) reduced the ethanol-induced increase in Gabra4 transcription, while transfection of cortical neurons with a constitutively active Hsf1 construct induced Gabra4 expression in the absence of alcohol. Transfection with a dominant-negative Hsf1 construct abolished Gabra4 induction by alcohol (Pignataro et al., 2007; Fig. 5A, B). Acute exposure to alcohol also rapidly increases the transcription of the heat shock protein (Hsp) genes Hsp27, Hsp40, Hsp70,Hsp90 and Cryab in cultured mouse cortical neurons. These data further confirm that the stimulation of Gabra4 by alcohol in the mouse cortex is dependent on the activation of Hsf1 and the heat shock pathway. Similar, although smaller, increases in Gabra4 were obtained in the brains of adult alcohol-naïve rodents after a single intra-peritoneal injection of ethanol (Liang et al., 2007; Pignataro et al., 2007; Werner and Morrow, unpublished observations).
Figure 5.
Hsf1 is required for the induction of Gabra4 by ethanol (EtOH). A, Knockdown of Hsf1 inhibits the activation of Gabra4 transcription. Treatment of neurons with Hsf1 siRNA dramatically reduced the activation of Gabra4 transcription by EtOH, while treatment with control (C) siRNA had no effect (by one-way ANOVA vs. control cells, with Dunnett's Multiple Comparison post hoc test, n ≥ 3). B, Cortical neurons transfected with the constitutively active Hsf1 (Hsf1-act) construct show an increase in Gabra4 mRNA expression similar to the induction seen with EtOH. The dominant-negative form of Hsf1 (Hsf1-inact) completely abolishes the induction of Gabra4. Hsf1-inact alone had no effect on Gabra4 expression (by one-way ANOVA vs control cells, with Dunnett's Multiple Comparison post hoc test, n ≥ 6). Data are the mean ± s.e.m. (*P < 0.05, **P < 0.01). [Adapted from Pignataro et al., 2007.]
3.5. Epigenetic mechanisms
Another effective mechanism of gene regulation is not dependent on the presence of cis-acting sequences or the binding of transcription factors binding, but is mediated by alterations in the physical packing of DNA within the chromatin by virtue of its interactions with histones. The degree of packing of the DNA influences its accessibility to the transcriptional machinery, and a host of biochemical mechanisms that influence chromatin structure are often collectively referred to as epigenetic mechanisms.
Increasing evidence has shown that epigenetic mechanisms, including the covalent modification of DNA and histones (acetylation, methylation, phosphorylation, ubiquitinylation, ADP-ribosylation and SUMOylation), regulate gene transcription via changes in chromatin structure (Grunstein, 1997; Turner, 2002; Egger et al., 2004; Hsieh and Gage, 2005; Verdone et al., 2005). The nucleosome, the fundamental unit of chromatin, contains 147 bp of DNA wrapped 1.7 times around a histone octamer core (a central H3-H4 tetramer and two H2A-H2B heterodimers). After packaging, the nucleosome is bound by a H1 histone and lines up in arrays to form a 30 nm chromatin fiber. Nucleosome remodeling complexes can alter accessibility via nucleosome motility; for example, the histone octamer can slide along the DNA, be transferred to a different DNA molecule, or change its binding interactions with the DNA. Modification of the histone N-terminus by serine phosphorylation, lysine acetylation or methylation can also allow DNA-binding proteins to gain intermittent access to the DNA.
Two enzymes in particular control this process of chromatin remodeling. These enzymes are known as histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Grunstein, 1997; Hsieh and Gage, 2005). HATs induce lysine acetylation at the N-terminal of histones, causing relaxation of the nucleosomes. In particular, acetylation is thought to promote active gene expression by interacting with proteins that increase accessibility and recruiting enzymes that acetylate adjacent nucleosomes, resulting in a positive feedback loop and enhanced transcription. HDACs reverse acetylation to allow histones to package DNA into a more condensed chromatin, thereby blocking access to transcriptional activators and decreasing gene transcription (Grunstein, 1997; Turner, 2002; Hsieh and Gage, 2005).
CREB is an important regulator of neuronal plasticity, and can be phosphorylated to form pCREB, which has intrinsic HAT activity and can regulate chromatin structure (Silva et al., 1998; Korzus et al., 2004; Hsieh and Gage, 2005). It was demonstrated recently that the anxiolytic effects of acute ethanol were concomitant with an increase in CREB, inhibition of HDAC activity and subsequent increase of H3-H4 acetylation and NPY expression in total cell lysates from the rat amygdala. Withdrawal (1 day) from a 15-day ethanol diet produced anxiety-like behavior and decreased H3-H4 acetylation. Treatment of the ethanol-withdrawn rats with the HDAC inhibitor, trichostatin A, rescued the deficits in H3-H4 acetylation and prevented anxiety as measured in the elevated plus maze. These withdrawal effects are therefore believed to stem in part from HDAC activation and CREB inhibition, leading to deacetylation of H3-H4, and a decrease in NPY expression in the amygdala (Pandey et al., 2008a).
In contrast, the methylation of the C5 position of the cytosine found on CpG dinucleotide ‘islands’ silences gene expression by inhibiting the binding of transcription factors or by directly binding repressors that can recruit HDACs. Of particular interest, the 5′-non-coding region of the NR2B mouse gene (Grin2b) contains several CpG islands that extend into exon 2 and exon 3. These CpG islands show strong sequence homology to similar regions of the human and rat Grin2b genes (Klein et al., 1998), and are methylated across the regions 287-461 and 1150-1873 bp downstream of the transcription initiation site in adult cerebral cortex (Ravindran and Ticku, 2005). Chronic ethanol treatment of cultured cortical neurons produced a concurrent decrease in this CpG island methylation and an increase in Grin2b expression, while acute ethanol treatment was ineffective (Ravindran and Ticku, 2005, 2006). These data suggest that chronic ethanol reduces DNA methylation to relax the nucleosome packing of Grin2b, making it more accessible to transcription factors and RNA polymerase, and thereby increasing Grin2b expression.
4. Genome profiling of alcohol-responsive genes (ARGs)
Recent advancements in technology have made it possible to study the effects of ethanol, not just on individual genes, but across the entire genome of an organism. The current state-of-the-art in this area involves direct sequencing technologies and a variety of gene microarray approaches, but this field actually began at the protein level.
Dr. Michael Miles was the first to study the promoter regions of ARGs, and also pioneered the application of global “proteomics” approaches to investigate the effects of ethanol, using 2D gel electrophoresis. In an early study, the NG108-15 hybrid cell line was treated with 100 mM ethanol for 48 h, mRNA was collected and translated in vitro and the resulting proteins resolved with 2D gel electrophoresis. This proteomics approach resolved over 400 proteins encoded by genes that were sensitive to ethanol. Among the up-regulated genes, some were induced exclusively by ethanol, while other genes were part of a group induced by heat shock. Within this last class, two members of the heat shock protein family, heat-shock cognate protein 70 (Hsc70) and Hsc110, were identified as ethanol-inducible genes (Miles et al., 1992).
Following these early experiments, a vast number of studies have been done at the level of the genome using microarray techniques. The first genome microarray study performed in post mortem alcoholic human brain tissue showed consistent down-regulation of myelin-related genes in the frontal cortex. Even though these genes are expressed in oligodendrocytes, they clearly impact neuronal function. The expression of M6 neuronal glycoprotein (GPM6A), myelin-associated glycoprotein (MAG), myelin-associated oligodendrocyte basic protein (MOBP), myelin basic protein (MBP), myelin proteolipid protein (PLP1), myelin-oligodendrocyte glycoprotein (MOG), myelin protein Po and oligiodendrocyte-myelin glycoprotein (OMG) was decreased in the brains of human alcoholics compared with brains from control individuals (Lewohl et al., 2000). A similar post mortem study found that genes involved in the ubiquitin-proteasome pathway were down-regulated in alcoholic brains (Sokolov et al., 2003; Liu et al., 2004) as well as eleven nuclear genes encoding mitochondrial proteins and twenty-one genes involved in gene expression and replication (Sokolov et al., 2003). Among the 12,000 genes investigated with a microarray study in individuals with a history of alcohol abuse, 54 genes were up-regulated and 24 down-regulated in the prefrontal cortex of alcohol abusers. Among the genes up-regulated were many members of the heat shock protein family, including HSP70-2, CRYAB, HSP27-1 and HSP40-1 (Iwamoto et al., 2004). In general, up-regulation of chaperone proteins seems to be a common finding in the brains of alcoholics. Genomic microarray analysis performed in C. elegans exposed to high concentrations of ethanol revealed 230 genes that were activated within 15 minutes. Among these genes were a novel carboxyl esterase-like protein with strong similarity to neuroligin (a protein involved in axon guidance), glr-2, a gene that encodes a subunit of the AMPA glutamate receptor subunit (homologous to mammalian GluR2), and five members of the heat shock protein family (Kwon et al., 2004). Miroarrays performed on cultured cortical neurons chronically exposed to ethanol showed that heat shock protein family genes Hsp105, Hsp70 and Hsp30 and genes involved in protein synthesis and the ubiquitin-proteasome pathway were down-regulated, while Grin2b expression was up-regulated by ethanol (Gutala et al., 2004). Recently, a microarray study showed that chronic ethanol treatment (75 mM, 5 days) of cultured cortical neurons produced increased gene expression of Hsp70, Hsp84 and Hspa8 (Wang et al., 2007).
A widely employed research strategy in rodents is the use of comparative microarray studies to search for genetic factors that predispose an animal towards alcoholism. In one particular paradigm, rats that have experienced several forced cycles of ethanol intoxication and withdrawal show a sustained increase in voluntary ethanol intake. Microarray analysis of brain cortex from these animals revealed higher gene expression of the metabotropic glutamate receptor GluR2 subunit (Grm2), stress-associated kinases (SAPK (Mapk9), p38 (Malpk14)), stress protein (Hsc-70 related protein), and insulin-like growth factor II (Igf2, Rimondini et al., 2002). Another study looked at expression in whole brain in two acutely-treated mouse strains (C57BL/6J and DBA/2J) with differing ethanol preferences, and revealed a down-regulation of three myelin-related genes, Erbb3, Mobp and Nkx2-2 after exposure to ethanol (Treadwell and Singh, 2004). A similar study investigated patterns of gene expression in different brain regions of mice from the same two different inbred strains acutely treated with alcohol. The results showed that ethanol induced a stronger response in DBA/2J mice relative to C57BL/6J in the prefrontal cortex. The gene clusters showing the most robust response to alcohol included a statistical over-representation of genes involved in neurogenesis, plasticity, myelination and metal ion homestasis (Kerns et al., 2005).
An alternative approach to investigating the genetic basis of ethanol predisposition is the analysis of quantitative trait loci (QTLs). Quantitative trait refers to the inheritance of a phenotypic characteristic or trait that varies in degree and can be attributed to the interactions between two or more genes and their environment. Individual QTL for a trait can be found on different chromosomes and statistical analysis is used to determine whether particular alleles lie within a certain locus and produce the expected phenotype. Since there is a wide variation in alcohol preference among strains of inbred mice, microarray studies have been used to identify possible QTLs for alcohol intake. A recent analysis of alcohol-naïve animals across inbred mouse strains identified a set of cis-regulated candidate genes for an alcohol preference QTL in chromosome 9. These genes include: Arhgef12, Carm1, Cryab, Cox5a, Dlat, Fxyd6, Limd1, Nicn1, Nmnat3, Pknox2, Rbp1, Sc5d, Scn4b, Tcf12, Vps11 and Zfp291 (Mulligan et al., 2006). It is quite remarkable that, once again, a heat shock protein family member (Cryab) was identified in this screen. The gene Arhgef12 encodes a Rho guanine nucleotide exchange factor (GEF), a molecule able to modulate the activity of Rho. These proteins are involved in cytoskeleton control and synaptic plasticity (Bonhoeffer and Yuste, 2002). Carm1 encodes for the coactivator-associated arginine methyltransferase 1, a protein capable of methylating histone H3 and regulating gene expression. Cox5a codifies for the subunit Va of the cytochrome C oxidase that is important for the normal function of the cell mitochondria and is involved in the production of inflammatory mediators. Dlat gene produces dihydrolipoamide S-acetyltransferase that is part of the Pyruvate Dehydrogenase Complex in the mitochondria and is crucial for energy production and has been implicated in cirrhosis. Fxyd6 encodes the protein phosphohippolin, which is primarily expressed in the brain. Phosphohippolin modulates the kinetic activity of Na+,K+-ATPase and has long-term physiological importance in maintaining cation homeostasis. The FXYD domain containing ion transport regulator 6 (Fxyd6) gene is located within a region of chromosome 11 (11q23.3) that has been shown by a number of genome scans to be one of the most well-established linkages to schizophrenia. Pknox2 encodes the Pbx/knotted 1 homeobox 2 while Scn4b gene produces the subunit β4 of the voltage-gated sodium channel.
Given the frequency of studies linking ethanol and heat shock proteins, our laboratory has been interested in studying the mechanism by which ethanol regulates genes involved in the heat shock cascade. We have already observed that acute ethanol treatment of cultured cortical neurons activated heat shock factor 1 (Hsf1), allowing its binding to the alcohol response element (ARE), and thereby increasing gene expression of Gabra4 (Pignataro et al., 2007). These initial results on the interaction of ethanol and the heat shock pathway via Hsf1 were further confirmed through the observation that acute alcohol exposure rapidly increases expression levels of heat shock protein (Hsp) genes Hsp27, Hsp40, Hsp70,Hsp90 and Cryab in cultured mouse cortical neurons. To further investigate this interaction at the genomic level, parallel gene microarray experiments were done on mouse cortical neurons exposed to ethanol (60 mM) or heat (42°C). The microarray data, not unexpectedly, revealed a large number of genes that were acutely up-regulated by ethanol (Pignataro et al., 2007, see Supplemental Table 1), and an even larger number that were activated by heat shock (Pignataro et al., 2007, see Supplemental Table 2). We were particularly interested in genes responding to both treatments and focused our analysis on the nine genes that showed dramatic response (>50% stimulation) to ethanol and heat shock (Pignataro et al., 2007; Table 1). The nine genes in the set are all specifically expressed in neurons and contain one or more ARE-like sequences, located either in the 5′-flanking domain (as in Hsp70) or downstream in an intron/exon region (as in Gabra4). This set of nine genes is especially interesting with respect to brain plasticity and adaptation to alcohol, since it includes genes that encode proteins involved in synaptic transmission: Syt1, encoding synaptotagmin I, a calcium-binding protein involved in neurotransmitter release, and Spnb2, encoding spectrin beta 2, a calcium sensor involved in vesicle docking to the plasma membrane. The other genes encode proteins that are important in synapse formation and plasticity, such as neurogranin (Nrgn), cadherin 13 (Cdh13) and glycoprotein m6a (Gpm6a); in microtubule assembly (microtubule-associated protein 1B; Mtap1b) or in protein trafficking (SEC23A; Sec23a) (Pignataro et al., 2007). Among the genes up-regulated by ethanol, but not by heat, was a variant of insulin-like growth factor 1 (Igf1), a gene previously reported by Rimondini et al (2002). In addition, ethanol exposure significantly decreased the expression of genes related to the ubiquitin-proteasome pathway (Ube2e1, Uqcrb, Ndufc2, Ndufa5, Ndufb4) and the TATA box binding protein (TBP)-associated factor 9 (Taf9) (Pignataro et al., unpublished observations).
Conclusions
In this review we have attempted to draw together the evidence gathered from a decade or more of research on alcohol regulation of gene expression both ex vivo and in vitro, in a variety of organisms (human, mouse, rat, C. elegans) using a combination of microscopic (promoter analysis) and macroscopic (genome screens) approaches to the problem. In reviewing the literature for this article, we were less overwhelmed by the ubiquity of alcohol-responsive genes than we expected, and more surprised by the consistency between studies employing quite different technical approaches. When considered in toto, the focused studies of individual genes and promoters together with the whole genome screens have started to deliver surprising new insights. The combination of several new technologies now in use promises to answer broad questions about the cellular pathways that are regulated by acute and chronic ingestion of ethanol.
While the data sets are obviously quite diverse, it is likely that many of the differences arise from the use of multiple ethanol treatment paradigms and various model systems. In particular, the use of single time points for measurements introduces an obvious source of variation. For example, we and others have identified genes that are rapidly up-regulated by alcohol within <30 minutes, but the expression reaches a peak within 1-2 hours and then declines, so that the changes in expression would be missed in other studies using an 8 or 24 hour time point. It is therefore unsurprising that the specifics of the data sets are different, and the possibility of spatial (cortex vs. thalamus etc…) and cell-type (principal cells, interneurons etc…) heterogeneity of gene regulation introduces further complications when comparing whole brain studies to studies on more homogeneous cell populations.
In spite of all of this heterogeneity, there are remarkable indications that parts of the puzzle have started to make sense, and consistent themes are emerging. In a variety of studies, ethanol treatment triggers changes (usually activation) in the expression of genes involved in the cell cycle, in oxidative stress and apoptosis. Genes that are frequently up-regulated by ethanol encode heat shock proteins, and proteins related to synaptic transmission and synaptic plasticity. Alcohol down-regulates genes related to myelination, the ubiquitin-proteasome pathway and protein synthesis.
Understanding the genomic or epigenetic basis of these effects of alcohol on highly regulated cellular pathways looks to be the next promising direction for the field. The identification of specific alcohol-sensitive transcription factors such as Hsf1, and specific sequence elements such as the ARE and the NRSE seems to be an important event. Further study of these novel cis-elements and trans-factors, along with the discovery of as yet unknown nuclear and cytoplasmic players, should continue to help decipher the transcriptional events underlying the changes in gene expression of the many ARGs that have been identified, and this in turn will yield new insights into the molecular mechanisms of the behavioral and physiological changes induced by alcohol.
Abbreviations
- AP-1
activator protein-1
- Arc
activity-regulated cytoskeleton-associated protein
- ARDS
alcohol-related developmental syndrome
- ARE
alcohol response element
- ARG
alcohol-responsive gene
- BDNF
brain-derived neurotrophic factor
- CAT
chloramphenicol acetyl-transferase
- CRE
cAMP-response element
- CREB
cAMP-response element binding protein
- E or EtOH
ethanol
- FAS
fetal alcohol syndrome
- G
gestational day
- GABA
γ-aminobutyric acid
- GABAA-R
GABAA receptor
- HAT
histone acetyltranferase
- HDAC
histone deacetylase
- HS
heat shock
- HSE
heat-shock regulatory element
- Hsf1
heat shock factor 1
- Hsp
heat shock protein
- Inr
initiator element
- luc
luciferase
- NMDA
N-methyl-D-aspartic acid
- NRSE
neuron-restrictive silencer element
- NRSF
neuron-restrictive silencer factor
- NT
neurotrophin
- QTL
quantitative trait locus
- RE-1
repressor element-1
- Sp1
specificity protein-1
- TFIID
transcription factor for RNA polymerase IID
- TGFβ
transforming growth factor β
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Bachtell RK, Wang YM, Freeman P, Risinger FO, Ryabinin AE. Alcohol drinking produces brain region-selective changes in expression of inducible transcription factors. Brain Res. 1999;847(2):157–165. doi: 10.1016/s0006-8993(99)02019-3. [DOI] [PubMed] [Google Scholar]
- Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G, et al. International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev. 1998;50(2):291–313. [PubMed] [Google Scholar]
- Becker HC. Kindling in alcohol withdrawal. Alcohol Health Res World. 1998;22(1):25–23. [PMC free article] [PubMed] [Google Scholar]
- Becker HC. Animal models of alcohol withdrawal. Alcohol Res Health. 2000;24(2):105–113. [PMC free article] [PubMed] [Google Scholar]
- Bessis A, Champtiaux N, Chatelin L, Changeux JP. The neuron-restrictive silencer element: a dual enhancer/silencer crucial for patterned expression of a nicotinic receptor gene in the brain. Proc Natl Acad Sci U S A. 1997;94(11):5906–5911. doi: 10.1073/pnas.94.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer T, Yuste R. Spine motility. Phenomenology, mechanisms, and function. Neuron. 2002;35(6):1019–1027. doi: 10.1016/s0896-6273(02)00906-6. [DOI] [PubMed] [Google Scholar]
- Brodie MS, Shefner SA, Dunwiddie TV. Ethanol increases the firing rate of dopamine neurons of the rat ventral tegmental area in vitro. Brain Res. 1990;508(1):65–69. doi: 10.1016/0006-8993(90)91118-z. [DOI] [PubMed] [Google Scholar]
- Budygin EA, Phillips PE, Wightman RM, Jones SR. Terminal effects of ethanol on dopamine dynamics in rat nucleus accumbens: an in vitro voltammetric study. Synapse. 2001;42(2):77–79. doi: 10.1002/syn.1101. [DOI] [PubMed] [Google Scholar]
- Cagetti E, Liang J, Spigelman I, Olsen RW. Withdrawal from chronic intermittent ethanol treatment changes subunit composition, reduces synaptic function, and decreases behavioral responses to positive allosteric modulators of GABAA receptors. Mol Pharmacol. 2003;63(1):53–64. doi: 10.1124/mol.63.1.53. [DOI] [PubMed] [Google Scholar]
- Calton JL, Wilson WA, Moore SD. Magnesium-dependent inhibition of N-methyl-D-aspartate receptor-mediated synaptic transmission by ethanol. J Pharmacol Exp Ther. 1998;287(3):1015–1019. [PubMed] [Google Scholar]
- Chandler LJ, Harris RA, Crews FT. Ethanol tolerance and synaptic plasticity. Trends Pharmacol Sci. 1998;19(12):491–495. doi: 10.1016/s0165-6147(98)01268-1. [DOI] [PubMed] [Google Scholar]
- Chandra D, Jia F, Liang J, Peng Z, Suryanarayanan A, Werner DF, et al. GABAA receptor alpha 4 subunits mediate extrasynaptic inhibition in thalamus and dentate gyrus and the action of gaboxadol. Proc Natl Acad Sci U S A. 2006;103(41):15230–15235. doi: 10.1073/pnas.0604304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton ME, Sweetnam PM, Fitzgerald LW, Terwilliger RZ, Nestler EJ, Duman RS. Chronic ethanol administration regulates the expression of GABAA receptor alpha 1 and alpha 5 subunits in the ventral tegmental area and hippocampus. J Neurochem. 1997;68(1):121–127. doi: 10.1046/j.1471-4159.1997.68010121.x. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80(6):949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Cotto J, Fox S, Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110(Pt 23):2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- Criswell HE, Ming Z, Griffith BL, Breese GR. Comparison of effect of ethanol on N-methyl-D-aspartate- and GABA-gated currents from acutely dissociated neurons: absence of regional differences in sensitivity to ethanol. J Pharmacol Exp Ther. 2003;304(1):192–199. doi: 10.1124/jpet.102.041590. [DOI] [PubMed] [Google Scholar]
- Crowder TL, Ariwodola OJ, Weiner JL. Ethanol antagonizes kainate receptor-mediated inhibition of evoked GABA(A) inhibitory postsynaptic currents in the rat hippocampal CA1 region. J Pharmacol Exp Ther. 2002;303(3):937–944. doi: 10.1124/jpet.102.038471. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11(3):327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Dar MS, Mustafa SJ, Wooles WR. Possible role of adenosine in the CNS effects of ethanol. Life Sci. 1983;33(14):1363–1374. doi: 10.1016/0024-3205(83)90819-6. [DOI] [PubMed] [Google Scholar]
- Demarest K, Hitzemann B, Phillips T, Hitzemann R. Ethanol-induced expression of c-Fos differentiates the FAST and SLOW selected lines of mice. Alcohol Clin Exp Res. 1999;23(1):87–95. [PubMed] [Google Scholar]
- Deng W, Roberts SG. Core promoter elements recognized by transcription factor IIB. Biochem Soc Trans. 2006;34(Pt 6):1051–1053. doi: 10.1042/BST0341051. [DOI] [PubMed] [Google Scholar]
- Devaud LL, Fritschy JM, Sieghart W, Morrow AL. Bidirectional alterations of GABA(A) receptor subunit peptide levels in rat cortex during chronic ethanol consumption and withdrawal. J Neurochem. 1997;69(1):126–130. doi: 10.1046/j.1471-4159.1997.69010126.x. [DOI] [PubMed] [Google Scholar]
- Devaud LL, Smith FD, Grayson DR, Morrow AL. Chronic ethanol consumption differentially alters the expression of gamma-aminobutyric acidA receptor subunit mRNAs in rat cerebral cortex: competitive, quantitative reverse transcriptase-polymerase chain reaction analysis. Mol Pharmacol. 1995;48(5):861–868. [PubMed] [Google Scholar]
- Dunwiddie TV. Acute and Chronic Effects of Ethanol on the Brain: Interactions of Ethanol with Adenosine, Adenosine Transporters and Adenosine Receptors. In: Deitrich RA, Erwin VG, editors. Pharmacological Effects of Ethanol on the Nervous System. CRC Press; Boca Raton: 1996. pp. 147–161. [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Eisenman LM, Donovan HS, Johnson TE. Alcohol differentially affects c-Fos expression in the supraoptic nucleus of long-sleep and short-sleep mice. Brain Res. 2002;935(12):114–117. doi: 10.1016/s0006-8993(02)02453-8. [DOI] [PubMed] [Google Scholar]
- Faingold CL, N'Gouemo P, Riaz A. Ethanol and neurotransmitter interactions--from molecular to integrative effects. Prog Neurobiol. 1998;55(5):509–535. doi: 10.1016/s0301-0082(98)00027-6. [DOI] [PubMed] [Google Scholar]
- Farrant M, Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat Rev Neurosci. 2005;6(3):215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- Follesa P, Ticku MK. Chronic ethanol-mediated up-regulation of the N-methyl-D-aspartate receptor polypeptide subunits in mouse cortical neurons in culture. J Biol Chem. 1996;271(23):13297–13299. doi: 10.1074/jbc.271.23.13297. [DOI] [PubMed] [Google Scholar]
- Follesa P, Biggio F, Caria S, Gorini G, Biggio G. Modulation of GABA(A) receptor gene expression by allopregnanolone and ethanol. Eur J Pharmacol. 2004;500(13):413–425. doi: 10.1016/j.ejphar.2004.07.041. [DOI] [PubMed] [Google Scholar]
- Follesa P, Biggio F, Mancuso L, Cabras S, Caria S, Gorini G, et al. Ethanol withdrawal-induced up-regulation of the alpha2 subunit of the GABAA receptor and its prevention by diazepam or gamma-hydroxybutyric acid. Brain Res Mol Brain Res. 2004;120(2):130–137. doi: 10.1016/j.molbrainres.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Forsythe ID, Westbrook GL. Slow excitatory postsynaptic currents mediated by N-methyl-D-aspartate receptors on cultured mouse central neurones. J Physiol. 1988;396:515–533. doi: 10.1113/jphysiol.1988.sp016975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayer GG, Gordon A, Miles MF. Ethanol increases tyrosine hydroxylase gene expression in N1E-115 neuroblastoma cells. J Biol Chem. 1991;266(33):22279–22284. [PubMed] [Google Scholar]
- Gessa GL, Muntoni F, Collu M, Vargiu L, Mereu G. Low doses of ethanol activate dopaminergic neurons in the ventral tegmental area. Brain Res. 1985;348(1):201–203. doi: 10.1016/0006-8993(85)90381-6. [DOI] [PubMed] [Google Scholar]
- Goodlett CR, Horn KH, Zhou FC. Alcohol teratogenesis: mechanisms of damage and strategies for intervention. Exp Biol Med (Maywood) 2005;230(6):394–406. doi: 10.1177/15353702-0323006-07. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Gulya K, Dave JR, Hoffman PL. Chronic ethanol ingestion decreases vasopressin mRNA in hypothalamic and extrahypothalamic nuclei of mouse brain. Brain Res. 1991;557(12):129–135. [PubMed] [Google Scholar]
- Gutala R, Wang J, Kadapakkam S, Hwang Y, Ticku M, Li MD. Microarray analysis of ethanol-treated cortical neurons reveals disruption of genes related to the ubiquitin-proteasome pathway and protein synthesis. Alcohol Clin Exp Res. 2004;28(12):1779–1788. doi: 10.1097/01.alc.0000148117.17707.b4. [DOI] [PubMed] [Google Scholar]
- Hassan S, Duong B, Kim KS, Miles MF. Pharmacogenomic analysis of mechanisms mediating ethanol regulation of dopamine beta-hydroxylase. J Biol Chem. 2003;278(40):38860–38869. doi: 10.1074/jbc.M305040200. [DOI] [PubMed] [Google Scholar]
- Hendricson AW, Maldve RE, Salinas AG, Theile JW, Zhang TA, Diaz LM, et al. Aberrant synaptic activation of N-methyl-D-aspartate receptors underlies ethanol withdrawal hyperexcitability. J Pharmacol Exp Ther. 2007;321(1):60–72. doi: 10.1124/jpet.106.111419. [DOI] [PubMed] [Google Scholar]
- Hitzemann B, Hitzemann R. Genetics ethanol and the Fos response: a comparison of the C57BL/6J and DBA/2J inbred mouse strains. Alcohol Clin Exp Res. 1997;21(8):1497–1507. [PubMed] [Google Scholar]
- Hoffman PL, Rabe CS, Moses F, Tabakoff B. N-methyl-D-aspartate receptors and ethanol: inhibition of calcium flux and cyclic GMP production. J Neurochem. 1989;52(6):1937–1940. doi: 10.1111/j.1471-4159.1989.tb07280.x. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17(6):664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Hsieh KP, Wilke N, Harris A, Miles MF. Interaction of ethanol with inducers of glucose-regulated stress proteins. Ethanol potentiates inducers of grp78 transcription. J Biol Chem. 1996;271(5):2709–2716. doi: 10.1074/jbc.271.5.2709. [DOI] [PubMed] [Google Scholar]
- Hu XJ, Ticku MK. Chronic ethanol treatment upregulates the NMDA receptor function and binding in mammalian cortical neurons. Brain Res Mol Brain Res. 1995;30(2):347–356. doi: 10.1016/0169-328x(95)00019-o. [DOI] [PubMed] [Google Scholar]
- Hu XJ, Follesa P, Ticku MK. Chronic ethanol treatment produces a selective upregulation of the NMDA receptor subunit gene expression in mammalian cultured cortical neurons. Brain Res Mol Brain Res. 1996;36(2):211–218. doi: 10.1016/0169-328x(95)00223-f. [DOI] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2(10):867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Imperato A, Di Chiara G. Preferential stimulation of dopamine release in the nucleus accumbens of freely moving rats by ethanol. J Pharmacol Exp Ther. 1986;239(1):219–228. [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004;49(4):379–385. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Jia F, Pignataro L, Schofield CM, Yue M, Harrison NL, Goldstein PA. An extrasynaptic GABAA receptor mediates tonic inhibition in thalamic VB neurons. J Neurophysiol. 2005;94(6):4491–4501. doi: 10.1152/jn.00421.2005. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, O'Brien C. Drug addiction as a pathology of staged neuroplasticity. Neuropsychopharm. 2008;33(1):1660–180. doi: 10.1038/sj.npp.1301564. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: Implications for the behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25(9):2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Fraser CM. A strong promoter element is located between alternative exons of a gene encoding the human gamma-aminobutyric acid-type A receptor beta 3 subunit (GABRB3) J Biol Chem. 1993;268(6):4420–4428. [PubMed] [Google Scholar]
- Klein M, Pieri I, Uhlmann F, Pfizenmaier K, Eisel U. Cloning and characterization of promoter and 5′-UTR of the NMDA receptor subunit epsilon 2: evidence for alternative splicing of 5′-non-coding exon. Gene. 1998;208(2):259–269. doi: 10.1016/s0378-1119(98)00005-5. [DOI] [PubMed] [Google Scholar]
- Koob GF. Alcoholism: allostasis and beyond. Alcohol Clin Exp Res. 2003;27(2):232–243. doi: 10.1097/01.ALC.0000057122.36127.C2. [DOI] [PubMed] [Google Scholar]
- Koob GF. A role for GABA mechanisms in the motivational effects of alcohol. Biochem Pharmacol. 2004;68(8):1515–1525. doi: 10.1016/j.bcp.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraner SD, Chong JA, Tsay HJ, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9(1):37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- Kumar S, Porcu P, Werner DF, Matthews DB, Diaz-Granados JL, Helfand RS, Morrow LA. The role of GABAA receptors in the acute and chronic effects of ethanol: a decade of progress. Psychopharmacol. 2009;205(4):529–564. doi: 10.1007/s00213-009-1562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M, Ticku MK. Ethanol and regulation of the NMDA receptor subunits in fetal cortical neurons. J Neurochem. 1998;70(4):1467–1473. doi: 10.1046/j.1471-4159.1998.70041467.x. [DOI] [PubMed] [Google Scholar]
- Kumari M, Ticku MK. Regulation of NMDA receptors by ethanol. Prog Drug Res. 2000;54:152–189. [PubMed] [Google Scholar]
- Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim S, et al. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics. 2004;83(4):600–614. doi: 10.1016/j.ygeno.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–1882. [PubMed] [Google Scholar]
- Li L, Suzuki T, Mori N, Greengard P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc Natl Acad Sci U S A. 1993;90(4):1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Bosron WF, Dafeldecker WP, Lange LG, Vallee BL. Isolation of pi-alcohol dehydrogenase of human liver: is it a determinant of alcoholism? Proc Natl Acad Sci U S A. 1977;74(10):4378–4381. doi: 10.1073/pnas.74.10.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Suryanarayanan A, Abriam A, Snyder B, Olsen RW, Spigelman I. Mechanisms of reversible GABAA receptor plasticity after ethanol intoxication. J Neurosci. 2007;27(45):12367–12377. doi: 10.1523/JNEUROSCI.2786-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Suryanarayanan A, Chandra D, Homanics GE, Olsen RW, Spigelman I. Functional consequences of GABAA receptor alpha 4 subunit deletion on synaptic and extrasynaptic currents in mouse dentate granule cells. Alcohol Clin Exp Res. 2008;32(1):19–26. doi: 10.1111/j.1530-0277.2007.00564.x. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhang N, Cagetti E, Houser CR, Olsen RW, Spigelman I. Chronic intermittent ethanol-induced switch of ethanol actions from extrasynaptic to synaptic hippocampal GABAA receptors. J Neurosci. 2006;26(6):1749–1758. doi: 10.1523/JNEUROSCI.4702-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90(5):1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Loftis JM, Janowsky A. The N-methyl-D-aspartate receptor subunit NR2B: localization, functional properties, regulation, and clinical implications. Pharmacol Ther. 2003;97(1):55–85. doi: 10.1016/s0163-7258(02)00302-9. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, White G, Weight FF. Ethanol inhibits NMDA-activated ion current in hippocampal neurons. Science. 1989;243(4899):1721–1724. doi: 10.1126/science.2467382. [DOI] [PubMed] [Google Scholar]
- Ma L, Song L, Radoi GE, Harrison NL. Transcriptional regulation of the mouse gene encoding the alpha-4 subunit of the GABAA receptor. J Biol Chem. 2004;279(39):40451–40461. doi: 10.1074/jbc.M406827200. [DOI] [PubMed] [Google Scholar]
- Martin D, Tayyeb MI, Swartzwelder HS. Ethanol inhibition of AMPA and kainate receptor-mediated depolarizations of hippocampal area CA1. Alcohol Clin Exp Res. 1995;19(5):1312–1316. doi: 10.1111/j.1530-0277.1995.tb01617.x. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL. Mixed-agonist action of excitatory amino acids on mouse spinal cord neurones under voltage clamp. J Physiol. 1984;354:29–53. doi: 10.1113/jphysiol.1984.sp015360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles MF, Diaz JE, DeGuzman V. Ethanol-responsive gene expression in neural cell cultures. Biochim Biophys Acta. 1992;1138(4):268–274. doi: 10.1016/0925-4439(92)90003-6. [DOI] [PubMed] [Google Scholar]
- Miles MF, Diaz JE, DeGuzman VS. Mechanisms of neuronal adaptation to ethanol. Ethanol induces Hsc70 gene transcription in NG108-15 neuroblastoma x glioma cells. J Biol Chem. 1991;266(4):2409–2414. [PubMed] [Google Scholar]
- Miles MF, Wilke N, Elliot M, Tanner W, Shah S. Ethanol-responsive genes in neural cells include the 78-kilodalton glucose-regulated protein (GRP78) and 94-kilodalton glucose-regulated protein (GRP94) molecular chaperones. Mol Pharmacol. 1994;46(5):873–879. [PubMed] [Google Scholar]
- Mori N, Schoenherr C, Vandenbergh DJ, Anderson DJ. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 1992;9(1):45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- Mori N, Stein R, Sigmund O, Anderson DJ. A cell type-preferred silencer element that controls the neural-specific expression of the SCG10 gene. Neuron. 1990;4(4):583–594. doi: 10.1016/0896-6273(90)90116-w. [DOI] [PubMed] [Google Scholar]
- Morimoto T, Wang XH, Poo MM. Overexpression of synaptotagmin modulates short-term synaptic plasticity at developing neuromuscular junctions. Neuroscience. 1998;82(4):969–978. doi: 10.1016/s0306-4522(97)00343-6. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103(16):6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy J, Horvath C, Farkas S, Kolok S, Szombathelyi Z. NR2B subunit selective NMDA antagonists inhibit neurotoxic effect of alcohol-withdrawal in primary cultures of rat cortical neurones. Neurochem Int. 2004;44(1):17–23. doi: 10.1016/s0197-0186(03)00100-1. [DOI] [PubMed] [Google Scholar]
- Nelson TE, Ur CL, Gruol DL. Chronic intermittent ethanol exposure enhances NMDA-receptor-mediated synaptic responses and NMDA receptor expression in hippocampal CA1 region. Brain Res. 2005;1048(12):69–79. doi: 10.1016/j.brainres.2005.04.041. [DOI] [PubMed] [Google Scholar]
- Nie Z, Madamba SG, Siggins GR. Ethanol inhibits glutamatergic neurotransmission in nucleus accumbens neurons by multiple mechanisms. J Pharmacol Exp Ther. 1994;271(3):1566–1573. [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307(5950):462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- Ogbourne S, Antalis TM. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331(Pt 1):1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18(4):1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Metsis M, Timmusk T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res Mol Brain Res. 1999;72(1):30–39. doi: 10.1016/s0169-328x(99)00196-5. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008a;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette AJ, Perez SE, Anderson DJ. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc Natl Acad Sci U S A. 2000;97(22):12318–12323. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie J, Sapp DW, Tyndale RF, Park MK, Fanselow M, Olsen RW. Altered gabaa receptor subunit and splice variant expression in rats treated with chronic intermittent ethanol. Alcohol Clin Exp Res. 2001;25(6):819–828. [PubMed] [Google Scholar]
- Pignataro L, Miller AN, Ma L, Midha S, Protiva P, Herrera DG, et al. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J Neurosci. 2007;27(47):12957–12966. doi: 10.1523/JNEUROSCI.4142-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, Sperk G. GABA(A) receptors: immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience. 2000;101(4):815–850. doi: 10.1016/s0306-4522(00)00442-5. [DOI] [PubMed] [Google Scholar]
- Qiang M, Ticku MK. Role of AP-1 in ethanol-induced N-methyl-D-aspartate receptor 2B subunit gene up-regulation in mouse cortical neurons. J Neurochem. 2005;95(5):1332–1341. doi: 10.1111/j.1471-4159.2005.03464.x. [DOI] [PubMed] [Google Scholar]
- Qiang M, Rani CS, Ticku MK. Neuron-restrictive silencer factor regulates the N-methyl-D-aspartate receptor 2B subunit gene in basal and ethanol-induced gene expression in fetal cortical neurons. Mol Pharmacol. 2005;67(6):2115–2125. doi: 10.1124/mol.104.010751. [DOI] [PubMed] [Google Scholar]
- Quinn JP. Neuronal-specific gene expression--the interaction of both positive and negative transcriptional regulators. Prog Neurobiol. 1996;50(4):363–379. doi: 10.1016/s0301-0082(96)00041-x. [DOI] [PubMed] [Google Scholar]
- Rani CS, Qiang M, Ticku MK. Potential role of cAMP response element-binding protein in ethanol-induced N-methyl-D-aspartate receptor 2B subunit gene transcription in fetal mouse cortical cells. Mol Pharmacol. 2005;67(6):2126–2136. doi: 10.1124/mol.104.007872. [DOI] [PubMed] [Google Scholar]
- Ravindran CR, Ticku MK. Methylation of NMDA receptor NR2B gene as a function of age in the mouse brain. Neurosci Lett. 2005;380(3):223–228. doi: 10.1016/j.neulet.2005.01.042. [DOI] [PubMed] [Google Scholar]
- Ravindran CR, Ticku MK. Tyrosine kinase phosphorylation of GABA(A) receptor alpha1, beta2 and gamma2 subunits following chronic intermittent ethanol (CIE) exposure of cultured cortical neurons of mice. Neurochem Res. 2006;31(9):1111–1118. doi: 10.1007/s11064-006-9124-9. [DOI] [PubMed] [Google Scholar]
- Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. FASEB J. 2002;16(1):27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- Roberto M, Madamba SG, Stouffer DG, Parsons LH, Siggins GR. Increased GABA release in the central amygdala of ethanol-dependent rats. J Neurosci. 2004;24(45):10159–10166. doi: 10.1523/JNEUROSCI.3004-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DS, Hu Y, Lund IV, Brooks-Kayal AR, Russek SJ. Brain-derived neurotrophic factor (BDNF)-induced synthesis of early growth response factor 3 (Egr3) controls the levels of type A GABA receptor alpha 4 subunits in hippocampal neurons. J Biol Chem. 2006;281(40):29431–29435. doi: 10.1074/jbc.C600167200. [DOI] [PubMed] [Google Scholar]
- Roberts DS, Raol YH, Bandyopadhyay S, Lund IV, Budreck EC, Passini MA, et al. Egr3 stimulation of GABRA4 promoter activity as a mechanism for seizure-induced up-regulation of GABA(A) receptor alpha4 subunit expression. Proc Natl Acad Sci U S A. 2005;102(33):11894–11899. doi: 10.1073/pnas.0501434102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhale P, Verma J, Naorem A. Basal transcription machinery: role in regulation of stress response in eukaryotes. J Biosci. 2007;32(3):569–578. doi: 10.1007/s12038-007-0056-6. [DOI] [PubMed] [Google Scholar]
- Sanna E, Mostallino MC, Busonero F, Talani G, Tranquilli S, Mameli M, et al. Changes in GABA(A) receptor gene expression associated with selective alterations in receptor function and pharmacology after ethanol withdrawal. J Neurosci. 2003;23(37):11711–11724. doi: 10.1523/JNEUROSCI.23-37-11711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasner M, Buonanno A. Distinct N-methyl-D-aspartate receptor 2B subunit gene sequences confer neural and developmental specific expression. J Biol Chem. 1996;271(35):21316–21322. doi: 10.1074/jbc.271.35.21316. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267(5202):1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72(6):756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Cloning and expression of a gene encoding hsc73, the major hsp70-like protein in unstressed rat cells. EMBO J. 1987;6(4):993–998. doi: 10.1002/j.1460-2075.1987.tb04850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik KK. Genesis of alcohol-induced craniofacial dysmorphism. Exp Biol Med (Maywood) 2005;230(6):366–375. doi: 10.1177/15353702-0323006-04. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Hoffman PL. Alcohol addiction: an enigma among us. Neuron. 1996;16(5):909–912. doi: 10.1016/s0896-6273(00)80113-0. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41(3):105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29(2):357–369. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Tsai G, Coyle JT. The role of glutamatergic neurotransmission in the pathophysiology of alcoholism. Annu Rev Med. 1998;49:173–184. doi: 10.1146/annurev.med.49.1.173. [DOI] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111(3):285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Urso T, Gavaler JS, Van Thiel DH. Blood ethanol levels in sober alcohol users seen in an emergency room. Life Sci. 1981;28(9):1053–1056. doi: 10.1016/0024-3205(81)90752-9. [DOI] [PubMed] [Google Scholar]
- Valenzuela CF, Cardoso RA. Acute effects of ethanol on kainate receptors with different subunit compositions. J Pharmacol Exp Ther. 1999;288(3):1199–1206. [PubMed] [Google Scholar]
- Veatch LM, Gonzalez LP. Repeated ethanol withdrawal produces site-dependent increases in EEG spiking. Alcohol Clin Exp Res. 1996;20(2):262–267. doi: 10.1111/j.1530-0277.1996.tb01638.x. [DOI] [PubMed] [Google Scholar]
- Verdone L, Caserta M, Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83(3):344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Wang J, Gutala R, Sun D, Ma JZ, Sheela RC, Ticku MK, et al. Regulation of platelet-derived growth factor signaling pathway by ethanol, nicotine, or both in mouse cortical neurons. Alcohol Clin Exp Res. 2007;31(3):357–375. doi: 10.1111/j.1530-0277.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- Wilke N, Sganga M, Barhite S, Miles MF. Effects of alcohol on gene expression in neural cells. EXS. 1994;71:49–59. doi: 10.1007/978-3-0348-7330-7_6. [DOI] [PubMed] [Google Scholar]
- Wilke N, Sganga MW, Gayer GG, Hsieh KP, Miles MF. Characterization of promoter elements mediating ethanol regulation of hsc70 gene transcription. J Pharmacol Exp Ther. 2000;292(1):173–180. [PubMed] [Google Scholar]
- Wood IC, Roopra A, Buckley NJ. Neural specific expression of the m4 muscarinic acetylcholine receptor gene is mediated by a RE1/NRSE-type silencing element. J Biol Chem. 1996;271(24):14221–14225. doi: 10.1074/jbc.271.24.14221. [DOI] [PubMed] [Google Scholar]
- Woodward JJ. Ionotropic glutamate receptors as sites of action for ethanol in the brain. Neurochem Int. 1999;35(2):107–113. [PubMed] [Google Scholar]
- Woodward JJ. Ethanol and NMDA receptor signaling. Crit Rev Neurobiol. 2000;14(1):69–89. doi: 10.1080/08913810008443548. [DOI] [PubMed] [Google Scholar]
- Worst TJ, Vrana KE. Alcohol and gene expression in the central nervous system. Alcohol Alcohol. 2005;40(1):63–75. doi: 10.1093/alcalc/agh119. [DOI] [PubMed] [Google Scholar]
- Wu CH, Frostholm A, De Blas AL, Rotter A. Differential expression of GABAA/benzodiazepine receptor subunit mRNAs and ligand binding sites in mouse cerebellar neurons following in vivo ethanol administration: an autoradiographic analysis. J Neurochem. 1995;65(3):1229–1239. doi: 10.1046/j.1471-4159.1995.65031229.x. [DOI] [PubMed] [Google Scholar]
- Zoeller RT, Fletcher DL. A single administration of ethanol simultaneously increases c-fos mRNA and reduces c-jun mRNA in the hypothalamus and hippocampus. Brain Res Mol Brain Res. 1994;24(14):185–191. doi: 10.1016/0169-328x(94)90131-7. [DOI] [PubMed] [Google Scholar]