Abstract
Purpose
Mutations in the BRCA2 gene are dominantly inherited but cause cancers when the wild-type allele has loss of heterozygosity (LOH) within the cancer. Because most disease-associated BRCA2 mutations are protein-truncating mutations, a test for truncated BRCA2 proteins should identify most BRCA2 hereditary cancers.
Methods
We have developed a tissue truncation test to identify truncated BRCA2 proteins in breast cancer tissue biopsies in vivo that does not use amplification or genetic manipulations. N-terminal and C-terminal antibodies are used to visualize protein truncation by demonstrating that the beginning of the protein is present but the end (ie, terminus) is absent.
Results
A quantitative C-terminal immunostaining score or a C-terminal to N-terminal truncation ratio correctly classified 20 of 21 breast cancers arising in BRCA2 mutation carriers and 57 of 58 cancers arising outside the context of a multiple-case breast cancer family. This represents a sensitivity of 95% and a specificity of 98%. Because of the presence of C-terminal BRCA2 protein and atypical clinical features of the misclassified cancer in a BRCA2 mutation carrier, we performed polymerase chain reaction and sequence analyses on this cancer. The results showed continued presence of the BRCA2 wild-type allele in the cancer, which indicated that intact BRCA2 protein was present in this cancer.
Conclusion
This immunohistochemistry-based test (which takes only 4 hours) appears to identify BRCA2 hereditary cancer with high accuracy. The test also appears to diagnose the biochemical loss of BRCA2 protein in cancers (ie, BRCA2-mutant genotype), which will usually but not always agree with the presence of a germline BRCA2 mutation found by susceptibility testing by DNA sequencing of blood samples.
INTRODUCTION
Germline mutations in the BRCA2 gene are dominantly inherited and are generally thought to cause cancer after somatic loss of the wild-type allele within the cancer.1–5 Because most disease-associated BRCA2 mutations are protein-truncating mutations,6–7 a test for truncated BRCA2 proteins should identify most BRCA2 hereditary cancers. It is important to know which breast cancer patients have cancers arising in BRCA2 mutation carriers, because they have a much greater risk of subsequent breast cancer recurrence or development of ovarian cancer than patients with sporadic cancer.8–9 Identification of these gene mutation carriers also is important because family members who inherited mutations have a lifetime breast cancer risk of 35% to 80%,10 and BRCA2 breast cancers respond differently to specific treatments.11–12 Present strategies for finding BRCA2 mutation carriers are cumbersome and are only applicable in high-risk families in whom less than half the mutant cancers are found.13
We have developed an antibody-based method to identify truncated BRCA2 proteins in breast cancer specimens. This method successfully classified 20 of 21 breast cancers from patients in whom the cancers were arising in BRCA2 mutation carriers; this method may represent a useful new screening method to identify these patients. Comparison of N-terminal and C-terminal BRCA2 immunostaining correctly classified 57 of 58 cancers arising outside the context of a multiple-case breast cancer family.
METHODS
Generation of C-Terminal Monoclonal Antibody
We generated a monoclonal BRCA2 antibody by using the C-terminal peptide 3,284 to 3,294 of sequence TFVSPAAKAGG. This peptide was conjugated to keyhole limpet hemocyanin and was used to immunize mice. As an initial test of the immune response, we screened immunized mice by enzyme-linked immunosorbent assay (ELISA) and then tested the highest-titer mouse sera by performing immunohistochemistry (IHC) on MCF7 cell pellets and cancer samples; we also used the antisera for Western blots. Two mice were chosen for splenectomy and cell fusion on the basis of the specificity for C-terminal BRCA2 protein by using the immune response initial testing strategy. Clonal supernatants were screened by Western blotting of cell samples and by IHC on a patient with breast cancer arising in a BRCA2 mutation carrier (ie, in a separate patient not included in the 21 patients with breast cancer for the study) and on another breast cancer from a patient with no family history of cancer. The best clones were selected by choosing those that were high titer by ELISA, that detected a single 220-kDa band on Western blot, and that differentiated the breast cancer arising in a BRCA2 mutation carrier from the other breast cancer sample cleanly. On the basis of this strategy, clone 575A15 was selected and cloned, and the supernatant from the cell line was affinity purified.
Western Blotting of Proteins
MCF7 cells were lysed in radioimmunoprecipitation assay buffer and were standardized for equal protein, and then samples were separated by Tris/acetate sodium dodecyl sulfate polyacrylamide gel electrophoresis on 3% to 8% Novex NuPage mini-gels (Invitrogen, Carlsbad, CA) for 1 hour and 160 volts. Proteins then were transferred to polyvinylidene difluoride membranes at 30 volts for 1 hour (Figs 1 and 2) in Tris/glycine transfer buffer that contained 8% methanol at 16°C. Membranes were blocked for 1 hour at room temperature in 5% dry, nonfat milk in phosphate-buffered saline (PBS). The primary 575A15 monoclonal antibody was incubated overnight at 4°C and was diluted 1:500 in 0.5% dry, nonfat milk/PBS-Tween 0.1%. After appropriate washes, antirabbit–horseradish peroxidase (GE Healthcare/Amersham Biosciences, Pittsburgh, PA) that was diluted 1:5,000 in 0.5% dry nonfat milk/PBS-Tween 0.1% was added and was incubated for 1 hour at room temperature. Detection was performed with ECL Plus reagents (GE Healthcare/Amersham Biosciences).
Fig 1.
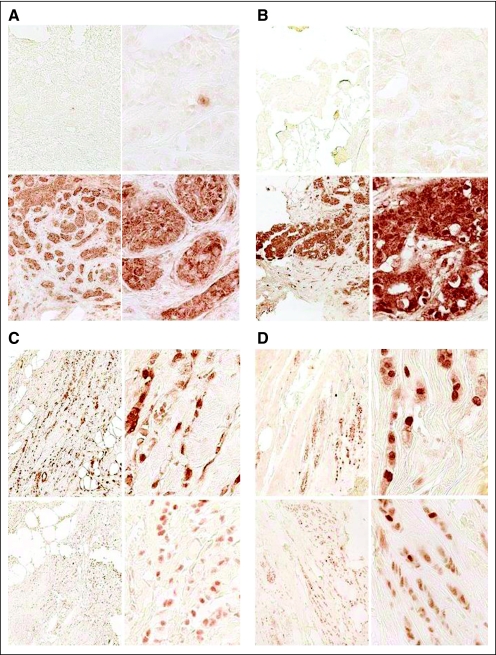
Dual C-terminal and N-terminal BRCA2 immunohistochemistry (IHC) of hereditary breast cancers (A and B, respectively) and of sporadic breast cancers (C and D, respectively). Magnification is (left) ×20 and (right) ×100 for all panels. The upper panel for each pair is C-terminal IHC, and the lower panel is N-terminal IHC. (A) Samples are from a BRCA2-mutant cancer with 7231del5. Top panel stained with C-terminal BRCA2 antibody, and second panel stained with N-terminal BRCA2 antibody. Only a lymphocyte in the top panel stains. (B) Samples are from a patient with a 9654delTT BRCA2 mutation at similar magnifications and similar staining. (C and D) The paired panels are from adjacent sections of the same sporadic breast cancer sample. Note the similar nuclear staining with both antibodies for the sporadic cancer samples.
Fig 2.
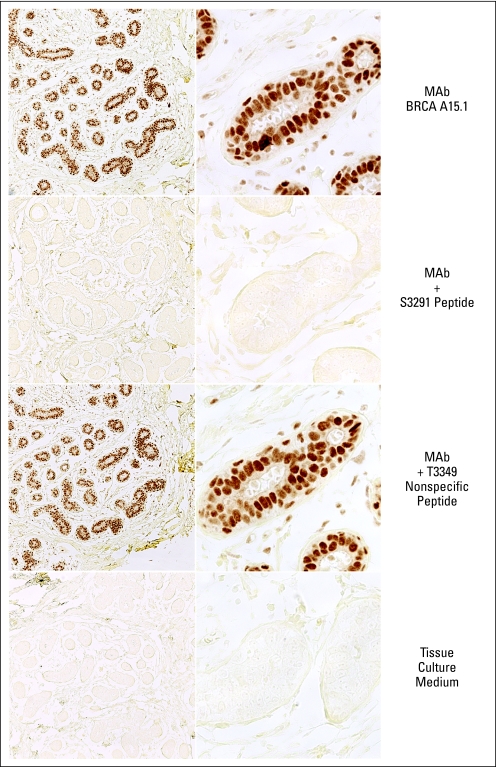
Immunohistochemistry with 575A15 C-terminal monoclonal antibody on normal breast epithelial lobules. Upper panels: untreated antibody. Middle panels: antibody mixed with excess of immunizing peptide (BRCA2 amino acids 3284 to 3294: TFVSPAAKAGG). Lower panels: antibody mixed with excess of control BRCA2 peptide (BRCA2 amino acids between 3300 and 3400, obtained from Abcam (Cambridge MA). Magnification is (left) ×20 and (right) ×100.
IHC
IHC analysis was performed on 4-micron sections of formalin-fixed, paraffin-embedded tissue. After deparaffinization and rehydration, heat-induced antigen retrieval was achieved in a 20 mmol/L citrate buffer at pH 6.0 for 10 minutes in a decloaking chamber (Biocare Medical, Concord, CA), and this was followed by a 25-minute, room temperature cooling period. Endogenous peroxidase was suppressed by incubating sections for 10 minutes in 3% hydrogen peroxide/PBS. Sections then were treated with a blocking solution (DAKO serum-free protein block; DAKO, Carpenteria, CA) for 1 hour. The N-terminal antibody used was from R&D systems MAB2476 (a commercially available mouse monoclonal antibody directed against an E coli–derived recombinant human BRCA2 protein that spans amino acids 1-200), which was used at 3 μg/mL. The C-terminal antibody was the newly generated mouse monoclonal 575A15 that is described in Methods and that was used at 4 μg/mL. Sections were incubated with the primary antibodies in a humidified chamber overnight at 4°C, were washed in buffer, and were treated with a horseradish peroxidase–labeled polymer detection system (DAKO Envision Plus, Dual Link) for 30 minutes at room temperature. Peptide block studies were performed by preincubating the C-terminal monoclonal antibody with a 50-fold excess of immunizing peptide antigen or a control peptide and staining as indicated. The chromagen used was DAKO DAB Plus (brown) for both the C-terminal and N-terminal antibodies. The cytoplasmic yellow counterstain was Metanil Yellow (ScyTek Laboratories, Logan, UT). The IHC results were read with the pathologist blinded to the carrier status of the cancer patients.
PCR Amplification of Tissue Samples and Genotyping by Restriction Digestion or DNA Sequencing
Paraffin sections on single slides were microdissected by our published method,14 were purified by xylene and ethanol extraction, and underwent proteinase K treatment and purification on a Qiagen column (Qiagen, Valencia, CA). PCR primers 5641F: 5′ ATGAAGATATTTGCGTTGAGGA and 6224R: 5′ CACTTGTCTTGCGTTTGTAATG were used to amplify DNA from tissue samples of normal breast, and normal and cancer samples from patients 13 and 21 were microdissected to obtain relative pure-normal versus cancer-cell populations. Amplified DNA then was purified on a Qiagen column and was restricted with the enzyme Hin4I, and samples were analyzed on a 2% agarose gel. PCR-amplified DNA also was used as a template for DNA sequencing with the nested primers 5753F: 5′ TGCATTTAGGATAGCCAGTG and 6000R: 5′ GAATGTCAGCAAAAACCTTAT.
RESULTS
Testing for protein-truncating mutations in patient DNA is a frequently used approach for breast cancer gene mutation screening. This is most commonly done by amplifying patient DNA with PCR and then assaying for truncation by in vitro translation—a test called the Protein Truncation Test.15 The goal in this research was to develop a method for identifying truncated proteins without DNA amplification or genetic manipulations of the tissue. Because there is a polymorphic DNA sequence variant found in 1% of the population in the C-terminus of BRCA2 that is not associated with breast cancer,16 we selected an epitope (amino acids 3284-3294) for immunization that would not identify that polymorphic truncated protein. Because of this design, all truncated BRCA2 proteins identified in breast cancers that used this antibody should be disease associated. To visualize protein truncation and to control for protein degradation or improper fixation, we employed an N-terminal antibody as a control to verify that full-length BRCA2 protein was present but truncated in breast cancers arising in BRCA2 mutation.
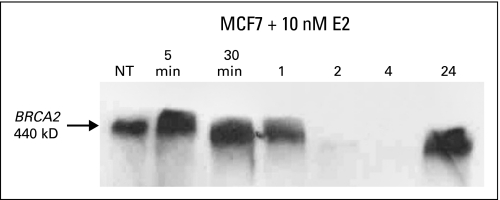
We selected the highest-titer monoclonal antibody that showed specificity for C-terminal BRCA2 protein in ELISA, Western blots, and IHC. Figure 3 shows that the 575A15 monoclonal antibody detects a single 440-kDa protein by Western blotting, which shows the expected biphasic expression pattern after estrogen treatment of MCF7 cells. To test the specificity of the monoclonal antibody for IHC, we performed peptide blocks with both the immunizing peptide and a control BRCA2 peptide. The results show complete inhibition of nuclear IHC staining by the immunizing peptide but no effect of the control peptide (Fig 2), which indicated that the monoclonal antibody is highly specific for BRCA2 protein even in tissue sections.
Fig 3.
MCF7 cells were cultured for 48 hours in 10% charcoal-stripped serum/phenol red–free DMEM and were treated with 10 nmol/L estrogen for 5 and 30 minutes and for 1, 2, 4, and 24 hours. Samples were blotted with the 575A15 BRCA2 C-terminal monoclonal antibody. The 440 kDa band is the only band on the blot.
To determine the sensitivity and specificity of this tissue truncation test for identifying BRCA2 hereditary cancers, we tested breast cancer samples from 21 different patients with BRCA2-truncating mutations and 58 sporadic breast cancers. The mutations for each patient are listed in Table 1. Figure 1A provides an example of such a tissue truncation test by demonstrating strong N-terminal BRCA2 protein in the lower panels but absent C-terminal staining in the upper panels within cancer cells. A lymphocyte within the tumor in Figure 1A provides strong nuclear staining, because the biallelic gene inactivation found in the cancer did not occur in nonmalignant cells such as inflammatory cells, which thus provides an internal control for the IHC process. Appendix Figure A1 (online only) also provides data to show that BRCA2 positivity is absent in tumors from mutations carriers but is present in adjacent normal breast tissue, because the wild-type allele is present. Figures 1B to 1D show protein truncation within cancers arising in BRCA2 mutation carriers but not within cancers arising outside the context of a multiple-case breast cancer family that show similar N-terminal and C-terminal immunostaining. By scoring the IHC as 0, 1+, 2+, or 3+ for both antibodies, we can determine N-terminal and C-terminal scores. Tables 1 and 2 show that this tissue truncation test can distinguish breast cancers arising in BRCA2 mutation carriers from likely sporadic breast cancers. The sensitivity for these tests for identifying these cancers was 95%, and the specificity was 98%.
Table 1.
BRCA2 Mutations Identified in Patients With Breast Cancer
| Patient | BRCA2 Mutation* | C-Terminal IHC | N-Terminal IHC | Truncation Ratio |
|---|---|---|---|---|
| 1 | 6503delTT | 0 and 0 | 2+ and 2+ | 0 |
| 2 | W2586X | 0 and 0 | 3+ and 3+ | 0 |
| 3 | W2586X | 0 and 0 | 3+ and 3+ | 0 |
| 4 | IVS7 + 2T>G | 0 and 0 | 2+ and 2+ | 0 |
| 5 | IVS7 + 2T>G | 0 and 0 | 2+ and 2+ | 0 |
| 6 | 9654delTT | 0 and 0 | 2+ and 3+ | 0 |
| 7 | 7231del5 | 0 and 0 | 2+ and 2+ | 0 |
| 8 | 7231del5 | 0 and 0 | 2+ and 3+ | 0 |
| 9 | 7231del5 | 0 and 0 | 3+ and 3+ | 0 |
| 10 | 7231del5 | 0 and 0 | 3+ and 3+ | 0 |
| 11 | 6503delTT | 0 and 0 | 3+ and 3+ | 0 |
| 12 | 1983del5 | 0 and 0 | 2+ and 2+ | 0 |
| 13 | S1882X | 3+ and 3+ | 3+ and 3+ | 1 |
| 14 | 5946delCT | 0 and 0 | 2+ and 2+ | 0 |
| 15 | 5849del4 | 0 and 0 | 2+ and 2+ | 0 |
| 16 | 983del4 | 0 and 0 | 2+ and 2+ | 0 |
| 17 | 983del4 | 0 and 0 | 3+ and 3+ | 0 |
| 18 | 4075delGT | 0 and 0 | 2+ and 2+ | 0 |
| 19 | 4075delGT | 0 and 0 | 2+ and 3+ | 0 |
| 20 | 3945delA | 0 and 0 | 3+ and 3+ | 0 |
| 21 | S1882X | 0 and 0 | 2+ and 2+ | 0 |
Abbreviation: IHC, immunohistochemistry.
Breast Cancer Information Core database designation, National Human Genome Research Institute.
Table 2.
Immunostaining Scores for Sporadic Breast Cancer Samples
| Patient | C-Terminal | N-Terminal | Patient | C-Terminal | N-Terminal |
|---|---|---|---|---|---|
| 1 | 3 and 3 | 2 and 2 | 30 | 2 and 2 | 2 and 2 |
| 2 | 3 and 2 | 3 and 2 | 31 | 2 and 3 | 2 and 2 |
| 3 | 2 and 3 | 2 and 2 | 32 | 1 and 1 | 1 and 1 |
| 4 | 3 and 2 | 3 and 2 | 33 | 2 and 2 | 2 and 2 |
| 5 | 3 and 3 | 3 and 2 | 34 | 2 and 3 | 2 and 3 |
| 6 | 3 and 3 | 3 and 2 | 35 | 3 and 3 | 3 and 3 |
| 7 | 2 and 3 | 2 and 2 | 36 | 3 and 3 | 2 and 2 |
| 8 | 3 and 2 | 3 and 2 | 37 | 3 and 3 | 2 and 2 |
| 9 | 3 and 3 | 3 and 2 | 38 | 3 and 3 | 2 and 3 |
| 10 | 3 and 3 | 2 and 3 | 39 | 3 and 2 | 2 and 2 |
| 11 | 3 and 3 | 2 and 2 | 40 | 3 and 3 | 3 and 3 |
| 12 | 3 and 3 | 3 and 2 | 41 | 2 and 2 | 2 and 2 |
| 13 | 3 and 3 | 2 and 3 | 42 | 3 and 3 | 3 and 3 |
| 14 | 2 and 3 | 2 and 2 | 43 | 2 and 2 | 2 and 3 |
| 15 | 3 and 3 | 3 and 2 | 44 | 2 and 2 | 2 and 2 |
| 16 | 3 and 2 | 2 and 2 | 45 | 2 and 3 | 3 and 2 |
| 17 | 3 and 3 | 3 and 2 | 46 | 2 and 2 | 2 and 2 |
| 18 | 2 and 3 | 3 and 2 | 47 | 2 and 2 | 1 and 2 |
| 19 | 3 and 3 | 2 and 2 | 48 | 1 and 1 | 1 and 1 |
| 20 | 2 and 3 | 3 and 3 | 49 | 2 and 2 | 2 and 2 |
| 21 | 2 and 3 | 2 and 2 | 50 | 3 and 2 | 2 and 2 |
| 22 | 3 and 3 | 2 and 2 | 51 | 2 and 2 | 2 and 2 |
| 23 | 3 and 3 | 2 and 3 | 52 | 3 and 3 | 2 and 3 |
| 24 | 3 and 2 | 2 and 2 | 53 | 3 and 2 | 2 and 2 |
| 25 | 2 and 3 | 2 and 2 | 54 | 2 and 3 | 2 and 2 |
| 26 | 3 and 3 | 3 and 3 | 55 | 2 and 2 | 2 and 2 |
| 27 | 2 and 2 | 2 and 2 | 56 | 2 and 2 | 2 and 2 |
| 28 | 3 and 3 | 3 and 3 | 57 | 1 and 1 | 1 and 1 |
| 29 | 2 and 2 | 2 and 2 | 58 | 3 and 3 | 2 and 2 |
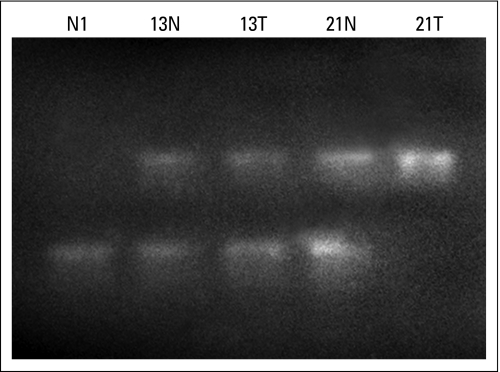
Because BRCA2-mutant cancer sample 13 had atypical clinical features (ie, estrogen-receptor negative) and expressed abundant C-terminal BRCA2 protein (Table 1), we hypothesized that this might represent an atypical cancer for a BRCA2 mutation carrier, which lacks the expected biallelic gene inactivation. Because inherited cancers normally undergo a somatic loss of wild-type BRCA2 sequence within the cancer, we PCR amplified this region of BRCA2 and then digested the amplified DNA with the restriction enzyme Hin4I, which digests the wild-type sequence but not the mutant-truncating 5873C→A DNA sequence. This occurs because Hin4I has a recognition sequence of 5′ 8(N)G A Py (N)5 (A/C/G) T C (N)13-3′, so the truncating mutation destroys the final C in the recognition sequence. Appendix Figure A2 (online only) shows that Hin4I digests normal tissue and patient 13 with cancer to a similar extent. However, cancer patient 13 has a 5873C→A mutation; however, in this instance, the enzyme does not digest tumor DNA, because the wild-type sequence is not present.
DISCUSSION
We have developed an IHC-based tissue truncation method to visualize protein truncations in BRCA2 hereditary breast cancer. This method was tested with both breast cancers arising in BRCA2 mutation carriers and sporadic breast cancers, and the method had a sensitivity of 95% and specificity of 98%. This strategy provides a direct demonstration that a truncating mutation is present within a sample, unlike a mere loss of C-terminal or internal immunostaining that could result from decreased expression, promoter methylation, protein degradation, or other reasons. Results listed in Table 2 demonstrate the importance of comparing IHC by using both antibodies, because analysis of only a C-terminal antibody would likely misclassify three samples (ie, patients 32, 48, and 57), which would reduce the test specificity to 93%.
There are two clear caveats for identifying BRCA2 mutations by searching for truncated proteins: first, this method cannot identify missense mutations or DNA sequence variants of unknown significance; second, truncated proteins must be relatively stable or the N-terminal IHC also would be negative. Because many BRCA2 missense variants have been identified as neutral (ie, not disease associated) and others have been identified as unknown significance (and therefore not reliable enough for genetic counseling at this time), this method should be able to rapidly identify most, but probably not all, disease-associated BRCA2 mutations.6,7 As missense mutations are identified that clearly are disease-associated, strategies need be developed to screen for these occurrences; this is also true for protein-truncation testing on DNA samples. If, in future samples, some truncated proteins are unstable, as has been reported,17 then the truncation test might be confusing, because both C-terminal and N-terminal IHC would be diminished or zero. A cellular process known as nonsense-mediated decay often produces instability of mRNAs that contain truncating (ie, nonsense) mutations that lead to protein truncation. The amount of mRNA instability often varies on the basis of the distance from the nonsense codon to the beginning of the exon,18 and this effect has reportedly decreased BRCA2 mRNA levels from 1.5-fold to four-fold.19 However, this finding differs from studies that have analyzed protein levels in transfected cells20 and that did not show marked decreases in protein levels in cells that expressed truncated BRCA2 proteins. We also did not observe decreased N-terminal IHC staining of BRCA2 proteins in the majority of patients, as listed in Table 2, although three patients with likely sporadic cancers did show 1+ staining with both N-terminal and C-terminal antibodies. Decreased total BRCA2 protein levels can be identified if BRCA2 hereditary cancers are identified as a result of regulatory mutations or large deletions. A summary of the strengths and weaknesses of this IHC strategy is listed in Table 3.
Table 3.
Strengths and Weaknesses of Immunohistochemistry Truncation Test
| Strengths | Weaknesses |
|---|---|
| Rapid, 4-hour turnaround | Cannot identify missense mutations |
| Excellent sensitivity and specificity | Nonsense-ediated decay could produce false negatives |
| More patients effectively screened | Does not identify the specific DNA mutation |
| Does not find missense variants | Possible false positives as a result of expression levels |
Because of the simplicity of this IHC test, it may be completed within 4 hours of receipt of tissue blocks and also may be used on small amounts of archival material. Because this method identifies the molecular lesion responsible for BRCA2 hereditary cancer (the loss of full-length functional BRCA2 protein), it is fundamentally a functional test that distinguishes cancers that lack wild-type BRCA2 protein from cancers that express wild-type BRCA2 protein. For this reason, these IHC results occasionally may differ from DNA sequence results if a sporadic-type cancer (without biallelic BRCA2 inactivation) occurs in a patient with a germline mutation. Because breast cancer is a common disease, it probably is not surprising to find some breast cancers in BRCA2 mutation carriers that appear to result from more sporadic breast cancer mechanisms rather than from the expected biallelic gene inactivation thought responsible for BRCA2 hereditary cancer. The data in Appendix Figure A2 suggest that cancer in patient 13 is such an occurrence. Because loss of heterozygosity in this region of chromosome 13 occurs in approximately 20% to 30% of sporadic breast cancers without germline mutations of BRCA2, this type of analysis is only informative in instances in which the wild-type allele is lost. The truncation testing strategy in this study clearly misclassified this patient for the purpose of identifying mutation carriers and family screening (hence, our 95% sensitivity, because this patient was misclassified). However, it may have provided the correct result for selecting targeted therapies aimed at BRCA2-defective cancers. Additional work is necessary to determine how frequently these types of patients are identified and whether the germline mutation or the loss of intact BRCA2 protein is a better predictor of therapeutic response.
An IHC test may be performed at most hospital laboratories and does not require complex or expensive instrumentation. Vaz et al21 have reported a C-terminal IHC test to detect BRCA1 mutations but have analyzed only a few patients and controls. IHC testing also may be performed on archived paraffin blocks that have been stored for some time. Several of our tested paraffin blocks were from patient samples that were more than 20 years old and still gave reliable data. One advantage of a successful IHC approach is that this could be done irrespective of family history, which is not a good indicator of carrier status, particularly in young women. The ability to perform a truncation test for hereditary cancer at time of biopsy and resection could be useful to identify appropriate therapies for these patients. BRCA2-mutant cancers reportedly have been more sensitive to ionizing radiation,22 poly (ADP-ribose) polymerase inhibitors,11–12 and cisplatin,23 so more complete identification of hereditary breast cancers could facilitate the appropriate targeted therapy for these patients. Although this truncation test cannot be used directly to screen family members, it does identify patients who likely have truncating mutations, so DNA sequencing of patients and subsequent screening of family members could be useful for cancer prevention.
Acknowledgments
The Acknowledgment and Appendix are included in the full-text version of this article; they are available online at www.jco.org. They are not included in the PDF version (via Adobe® Reader®).
We thank Jennifer Malone for performing the Western blot, Lisa Litzenberger for photography, and Kimberly Gibson for editorial assistance.
Appendix
Fig A1.
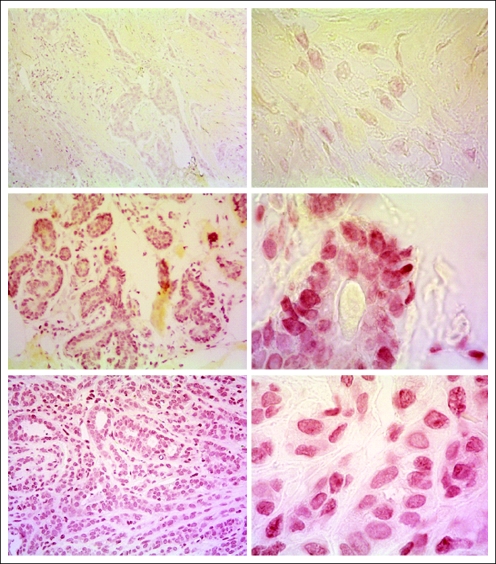
Immunohistochemistry performed on human breast tissue by using the C-terminal BRCA2 antibody provided by EMD Chemicals (Gibbstown, NJ). The red color represents specific staining of the antibody peroxidase reaction, and the yellow color represents a neutral counter stain, Metanil Yellow. Left panels are ×20, and right panels ×100, magnification. Tissue sections show BRCA2 hereditary cancer (upper panels); normal breast from patient with BRCA2 hereditary cancer (middle panels); and sporadic cancer (lower panels). The absence of staining in the hereditary cancer sections (upper right and left panels) are caused by the 6174delT truncation mutant in this patient.
Fig A2.
Relative presence of wild-type and mutant 5873C→A DNA sequences in cancers and normal tissues. Lane 1 is normal breast tissue from an individual with nonmutant tissue; lane 2 is normal breast tissue; lane 3 is breast cancer tissue from patient 13, who had 5873C→A BRCA2 mutation; lane 4 is normal breast tissue; and lane 5 is breast cancer tissue from patient 21, who had 5873C→A BRCA2 mutation. The normal tissue in lane 1 is entirely digested (583 base pairs), and cancer tissue in lane 5 is entirely undigested (351 base pairs), as would be expected if only the 5873C or the 5873A allele is present, respectively.
Footnotes
Supported by Grants No. R41CA128233 (J.T.H.) and CA086389 (H.T.L.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Vanessa J. Clark, Tissue Genetics (C); Jeffrey T. Holt, Tissue Genetics (U) Consultant or Advisory Role: None Stock Ownership: Jeffrey T. Holt, Tissue Genetics Honoraria: None Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Patrice Watson, Rita Lieberman, Carrie Snyder, Vanessa J. Clark, Henry T. Lynch, Jeffrey T. Holt
Financial support: Henry T. Lynch, Jeffrey T. Holt
Administrative support: Patrice Watson, Rita Lieberman, Carrie Snyder, Vanessa J. Clark, Henry T. Lynch, Jeffrey T. Holt
Provision of study materials or patients: Patrice Watson, Carrie Snyder, Henry T. Lynch, Jeffrey T. Holt
Collection and assembly of data: Patrice Watson, Rita Lieberman, Carrie Snyder, Vanessa J. Clark, Henry T. Lynch, Jeffrey T. Holt
Data analysis and interpretation: Patrice Watson, Rita Lieberman, Carrie Snyder, Vanessa J. Clark, Henry T. Lynch, Jeffrey T. Holt
Manuscript writing: Patrice Watson, Rita Lieberman, Carrie Snyder, Vanessa J. Clark, Henry T. Lynch, Jeffrey T. Holt
Final approval of manuscript: Patrice Watson, Rita Lieberman, Carrie Snyder, Vanessa J. Clark, Henry T. Lynch, Jeffrey T. Holt
REFERENCES
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Lancaster JM, Wooster R, Mangion J, et al. BRCA2 mutations in primary breast and ovarian cancers. Nat Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]
- 3.Kwiatkowska E, Teresiak M, Breborowicz D, et al. Somatic mutations in the BRCA2 gene and high frequency of allelic loss of BRCA2 in sporadic male breast cancer. Int J Cancer. 2002;98:943–945. doi: 10.1002/ijc.10289. [DOI] [PubMed] [Google Scholar]
- 4.Collins N, McManus R, Wooster R. Consistent loss of the wildtype allele in families from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- 5.Osorio A, De la Hoya M, Rodriguez-Lopez R. Loss of heterozygosity at the BRCA loci in tumor samples from patients with familial breast cancer. Int J Cancer. 2002;99:305–309. doi: 10.1002/ijc.10337. [DOI] [PubMed] [Google Scholar]
- 6.Narod SA, Offit K. Prevention and management of hereditary breast cancer. J Clin Oncol. 2005;23:1656–1663. doi: 10.1200/JCO.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Vink GR, van Asperen CJ, Devilee P, et al. Unclassified variants in disease-causing genes: Nonuniformity of genetic testing and counselling, a proposal for guidelines. Eur J Hum Genet. 2005;13:525–527. doi: 10.1038/sj.ejhg.5201379. [DOI] [PubMed] [Google Scholar]
- 8.Rebbeck TR, Lynch HT, Neuhausen SL, et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N Engl J Med. 2002;346:1616–1622. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- 9.Metcalfe KA, Lynch HT, Ghadirian P, et al. The risk of ovarian cancer after breast cancer in BRCA1 and BRCA2 carriers. Gynecol Oncol. 2005;96:222–226. doi: 10.1016/j.ygyno.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 10.King MC, Marks JH, Mandell JB. The New York Breast Cancer Study Group: Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 11.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 12.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 13.Simchoni S, Friedman E, Kaufmann B. Familial clustering of site specific cancer risks associated with BRCA1 and BRCA2 mutations in the Ashkenazi Jewish population. Proc Natl Acad Sci U S A. 2006;103:3770–3774. doi: 10.1073/pnas.0511301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen RA, Page DL, Holt JT. Identification of genes expressed in premalignant breast disease by microscopy-directed cloning. Proc Natl Acad Sci U S A. 1994;91:9257–9261. doi: 10.1073/pnas.91.20.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roest PA, Roberts RG, Sugino S, et al. Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum Mol Genet. 1993;2:1719–1721. doi: 10.1093/hmg/2.10.1719. [DOI] [PubMed] [Google Scholar]
- 16.Mazoyer S, Dunning AM, Serova O, et al. A polymorphic stop codon in BRCA2. Nat Genet. 1996;14:253–254. doi: 10.1038/ng1196-253. [DOI] [PubMed] [Google Scholar]
- 17.Walsh T, Casadie S, Coats KH, et al. Spectrum of mutations in BRCA1, BRCA2 CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 18.Kuzmiak HA, Mauqat LE. Applying nonsense-mediated mRNA decay research to the clinic: Progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Ware MD, DeSliva D, Sinilnikova OM, et al. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene. Oncogene. 2006;25:323–328. doi: 10.1038/sj.onc.1209033. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Silver DP, Walpia D, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 21.Vaz FH, Machado PM, Brandao RD, et al. Familial breast/ovarian cancer and BRCA1/2 genetic screening: The role of immunohistochemistry as an additional method in the selection of patients. J Histochem Cytochem. 2007;55:1105–1113. doi: 10.1369/jhc.7A7209.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Nat Cancer Inst. 1998;90:978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 23.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]