Abstract
bHLH-O proteins are a subfamily of the basic-helix-loop-helix transcription factors characterized by an ‘Orange’ protein-protein interaction domain. Typical members are the Hairy/E(spl), or Hes, proteins, well studied in their ability, among others, to suppress neuronal differentiation in both invertebrates and vertebrates. Hes proteins are often effectors of Notch signalling. In vertebrates, another bHLH-O protein group, the Hey proteins, have also been shown to be Notch targets and to interact with Hes. We have studied the single Drosophila Hey orthologue. We show that it is primarily expressed in a subset of newly born neurons, which receive Notch signalling during their birth. Unlike in vertebrates, however, Hey is not expressed in precursor cells and does not block neuronal differentiation. It rather promotes one of two alternative fates that sibling neurons adopt at birth. Although in the majority of cases Hey is a Notch target, it is also expressed independently of Notch in some lineages, most notably the larval mushroom body. The availability of Hey as a Notch readout has allowed us to study Notch signalling during the genesis of secondary neurons in the larval central nervous system.
Keywords: Drosophila, Hey, Notch, Asymmetric cell division, bHLH-O, Cell fate
INTRODUCTION
Among the superfamily of basic-helix-loop-helix (bHLH) transcription factors, several structurally distinct classes are discerned. One of these, the bHLH-Orange (bHLH-O) class (Fischer and Gessler, 2007; Iso et al., 2003), is characterized by the ‘Orange’ domain, a protein interaction domain perhaps serving as an extended dimerization surface (Taelman et al., 2004). bHLH-O proteins are important developmental and physiological regulators in processes ranging from neurogenesis to circadian rhythm control.
In a number of invertebrate and vertebrate species, bHLH-O repressors are known to inhibit neural differentiation. In Drosophila, the seven E(spl) bHLH-O proteins are expressed in the neuroectoderm, where they inhibit cells from differentiating as neuroblasts (NBs) (Jennings et al., 1994; Nakao and Campos-Ortega, 1996). In vertebrates, a number of Hes bHLH-O proteins, notably Hes1, Hes3 and Hes5 in the mouse, are also expressed in the neuroectoderm; in this case it is the neural stem cells that express the Hes genes, which are subsequently downregulated in the differentiating neuronal progeny (Kageyama et al., 2008). Triple Hes1, Hes3, Hes5 knock-out causes premature neural differentiation, disruption of the neuroepithelium and a hypoplastic nervous system owing to stem cell depletion (Hatakeyama et al., 2004). In Drosophila, loss of the entire E(spl) locus results in supernumerary NB specification from the neuroectoderm and a hyperplastic nervous system (Lehman et al., 1983). Despite these differences, owing to the different mode of neural precursor specification between vertebrates and insects, the generalization can be made that E(spl)/Hes proteins antagonize neuronal differentiation. At most developmental settings across metazoan phylogeny, neural expression of E(spl)/Hes genes is a direct response to Notch signalling (Bailey and Posakony, 1995; Lecourtois and Schweisguth, 1995; Ohtsuka et al., 1999).
Expression of another subfamily of bHLH-O genes has been detected in the progenitor cell zones of the developing vertebrate central nervous system (CNS). These genes encode the three Hey proteins, so named after a characteristic tyrosine residue in their C-terminal domain (Hairy/enhancer-of-split like with a Y); they are also known as Hrt, Herp, Hesr, Chf or Gridlock (Kokubo et al., 1999; Leimeister et al., 1999). Although neural defects are minor in Hey knock-out mice, overexpression studies have suggested that Hey and Hes proteins might synergize with each other in suppressing neural differentiation and maintaining the neural stem cell fate (Sakamoto et al., 2003). Hey1 has even been linked to the pathogenesis and aggressiveness of gliomas (Hulleman et al., 2009). Hey knock-out mice have highlighted their roles in developmental processes outside the nervous system, in particular, heart and vasculature development (Fischer et al., 2004; Kokubo et al., 2005). In these contexts, all three mammalian Hey genes appear to respond to Notch signalling, similar to E(spl)/Hes genes in neurogenesis. Biochemical data support Hes-Hey heterodimer formation (Iso et al., 2001; Taelman et al., 2004), raising the possibility that these two subclasses of bHLH-O proteins might synergize in some developmental contexts as Notch effectors.
The Drosophila genome contains a single Hey orthologue (Kokubo et al., 1999), which had not been studied to date. We decided to characterize it in the hope of better understanding the process of neural precursor specification, based on the assumption that, by analogy to vertebrates, Hey might display protein-protein interactions with E(spl). To our surprise, Hey was not co-expressed with the E(spl) proteins in the neuroectoderm, rather was restricted to differentiating neurons, suggesting a radically different role in neurogenesis than we had assumed. Once NBs are specified in Drosophila, they undergo cycles of asymmetric cell divisions that give rise to a secondary precursor, called a ganglion mother cell (GMC), in addition to self-renewing. GMCs divide once to give rise to two neurons or, less often, glia. The majority of GMC divisions are asymmetric, with the fates of the two daughters dictated by unequal levels of Notch signalling. The ‘A’ sibling neuron requires high Notch signalling, whereas the ‘B’ sibling neuron downregulates Notch reception, which is usually achieved by asymmetric segregation of a Notch inhibitor, Numb, into the nascent ‘B’ neuron (Skeath and Thor, 2003). We describe a complex pattern of Hey expression in relation to these divisions during both neurogenic phases of the animal, early embryogenesis and larval life, where thousands of new neurons are added to generate the adult CNS (Maurange and Gould, 2005). In all sibling pairs that we could identify, Hey was expressed in the ‘A’ neuron. Genetic analysis confirmed that Hey is a Notch target gene in most instances. Our results extend the Hey-Notch relationship to Drosophila in support of an ancient connection between bHLH-O genes and Notch activity and, for the first time, implicate a bHLH-O protein in the process of GMC asymmetric division.
MATERIALS AND METHODS
DNA constructions
Cloning of Drosophila Hey was done by PCR amplification from an embryonic cDNA library using gene-specific primers: Forward (EcoRI): gccgaattcATGGATCACAACATG; Reverse (XhoI): taactcgagTCAATAGGCCATCTC. Amplified sequences were cloned into Bluescript and pGEM-T easy vectors. To ectopically express Hey in vivo, we subcloned the cDNA (EcoRI/XhoI) into the pUAST vector (Brand and Perrimon, 1993) and used it to transform flies.
Fly strains
All fly stocks were obtained from the Bloomington Stock Centre, the Exelixis Collection at Harvard or individual laboratories (see acknowledgements). The enhancer trap line AJ96-lacZ was used to identify the dMP2/vMP2 neurons (Menne and Klambt, 1994). K33-lacZ, an enhancer trap in E(spl)mγ (HLHmγ – FlyBase) (Cooper et al., 2000) was used to mark larval NBs. Mutant backgrounds were: Hey f06656/CyO; w; Df(2R)ED1735/CyO; w; Df(2R)Exel6055/CyO; numb2 pr cn Bc/CyO ftz-lacZ; th st cu e spdoc55 ca/TM3 ftz-lacZ; mam-lacZ04615/CyO; e Df(3R)E(spl)b32.2/TM3; w; FRT82B Dlrev10 e SerRX106/TM6B; w; FRT82B Dlrev10/TM6B; y wa N54l9 FRT19A/FM7; w; Su(H)Δ47 FRT40A/CyO; h th st FRT82B neur1 cu e/TM6B; w; mib1EY9780 FRT2A/TM6B; w; FRT82B es spdoG104/TM3. For mosaic generation using the MARCM technique (Lee and Luo, 2001), appropriate FRT counter-chromosomes were used bearing αtubGal80 transgenes in the background of hs-FLP, atub-Gal4 and UAS-GFP. For ectopic expression studies we used ftz.ng-Gal43 and 253-Gal4 drivers combined with the UAS-Hey responders generated in this work. Larval and embryonic over-expressions were performed at 25°C and 30°C, respectively.
Antibody production
Full length Hey cDNA was cloned into the bacterial expression vector pET16b (Novagen) in frame with the 6×His tag, under the T7 promoter. Cultures of transformed BL21(DE3) E. coli were induced with 1 mM IPTG for 3 hours at 37°C and recombinant protein was purified under denaturing conditions (8 M urea), on a Ni-NTA resin (Qiagen) according to the manufacturer's instructions. The purified protein was used to immunize mice and guinea pigs at Davids Biotechnologie (www.dabio.de).
In situ hybridization and immunohistochemistry
Fixation and subsequent in situ hybridization or immunohistochemistry of embryos and dissected larval tissues was performed according to standard protocols. Primary antibodies were rabbit anti-β-gal, 1:10000 (Cappel); mouse anti-β-gal, 1:1000, (Promega); rabbit anti-Eve, 1:2000 (gift from M. Frasch) (Frasch et al., 1987); mouse anti-Eve 2B8, 1:30 (Patel et al., 1994); guinea pig anti-Odd, 1:200 (gift from Dave Kosman); rat anti-Elav 7E8A10, 1:100 [Developmental Studies Hybridoma Bank (DSHB)]; mouse anti-Prospero MR1A, 1:20 (DSHB); mouse anti-Hnt, 1:50 (DSHB); rabbit anti-GFP, 1:30,000 (Minotech); mouse anti-Repo 8D12, 1:25 (DSHB); guinea pig or mouse anti-Hey 1:1000 (this study); rat anti-Dpn, 1:1 (gift from C.Doe); rat anti-E-cadherin 1:20 (DSHB); rabbit anti-Ase 1:1000 (gift from A. Jarman); and rabbit anti-Numb 1:1000/1:100 (gifts from Y.-N. Jan and J. Knoblich). Detection was done using secondary antibodies conjugated to Alexa 488, 555, 568, 633 or 647 (Molecular Probes), or Cy3 (Jackson ImmunoResearch). Embryos and tissues were imaged on a BioRad Radiance 2100 or Leica SP2 confocal microscope.
RESULTS
Hey is transiently expressed in a subset of embryonic neurons and glia
We amplified a full-length Hey cDNA from a Drosophila cDNA library, which we used as a probe for in situ hybridization (Fig. 1A), and for cloning in prokaryotic expression vectors. We then used bacterially expressed full-length Hey protein to raise anti-Hey antibodies. There were no obvious differences between the RNA and protein patterns. Hey protein showed nuclear accumulation, as expected for a transcription factor, and was primarily detected in a segmentally repeated pattern within the CNS (Fig. 1A,B) starting at stage 10. Later, more Hey-positive cells gradually appear in the CNS. The neuroectodermal epithelium, where the related E(spl) bHLH-O proteins are expressed already starting at stage 8 (Jennings et al., 1994), is devoid of Hey expression, which instead is detected at deeper levels overlapping with the GMC/immature neuron marker Pros (Fig. 1C,D) (Vaessin et al., 1991). From double-staining with the neuronal antigen Elav (Robinow and White, 1991) it was clear that the vast majority of Hey-positive cells represent neurons (Fig. 1E,F) rather than GMCs, confirmed as lack of colocalization with the NB/GMC marker Asense (Brand et al., 1993) (Fig. 1G,G′). Besides neurons, Hey expression was detected in a subset of Repo-positive glia of the CNS (Halter et al., 1995) (Fig. 1H,H′) and peripheral nervous system (PNS; not shown). Of note, Eve staining, which was used to visualize particular neurons (see below), also marks the dorsally located pericardial cells (Frasch et al., 1987; Su et al., 1999). No Hey immunoreactivity was detected within or near these heart precursors (Fig. 1C), contrary to the strong expression of mammalian Hey genes during cardiogenesis. Finally, a few Hey-positive cells per segment were detected in the embryonic PNS (Fig. 1A,B). Most of these were also neurons, by virtue of being Elav-positive (data not shown), but were not characterized further.
Fig. 1.
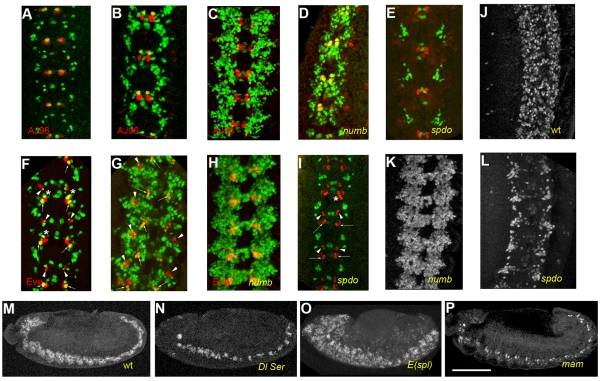
Expression of Hey in the embryonic CNS. (A) Hey mRNA and (B) Hey protein patterns in stage 12 embryos. Both RNA and protein are detected in the CNS (most of the signal) and PNS (arrows). Anterior is left and ventral is down. (C) Ventral view of a stage 11 embryo stained for Pros (blue), Eve (green) and Hey (red). Hey-positive cells are a subset of the Pros-positive cells (GMCs and immature neurons). Eve marks a small subset of neurons and pericardial cells (arrows). (D) Sagittal view of stage 12 embryo stained for β-galactosidase (green) to image E(spl)-m8-lacZ expression in epithelial cells and Hey (red). Anterior is left and ventral is down. Note the lack of Hey staining in the superficial neuroepithelium (arrow). Weak β-galactosidase staining in the deeper neuronal layers (arrowhead) is probably perduring protein from earlier neuroepithelial expression. (E-H′) Ventral views of embryos, anterior is up. (E,F) Images of two different focal planes (E is more superficial) of a stage 15 embryo stained for Hey (green) and Elav (red). Elav marks neurons. Only a few Hey-positive cells (white arrows) are Elav-negative. (G,G′) Stage 10 embryo stained for Hey (green) and Asense (red, grey in G′). Asense marks GMCs and neuroblasts. Arrowheads in G′ indicate rare cases of GMCs that express Hey. (H,H′) Hey (green) and Repo (red, grey in H′) in a stage 15 embryo. Arrowheads in H′ mark examples of Repo-positive cells expressing Hey. Scale bars: 100 μm in A-C; 16 μm in D; 50 μm in E-H.
We used lineage-specific markers to characterize Hey expression in more detail. One was Even skipped, which marks a subset of neurons (Skeath and Doe, 1998): the aCC/pCC sibling pair, the RP2 motoneuron, the cluster of U motoneurons and the cluster of EL interneurons. Another was the AJ96-lacZ enhancer trap (Menne and Klambt, 1994), which marks the MP2 precursor and its progeny, the dMP2/vMP2 neurons. With AJ96-lacZ, we detected strong Hey accumulation in vMP2 but not in dMP2 (Fig. 2B). We could even detect weak Hey expression shortly before mitosis of the MP2 progenitor during late stage 10 (Fig. 2A). Among the Eve-positive neurons (Fig. 2G,H), pCC and the U neurons expressed Hey. aCC, RP2 and the EL neurons were Hey-negative. At stage 11, the sibling of RP2, RP2sib, a smaller cell, which only transiently expresses Eve, was Hey-positive (Fig. 2G). Hey expression in all these neurons appeared transient. For example, whereas immunoreactivity in vMP2 was strong at stage 12, it was downregulated and barely detectable by stage 14 (Fig. 2C). Similarly, by stage 14 no Hey could be detected in pCC cells, although it was still expressed strongly in some of the later-born U motorneurons (Fig. 2H). Transient Hey expression was also observed in the two identical progeny of MP1, a midline precursor, which are marked by Odd (see Fig. S1 in the supplementary material).
Fig. 2.
Hey expression in wild-type and mutant backgrounds. (A-E) AJ96-lacZ line stained for Hey (green) and β-gal (red) to mark the vMP2/dMP2 cells in wild-type (A-C) and mutant backgrounds (D,E). (A) Hey expression in the MP2 lineage starts at stage 10 within the undivided MP2 neuroblast. (B) In stage 12 embryos Hey is expressed in vMP2s, the anteriorly located AJ96-positive cells, but not in dMP2s. (C) In stage 15 embryos Hey expression is turned off. (D) In the numb genetic background, which induces transformation of dMP2 into vMP2, Hey is expressed in both AJ96-positive cells (stage 12). (E) The opposite (Hey absence) is observed in spdo embryos, in which vMP2 is transformed into dMP2 (stage12). Note the scarcity of Hey-positive cells (also in I) compared with equivalently staged wild-type (B) or numb (D) embryos. (F-I) Hey expression in Eve-positive lineages in wild-type (F,G) and mutant (H,I) backgrounds. (F) A stage 11 embryo montage showing deep focal planes with aCC/pCC and RP2/RP2sib pairs marked with Eve (red). Hey is expressed in pCC (arrow) but not aCC. RP2 does not express Hey in contrast to the smaller RP2sib (arrowhead) which is still Eve-positive at this stage. In some segments a Hey/Eve-positive U cell (asterisk) is evident. (G) A stage 15 embryo montage of superficial focal planes to visualize U and EL lineages. Eve marks the U and EL cells. Hey is expressed in the U cells (arrows) but not in the EL cells (arrowheads). (H) In a stage 15 numb embryo more cells within the U-cluster are labelled with Eve, as Usib is transformed into U. (I) Loss of Notch signalling in a spdo embryo (stage 11) results in two RP2s (arrowheads) and two aCCs (arrows) per hemisegment. Eve-positive U cells are transformed to Usib cells (Eve-negative or weakly positive near the aCC pairs). Persistent Eve-positive cells at the U position (asterisk) are either undivided GMCs or Usib cells that have not yet extinguished Eve expression. None of these cells express Hey. Hey expression is limited to a few midline cells and a cluster of lateral cells. (J-P) Ventral views of stage 15 embryos (J-L) and sagittal views of stage 12 embryos (M-P) of different genetic backgrounds stained for Hey. (J) Wild-type; (K) numb2; (L) spdoc55; (M) wild-type; (N) Dlrev10 SerRX106; (O) Df(3R)E(spl)b32.2; (P) mam04615. Note the increased number of Hey-positive neurons in the E(spl) and numb embryos compared with that of wild type embryos. Conversely, there are fewer Hey-positive neurons in Dl Ser, mastermind and spdo embryos. Anterior is up (A-L) or to the left (M-P). Scale bars: 55 μm in A-L; 130 μm in M-P.
Most of the neurons described above belong to well-characterized lineages, in which sibling fates arise through differential Notch signalling. In each of the RP2/RP2sib, aCC/pCC and dMP2/vMP2 pairs, the second cell requires Notch signalling in order to acquire the ‘A’ fate, distinct from that of its sibling cell (‘B’ fate) (Skeath and Doe, 1998; Spana and Doe, 1996). Also in the U lineages, which arise from sequential GMCs from neuroblast NB7-1 (Cleary and Doe, 2006), the U neurons require Notch, whereas their Eve-negative Usib neurons do not. All Notch-requiring cells, namely RP2sib, pCC, vMP2 and the U cells, robustly express Hey, whereas none of their ‘B’-fate siblings do so. This raises the possibility that Hey is expressed in response to Notch.
Hey is a target of Notch signalling in the embryonic CNS
We used mutations that perturb Notch signalling to address whether Hey expression is regulated by Notch. As Notch is involved in a number of developmental decisions before neuron birth, most notably NB lateral inhibition (Lehman et al., 1983), it is expected that Notch-null embryos will exhibit a complex phenotype, which might obscure a later effect on Hey. We therefore turned our attention to mutations that have no defects in lateral inhibition, but disrupt Notch signalling specifically at later asymmetric cell divisions. Although Mastermind is an essential nuclear cofactor in Notch signalling (Bray, 2006), the hypomorphic mam04615 allele has sufficient activity to carry out lateral inhibition normally, but fails during asymmetric cell divisions (Skeath and Doe, 1998). spdo is dispensable for lateral inhibition, but its disruption abolishes Notch signalling, specifically in asymmetric cell divisions (O'Connor-Giles and Skeath, 2003; Skeath and Doe, 1998). In homozygous embryos for either mam04615 or spdoc55 we detected a dramatic loss in Hey immunostaining (Fig. 2E,I,L,P) compared with wild-type embryos of the same stage. In an AJ96-lacZ background, we could not detect Hey in either of the MP2 progeny neurons, in agreement with fact that this division is now symmetric, producing two dMP2 (Hey-negative) cells (Fig. 2E). Staining spdoc55 embryos with Eve reveals that Hey is absent from the symmetric aCC/aCC pair, that arises because of pCC-to-aCC cell fate switching. Finally, transformations of RP2sib into RP2 (two Eve-positive cells instead of one) and U into Usib (Eve-negative) result in the disappearance of Hey expression in RP2/RP2sib and U/Usib lineages as well (Fig. 2I). Similar Notch dependence of Hey expression is likely to occur in most other neuronal lineages, explaining the global reduction in Hey-positive cells.
In a converse experiment, we elicited ectopic Notch activity in ‘B’ neurons by using loss-of-function alleles of numb, which normally inhibits Notch signalling within ‘B’ neurons (Spana and Doe, 1996). In the severe numb2 mutant (Skeath and Doe, 1998), asymmetric GMC divisions become symmetric, giving rise to two ‘A’-type neurons, a phenotype opposite to that of mam and spdo. In agreement with Hey being a target of Notch, numb2;AJ96-lacZ embryos contain two Hey-positive vMP2 cells at the expense of the dMP2 siblings (Fig. 2D). Examining Eve-positive lineages, a ‘B’- to ‘A’-type switch is evident with loss of Eve staining at the RP2 (‘B’-type) position and an increase in Eve staining at the U (‘A’-type) location (Fig. 2I). All of the supernumerary U cells are Hey-positive. Overall, we note a clear increase in the number of Hey-expressing cells throughout the CNS of numb2 embryos compared with wild-type ones of the same stage. Taken together with the results from mutants with reduced Notch pathway activity, the numb phenotype confirms the responsiveness of Hey expression to Notch signalling in most CNS lineages.
We also studied a genotype with more severe disruption of Notch signalling. A double-null mutant for Dl and Ser, the genes encoding the only two Notch ligands in Drosophila, displays a strong neurogenic phenotype (Lehman et al., 1983). This refers to a severe hyperplasia of CNS neurons, resulting from the inability of nascent NBs to laterally inhibit their neighbours. Despite displaying a large increase in total neuronal numbers, Dl Ser embryos show a decrease in Hey-positive neurons, consistent with their Notch signalling defect (Fig. 2N). As a comparison, we used Df(3R)E(spl)b32.2, a deficiency for the entire E(spl) complex. This harbours seven bHLH-O genes that are crucial targets of Notch in lateral inhibition but are not needed for Notch signal transduction per se. Indeed, Df(3R)E(spl)b32.2-homozygous embryos display a neurogenic phenotype as severe as that of Dl Ser embryos; however, in this case the number of Hey-positive cells is increased, paralleling the global increase in neurons (Fig. 2O). This supports the notion that Notch signalling goes on in GMC divisions in the absence of E(spl), resulting in Hey expression. The fact that a few Hey-positive cells persist in Dl Ser, mam or spdo mutant embryos could be either due to residual Notch signalling in these mutant backgrounds, or to a Notch-independent mode of Hey expression in specific cells.
Hey is expressed in the larval CNS in response to Notch
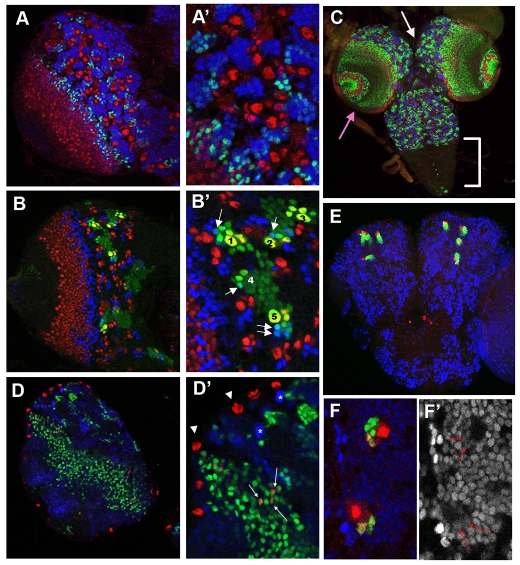
During larval stages, many NBs resume asymmetric divisions to produce large numbers of additional neurons in both the central brain and ventral nerve cord (VNC). This second burst of proliferation gradually ceases by pupariation, after which the post-embryonic (or secondary) neurons fasciculate with the pre-existing embryonic ones and remodel their projections in the process of building the adult CNS (Maurange and Gould, 2005; Truman et al., 2004). In third-instar larval CNS, we detected strong Hey accumulation in groups of cells positive for the neuronal marker Elav, but negative for the NB/GMC marker Ase (Fig. 3A). The Hey-positive cells are among the secondary neurons based on the following additional criteria, besides their lack of Ase and expression of Elav: (1) They are near the surface of the CNS; a group of Hey-positive cells was found adjacent to each NB/GMC cluster (Fig. 3A,B); (2) They have short axonal projections visualized by anti E-Cadherin/Shg (Fig. S2); and (3) They are GFP-positive when a NB lineage is positively marked as little as one day before fixation, indicating recent descent from the marked NB (Fig. 3B; see Fig. S2 in the supplementary material). Hey-positive immature neurons were also seen in large numbers within the optic lobe proliferation centres in two broad swaths below Ase-positive (Hey-negative) progenitors (Fig. 3C,D). As in the embryo, larval Hey expression is transient. Very few Hey-positive neurons were detected in the abdominal ganglion at late third instar (Fig. 3C), where imaginal neurogenesis has already ceased, although younger feeding larvae do contain Hey-positive cells in that region (data not shown). Furthermore, as neurogenesis ceases in other regions, such as the central brain after pupariation, the number of Hey-positive neurons decreases dramatically. By two days into pupation, only four mushroom body (MB) NBs per brain hemisphere continue to produce Hey-positive cells (Ito and Hotta, 1992) (Fig. 3E,F).
Fig. 3.
Expression of Hey in the larval CNS. (A,B,D) Single confocal sections of third instar larval brain hemispheres, anterior top, lateral left. (A′,B′,D′) are higher magnifications. (A,A′) Ase (red) marks neuroblasts (large nuclei) and GMCs (small nuclei). Hey (green) does not overlap with Ase. All Hey-positive cells are also positive for Elav (blue), a neuronal marker. (B,B′) GFP (green) highlights lineages marked 5 days before fixation. Ase (red) and Hey (blue) are visualized. In B′, five neuroblasts (NBs) are numbered; NB4 is not visible in this focal plane. Hey-positive (Ase-negative) cells exist in all five lineages, evident as green-blue nuclei (arrows) in lineages 1, 2, 4 and 5 in this focal plane. (C) Low magnification view (confocal projection) of a third larval instar CNS showing neuroblasts (Dpn, red), GMCs and secondary neurons (Pros, blue) and Hey (green). The abdominal ganglion (bracket) has ceased neurogenesis and is devoid of NBs, GMCs and young neurons, as well as Hey immunoreactivity, with the exception of a few midline cells. Note the different cellular organization of the optic lobe (pink arrow) versus central brain (white arrow). (D,D′) Repo (red) marks glia nuclei, which are predominantly found on the surface of the brain hemisphere (arrowheads) and are negative for Hey (green). A number (∼60) of glia (arrows) in the outer optic proliferation centre are found among the band of hundreds of Hey-positive cells, which are mostly neurons. K33-lacZ (blue) marks the NBs strongly (asterisks), but β-galactosidase perdures in GMCs and newly born neurons at lower levels. This section is deeper than the ones shown in A and B, thus containing fewer NBs in the central brain and more Hey-positive cells in the optic lobe. (E) Confocal projection of pupal brain, 2 days after pupariation. Posterior view, dorsal up. The majority of NBs have disappeared; only four mushroom body NBs are detectable by Ase staining (red). Each of these is accompanied by a cluster of Hey-positive cells (green), no other Hey-positive cells are detected. Prospero is detected in blue. (F,F′) Higher magnification of E. Single optical section showing two Ase-positive NBs accompanied by a few Hey-positive (green) nuclei. Two of the Hey-positive cells in each lineage are GMCs as they still express Ase (red). These are marked by red arrows in F′, which shows the Pros immunoreactivity (blue in F).
Although the majority of Hey-positive cells in the larval CNS are neurons, we also detected two instances of Hey-positive non-neuronal cells. One was in the dorsolateral brain, where a few cells were found positive for both Hey and Ase (see Fig. S3 in the supplementary material). These Hey/Ase double-positive cells were occasionally seen to label with phospho-histone H3-Ser10 (PH3), a mitotic marker. By virtue of their small size and characteristic anatomical location, we propose that these are the mushroom body (MB) GMCs. Indeed, in two day old pupal brains, where, as noted before, only the four MB NBs are actively proliferating, each of the four MB Hey-positive clusters includes 2-4 Ase-positive GMCs, in addition to many Ase-negative neurons (Fig. 3F). The second exception is a number of glial cells, which were revealed by Repo staining. Whereas surface glia are Hey-negative, many optic lobe glia located in the inner and outer proliferation centres are Hey-positive (Chotard and Salecker, 2007) (Fig. 3D).
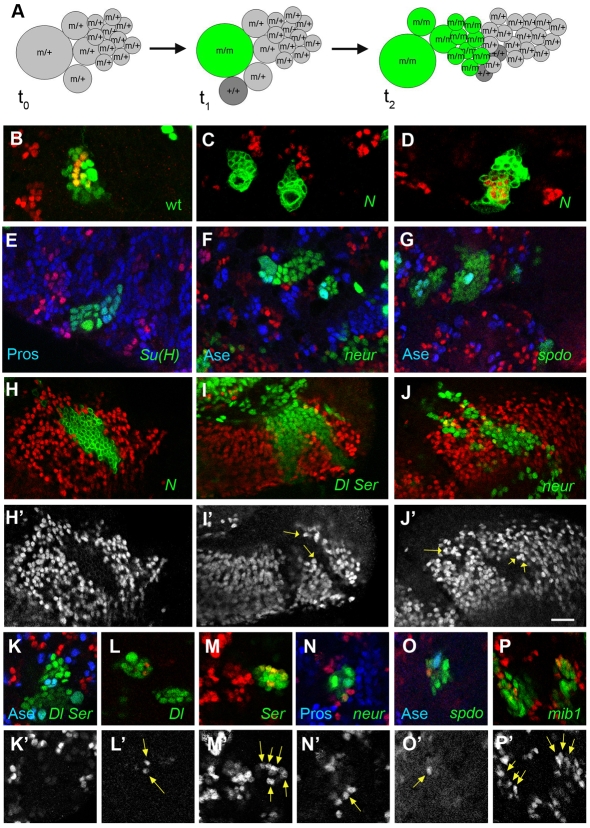
To address the dependence of Hey on Notch signalling in the larval CNS, we generated MARCM mosaic clones (Fig. 4A) mutant for various Notch pathway components. In lineages null for the Notch receptor, no Hey immunoreactivity was observed (Fig. 4C). The same was true for clones for a null Su(H) allele (Fig. 4E). Su(H) is the transcription factor via which intracellular cleaved Notch is targeted to its downstream genes (Bray, 2006); therefore, Hey expression in post-embryonic neurons of the central brain, VNC and optic lobe is activated via the canonical Su(H)-dependent Notch pathway. Despite the fact that Notch signalling was abolished by these null alleles of Notch or Su(H), NB proliferation was not markedly perturbed and mutant GMCs (marked by Pros or Ase) and neurons (marked by Pros or Elav) were formed in apparently normal numbers (Almeida and Bray, 2005) (Fig. 4E-G). Strikingly, in MB lineages, Hey in both GMCs and neurons was unaffected in Notch or Su(H) loss-of-function genotypes (Fig. 4D; see Fig. S3 in the supplementary material), making the MB a region that expresses Hey in a Notch-independent manner. The newly characterized PAN lineages (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008), however, behaved as most NBs do, displaying loss of Hey upon disruption of the Notch pathway (see Fig. S4 in the supplementary material).
Fig. 4:
Analysis of Notch signalling in the larval CNS. (A) Schematic of MARCM clone (Lee and Luo, 2001) progression in the CNS. A single neuroblast (NB) lineage is shown, where the NB is depicted large, the GMCs are intermediate in size and the neurons are small circles. FLP recombinase is induced at time t0 in a m/+ genetic background (m is any mutation). Mitotic recombination in the NB results in two progeny cells of different genotype after the next NB mitosis, at time t1. The renewed NB has become m/m and has turned on GFP expression (green), whereas its new GMC progeny is +/+ (dark grey) and does not express GFP, the same as all the unrecombined m/+ cells. As the NB and GMCs continue to proliferate, more m/m GFP-positive (green) cells are produced (time t2). (B-P′) All images shown are single confocal sections with examples of GFP-marked clones (green) stained for Hey (red) and other markers, as indicated in blue. GFP is nuclear in all panels, except in Notch clones (C,D,H), where a membrane-targeted GFP was used. (B) a neutral (wild-type) clone with several Hey-positive cells. (C) Two Notch54l9 clones in the central brain are devoid of Hey-positive cells, whereas (D) a clone of the same genotype in a mushroom body lineage contains Hey-positive cells. (E) A Su(H)Δ47 clone in the ventral nerve cord, also stained for Pros (blue), which persists in mutant cells, although Hey is lost. (F) Two neur1 clones, also stained for Ase (blue), which is not affected by the mutation. (G) Two spdoG104 clones marked as in F. (H-J) Examples of clones in the outer proliferation centre of the optic lobe. In all cases, Hey expression is lost from the clone, best seen as unstained patches in H′-J′ (Hey channel alone). In a Notch54l9 clone (H), no Hey expression is detected in any mutant cells, whereas in Dlrev10 SerRX106 (I) and neur1 (J) clones, several mutant cells near the clone borders are Hey-positive (arrows in H′-J′). (K-P) MARCM clones in the central brain mutant for Dlrev10 SerRX106 (K), Dlrev10 (L), neur1 (N) or spdoG104 (O) lack Hey-positive cells; exceptions are marked by arrows in the Hey-only channel (K′-P′). SerRX106 (M) or mib1EY9780 (P) clones contain Hey-positive cells (arrows). Scale bar: 10 μm.
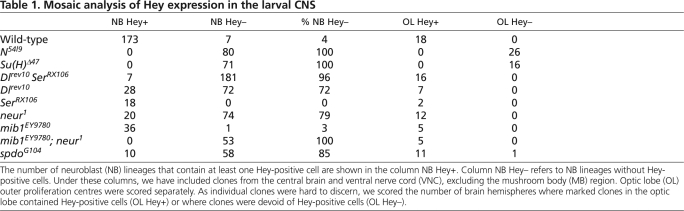
To gain a more complete picture of Notch signalling in secondary neurons we analyzed null mutant clones of the signal-sending machinery, namely Dl, Ser, neur and mib1, as well as the accessory factor spdo (Table 1). In the central brain and ventral nerve cord, clones doubly mutant for Dl Ser displayed a marked absence of Hey-positive cells (Fig. 4K), with only 4% of the clones containing a small number of Hey-positive cells. The incidence of Hey-positive mutant cells is probably due to the non-autonomy of the Dl Ser mutations, namely the fact that mutant cells can still receive a signal sent from adjacent wild-type cells. The fact that this only happens rarely suggests that secondary neurons, in most cases, receive the signal from adjacent neurons of the same lineage, possibly even their immediate siblings. Contrary to the central brain/VNC, the medulla precursor neurons in the optic lobe displayed more widespread non-autonomous behaviour in Dl Ser clones. In all of these clones there were a few Hey-positive mutant cells, in contrast to Notch or Su(H) clones, where none of the mutant cells expressed Hey (Fig. 4H-J). Mosaics null for Dl or neur, were qualitatively similar to Dl Ser, lacking Hey-positive neurons (Fig. 4F,J,L,N). One difference was in the central brain/VNC, where clones with Hey-positive cells were encountered more frequently, 28% and 21% for Dl and neur, respectively. This might be owing to low level residual signalling in these genetic backgrounds. In the case of the Dl mutation, this is probably due to Ser, considering the result from the Dl Ser double mutation. In the case of neur, the most likely candidate for supplying residual activity is Mib1, as these two ubiquitin ligases have been shown to have overlapping functions (Pitsouli and Delidakis, 2005; Wang and Struhl, 2005). Ser- or mib1-null mutations did not affect Hey expression (Fig. 4M,P), suggesting that their role is only a minor supportive one in the present context.
Table 1.
Mosaic analysis of Hey expression in the larval CNS
Finally, we tested clones mutant for the tetraspannin Spdo, an accessory protein required only in instances of Notch signalling associated with asymmetric cell divisions (O'Connor-Giles and Skeath, 2003; Skeath and Doe, 1998). These clones also lacked Hey immunoreactivity, implicating Spdo in the genesis of secondary neurons (Fig. 4G). Fifteen percent of the brain/VNC clones did contain a few Hey-positive cells (Fig. 4O), as did most optic lobe clones. As the spdoG104 allele is a probable null, the non-autonomy observed might reflect a requirement for spdo in signal emission, similar to Dl and neur, although evidence to date places Spdo function in the signal receiving cell (Hutterer and Knoblich, 2005; Langevin et al., 2005; O'Connor-Giles and Skeath, 2003; Roegiers et al., 2005), making such an interpretation unlikely. Alternatively, escapers could be explained by perdurance of the wild-type spdo RNA and protein that were present in the progenitor cell that was still spdo+/– before clone induction.
In conclusion, our mosaic analysis lends support to a mode of Notch-dependent asymmetric cell division in the birth of secondary neurons for most CNS lineages. This cannot be unequivocally demonstrated because, unlike in the embryo, specific ‘A versus B’ neuron markers are lacking for the larval stage. Yet, the Notch-dependence, and especially the Spdo-dependence, of Hey expression is consistent with asymmetric GMC division, where one cell escapes Notch signalling by inheriting Numb, whereas the other receives a Spdo-aided Notch signal and, as a result, adopts a different fate. Indeed, most Hey-positive cells in the larval CNS display low or undetectable levels of Numb accumulation (see Fig. S5 in the supplementary material).
Hey participates in asymmetric neuron fate establishment in the embryo
A piggyBac recessive lethal insertion, WH-f06656, was isolated in the first intron of Hey during the Exelixis screen (Thibault et al., 2004). Using our antisera, we were unable to detect any Hey protein in homozygous embryos (Fig. 5A) and we therefore refer to this insertion as Heyf06656. To determine whether the lethality is due to disruption of Hey or some distantly linked secondary mutation, we tested the Heyf06656 chromosome for complementation against two deficiencies in the 44A region, Df(2R)exel6055 and Df(2R)ED1735. In both cases, no heterozygotes were obtained confirming that lethality maps at or near the Hey locus.
Fig. 5.

Loss- and gain-of-function analysis of Hey. (A) Low magnification image of two embryos stained for Hey (red) and β-galactosidase (green), which reveals wg-lacZ from the balancer chromosome (Hey+) over which Heyf06656 is kept. The homozygous mutant embryo (white arrowhead), identified by the absence of the wg-lacZ pattern, does not express Hey. (B-I) High magnification images. Anterior is up. (B) Eve expression pattern in a stage 15 Heyf06656-homozygous embryo is similar to that in wild type embryos (see F; Fig. 2G). (C) Odd expression pattern (green) in a stage 15 Heyf06656;AJ96-lacZ embryo. β-galactosidase (red) marks vMP2/dMP2 neurons. Besides MP1s (arrow), Odd is expressed only in dMP2s (arrowheads) as in wild-type embryos (see Fig. S1 in the supplementary material). (D-E) Pattern of UAS-GFP expression (red) driven by ftz.ng-Gal43 versus Eve staining in blue. (D,D′) Montage of deep focal planes, showing aCC/pCC pairs (arrowhead) and RP2 (asterisk). GFP is detected in all three cells and additional neighbouring neurons, but not in the occasional U-cell that lies near the aCC/pCC pair and is also positive for Eve (blue arrows in D′, Eve channel). (E) Superficial focal plane montage showing the U (arrowhead) and EL (asterisk) clusters, both of which are devoid of GFP. (F) Eve expression pattern in a stage 16 wild-type embryo. Note the solo RP2 neurons (asterisk) between the clusters containing EL (lateral) and U/aCC/pCC (medial) neurons. (G) Eve staining of a stage 16 Hey-overexpressing embryo is similar to that of a numb mutant. RP2 cells are absent; two persisting ones are marked by asterisks. The EL and U clusters are not affected, as ftz.ng-Gal43 is not expressed there. (H) In wild-type embryos, two dMP2 (lateral to midline) and two MP1 (midline) neurons per segment are marked with Odd. (I) In Hey-overexpressing embryos, Odd expression persists in the two MP1 neurons, but it is extinguished from most dMP2s. White arrows show two dMP2s that are still labelled with Odd. Scale bar: 25 μm.
The lethal phase of Heyf06656/Df individuals was determined to be late embryonic/early larval. Between 40% and 85% (depending on the experiment) of these individuals hatched, but most died within the first instar. The hatched larvae were less active than their wild-type (Hey/+) siblings, and a small number even went through the first larval moult before dying. No obvious cuticular defects were detected in the dead embryos and larvae. We stained homozygous mutant embryos for various neuronal markers to assess CNS integrity. No consistent defects in the anatomy of longitudinal, commissural and peripheral nerve tracts were seen (data not shown). We further determined whether particular neuronal fates might be affected. If Hey is needed to transduce the Notch signal which distinguishes ‘A’- from ‘B’-type neurons, we would expect the Hey mutant embryos to have a phenotype similar to spdo mutant ones, namely vMP2>dMP2, RP2sib>RP2 and U>Usib cell fate switches. Yet, Eve staining of Hey mutants revealed the presence of a single RP2 and a normal looking U-cluster per hemisegment (Fig. 5B). Similarly, 22C10 staining, which reveals the pioneer axonal tracts of dMP2 (points anteriorly) and vMP2 (points posteriorly), did not detect any defect in Hey mutants (data not shown). This was confirmed by staining for Odd, a protein expressed specifically in the dMP2 neurons and the two MP1 midline neurons (Spana et al., 1995). Besides the MP1s, only one cell per hemisegment (dMP2) was Odd-positive in Heyf06656 embryos and not two as we would expect had there been a vMP2>dMP2 fate switch (Fig. 5C). We conclude that Hey is not strictly required to realize the ‘A’ cell fate, despite its ‘A’-specific expression pattern. Alternatively, Hey could be required for ‘A’ versus ‘B’ fate determination but it might act redundantly with another Notch target so that the single Hey knockout produces only a slight defect, consistent with the variable embryo-larval lethality observed.
If the latter hypothesis is correct, we would expect that ectopic overexpression of Hey might cause an opposite ‘B>A’ fate switch, similar to those seen in numb loss-of-function embryos, where endogenous Hey is overexpressed (Fig. 2). To test our hypothesis, we generated UAS-Hey transgenic flies and induced overexpression in various GMCs and their progeny by using the ftz.ng-Gal43 line (Lin et al., 1995). This driver is expressed, among other neurons, in aCC/pCC, RP2/RP2sib and dMP2/vMP2 cells, but not in the U or EL lineages (Fig. 5D,D′,E). This would place Hey ectopically in aCC, RP2 and dMP2, in addition to bolstering its levels in pCC, RP2sib and vMP2. Eve staining of ftz.ng-Gal43;UAS-Hey embryos revealed a loss of RP2 in 52% of the hemisegments (73/140 hemisegments from 9 embryos) suggesting a fate switch RP2 (Eve-positive)>RP2sib (Eve-negative) (Fig. 5G). Odd staining revealed loss of dMP2 in 70% of hemisegments (35/50 hemisegments from 2 embryos), consistent with a dMP2 (Odd-positive)>vMP2 (Odd-negative) transformation (Fig. 5I). Although Hey expression is also driven ectopically in the aCC cell, its putative transformation into a pCC cell fate could not be detected as Eve labels both siblings. From the above results, we conclude that Hey has the ability to switch neuronal fates from the ‘B’ to the ‘A’ fate in the absence of Notch signalling. Therefore, at least in these asymmetric GMC divisions (MP2 and GMC4-2), Hey expression appears sufficient to implement the Notch fate-determination effect.
In the process of the above gain-of-function experiments, we explored different Gal4 drivers to ectopically express Hey. We never observed any suppression of neurogenesis in either the embryo or the larva/adult using neur-Gal4, dpp-Gal4, 253-Gal4 or Eq-Gal4 (see Fig. S6 in the supplementary material; data not shown). This is contrary to the amply documented loss of neural elements obtained when bHLH-O proteins of the E(spl)/Hes family are overexpressed. In our controls, we ectopically expressed E(spl)m7 and hairy and, in both cases, we observed significant suppression of neurogenesis. We conclude that Drosophila Hey has significantly diverged from the Hairy/E(spl) family in both structure and function and is involved in neuronal fate decisions rather than in regulating the number of neural precursors.
DISCUSSION
Hey is a transducer of the Notch signal in GMC asymmetric cell division
We have analyzed the expression pattern and function of the single Hey gene in Drosophila. We detected Hey almost exclusively in the CNS in young postmitotic neurons and glia, specifically those that receive a Notch signal at birth. It has long been appreciated (Buescher et al., 1998; Skeath and Doe, 1998; Spana and Doe, 1996; Udolph et al., 2001) that Notch signalling plays an important role in the acquisition of neuronal/glial cell fate after GMC division, with most GMCs producing two different progeny, an ‘A’ cell with high Notch activity and a ‘B’ cell with no Notch activity. Still, no Notch target genes had been identified in this process. We now show that Hey is such a target gene in many, and perhaps all, GMC asymmetric divisions. Our conclusions are based on the expression pattern of Hey, its response to Notch pathway perturbation and on the ability of ectopic Hey to block development of RP2 and dMP2, two ‘B’-type neurons.
Although we have good evidence that Hey expression can recapitulate the effect of Notch signalling, Hey loss-of-function has only a mild phenotype. The trivial possibility that the transposon insertion allele used has residual activity is unlikely as (1) no Hey protein is detectable in homozygous mutants and (2) the Heyf06656 allele results in recessive lethality. Nevertheless, the issue will be permanently decided with the generation and analysis of more Hey alleles. The alternative hypothesis, which seems more probable, is that one or more additional factors besides Hey can also act as nuclear effectors downstream of Notch in the ‘A’ GMC progeny. No Hey paralogues exist in the D. melanogaster genome, but structurally divergent proteins, even outside the bHLH-O family, could share similar functional characteristics. At the moment, we have no good candidate for such a factor; however, we have excluded a number of bHLH-O factors that do not seem to be co-expressed with Hey in neurons, namely E(spl)mγ and m8, Hairy and Dpn (Fig. 1D; data not shown).
Besides GMCs, a number of other neural progenitors, namely NBs, sensory organ precursors (SOPs) and SOP progeny cells, all undergo asymmetric cell divisions with Notch involvement (Knoblich, 2008; Lai and Orgogozo, 2004). We could not detect Hey expression in either the NB/GMC pair or in the SOP progeny cells of external sensory organs (Fig. 1; Fig. 3; Fig. 4; data not shown), suggesting that Hey expression is turned on exclusively in GMC asymmetric divisions. Hey-positive glia could also be the progeny of asymmetrically dividing GMCs (Udolph et al., 2001). It is yet unclear which cells might be the immediate progenitors of the few Hey-positive PNS neurons.
Notch signalling is intimately related with bHLH-O genes
Until the present work and the recent paper by Krejci et al. (Krejci et al., 2009), the only Drosophila bHLH-O genes known to be targets of Notch were the seven of the E(spl) complex. Hey and two other bHLH-O genes, dpn and Her, had been predicted as candidate Notch targets based on nearby clustering of putative Su(H) binding sites, the DNA elements via which activated Notch is tethered to its target genes (Rebeiz et al., 2002). Although Her does not seem to be a true Notch target (Rebeiz et al., 2002), Krejci et al. (Krejci et al., 2009) have shown that dpn is a Notch target in the muscle-progenitor-like Drosophila DmD8 cell line; an in vivo context for such a response has yet to be determined. Together with Hey, this makes a total of 9 out of 13 bHLH-O genes in the Drosophila genome which are regulated by Notch. It should be stressed that Notch has a number of additional (non-bHLH-O) targets, depending on the species and cellular context, but few, if any, show such widespread association as the bHLH-O genes. The latter are activated by Notch in a multitude of unrelated contexts, such as neuroectoderm, mesoderm, wing epithelium, leg segmentation (Bray, 2006; Lai, 2004) and now GMC asymmetric cell divisions in Drosophila, and in neural progenitors, presomitic mesoderm, cardiogenesis and vasculogenesis in vertebrates (Aulehla and Pourquie, 2008; High and Epstein, 2008; Louvi and Artavanis-Tsakonas, 2006).
In addition to its widespread Notch-dependent expression, we have detected a clear instance of Notch-independent expression of Hey within the GMCs and neurons of the MB precursors (Fig. 4D; see Fig. S3 in the supplementary material). Other examples where Hey expression does not correlate with known events of Notch signalling are the MP2 NB and the two MP1 midline neurons (Fig. 2A; see Fig. S1 in the supplementary material). It is also clear that in embryos with severe Notch signalling defects we still observe a small number of Hey-positive cells in the CNS (Fig. 2N), suggesting that there are additional neural lineages, where Hey is likely to be expressed independently of Notch. Analysis of the cis regulatory regions of Hey should shed light on Notch-dependent and Notch-independent enhancer elements.
Hey function has diverged despite structural conservation
The bHLH-O family has undergone considerable diversification during evolution (Simionato et al., 2007). Although sequence analysis can unambiguously assign genes to this family, it cannot identify orthologues in distantly related species. A classic example is the Drosophila to mammals comparison, where no clear orthologue relationships exist between Hairy, Dpn and the seven E(spl) in Drosophila and Hes1, 2, 3, 5, 6 and 7 in mammals, suggesting that the diversification of these proteins occurred separately after divergence of protostomes and deuterostomes. Hey proteins are the singular exception, being particularly well conserved. The bHLH domain of Drosophila Hey shows 97-98% similarity to that of its mammalian counterparts. This might lead one to expect substantial conservation of Hey function, which, strangely enough, was not observed.
First, mammalian Hey genes have a very broad expression pattern, including presomitic mesoderm, embryonic heart, vascular precursors, developing brain and spinal cord, neural crest etc (Kokubo et al., 1999; Leimeister et al., 1999). Fly Hey, by contrast, seems confined within the CNS and PNS. Although there is complexity in its expression, as documented here with its contextual Notch dependence/independence, the great majority of its expression pattern seems to be in the newly born Notch-dependent ‘A’-type neurons. The absence of Hey expression from the developing Drosophila heart is most striking, given the foremost importance of Hey genes in vertebrate cardiogenesis. A second indicator of functional non-conservation comes from comparing the role of Hey within the nervous systems of mammals versus Drosophila. In the former, Hey has been proposed to act in the maintenance of progenitor fate and to antagonize neuronal differentiation, similar to Hes proteins (Sakamoto et al., 2003). In fact, it has been proposed that Hey-Hes heterodimers mediate these effects. In the fly, we could not detect Hey expression within progenitor cells, with the few rare GMC exceptions, noted above. We could not even detect Hey-E(spl) or Hey-Dpn co-expression, although we did not test all seven E(spl) genes for lack of specific reporter lines. To overcome any doubt, we made functional tests by ectopically expressing Hey. Instead of suppressing sensory organ formation, it mildly increased the number of bristles, showing an opposite phenotype from that of E(spl) or hairy ectopic expression (see Fig. S6 in the supplementary material). We are therefore confident that Hey does not antagonize neural differentiation in the fly.
This leaves us with the puzzle of why Hey is so strongly conserved. Perhaps some yet uncharacterized molecular aspect of its role in chromatin recognition/transcriptional regulation is conserved, despite considerable diversification in cellular and developmental contexts. These contexts have diverged greatly between insects and vertebrates, the only unifying theme being their regulation by Notch signalling. A homologous function might be that of promoting gliogenesis, as Hey2 was shown to promote Müller glia formation in the murine retina (Satow et al., 2001). Further comparative studies encompassing more species will no doubt shed light on the function of this highly conserved bHLH-O protein.
Supplementary Material
Acknowledgements
We are grateful to S. Parkhurst, S. Artavanis-Tsakonas, T. Klein, Y.-N. Jan, H. Bellen, C. Doe, D. Kosman, M. Frasch, J. Knoblich and A. Jarman for their kind gifts of antibodies and fly stocks. We thank I. Livadaras for fly injections and K. Trahana for help with the mosaic analysis experiments. We acknowledge the scholarship support of E.Z. by T. and M. Martinos. Work was funded by the PENED programme of the GSRT, Greece (03ED555) and by a programme grant from the Medical Research Council to S.B.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.043604/-/DC1
References
- Aulehla A., Pourquie O. (2008). Oscillating signaling pathways during embryonic development. Curr. Opin. Cell Biol. 20, 632-637 [DOI] [PubMed] [Google Scholar]
- Bailey A. M., Posakony J. W. (1995). Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes & Development 9, 2609-2622 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E., Reichert H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q., Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185-1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G., Knoblich J. A. (2008). The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 [DOI] [PubMed] [Google Scholar]
- Brand M., Jarman A. P., Jan L. Y., Jan Y. N. (1993). asense is a Drosophila neural precursor gene and is capable of initiating sense organ formation. Development 119, 1-17 [DOI] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689 [DOI] [PubMed] [Google Scholar]
- Buescher M., Yeo S. L., Udolph G., Zavortink M., Yang X., Tear G., Chia W. (1998). Binary sibling neuronal cell fate decisions in the Drosophila embryonic central nervous system are nonstochastic and require inscuteable-mediated asymmetry of ganglion mother cells. Genes Dev. 12, 1858-1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C., Salecker I. (2007). Glial cell development and function in the Drosophila visual system. Neuron Glia Biol. 3, 17-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M. D., Doe C. Q. (2006). Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 20, 429-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M. T., Tyler D. M., Furriols M., Chalkiadaki A., Delidakis C., Bray S. (2000). Spatially restricted factors cooperate with Notch in the regulation of Enhancer of split genes. Dev. Biol. 221, 390-403 [DOI] [PubMed] [Google Scholar]
- Fischer A., Gessler M. (2007). Delta-Notch-and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 5, 4583-4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. (2004). The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18, 901-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H., Levine M. (1987). Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6, 749-759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter D. A., Urban J., Rickert C., Ner S. S., Ito K., Travers A. A., Technau G. M. (1995). The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development 121, 317-332 [DOI] [PubMed] [Google Scholar]
- Hatakeyama J., Bessho Y., Katoh K., Ookawara S., Fujioka M., Guillemot F., Kageyama R. (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539-5550 [DOI] [PubMed] [Google Scholar]
- High F. A., Epstein J. A. (2008). The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 9, 49-61 [DOI] [PubMed] [Google Scholar]
- Hulleman E., Quarto M., Vernell R., Masserdotti G., Colli E., Kros J. M., Levi D., Gaetani P., Tunici P., Finocchiaro G., et al. (2009). A role for the transcription factor HEY1 in glioblastoma. J. Cell Mol. Med. 13, 136-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A., Knoblich J. A. (2005). Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 6, 836-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T., Sartorelli V., Poizat C., Iezzi S., Wu H. Y., Chung G., Kedes L., Hamamori Y. (2001). HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 21, 6080-6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T., Kedes L., Hamamori Y. (2003). HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 194, 237-255 [DOI] [PubMed] [Google Scholar]
- Ito K., Hotta Y. (1992). Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 149, 134-148 [DOI] [PubMed] [Google Scholar]
- Jennings B., Preiss A., Delidakis C., Bray S. (1994). The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120, 3537-3548 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. (2008). Roles of Hes genes in neural development. Dev. Growth Differ. 50Suppl. 1, S97-S103 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597 [DOI] [PubMed] [Google Scholar]
- Kokubo H., Lun Y., Johnson R. L. (1999). Identification and expression of a novel family of bHLH cDNAs related to Drosophila hairy and enhancer of split. Biochem. Biophys. Res. Commun. 260, 459-465 [DOI] [PubMed] [Google Scholar]
- Kokubo H., Miyagawa-Tomita S., Nakazawa M., Saga Y., Johnson R. L. (2005). Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev. Biol. 278, 301-309 [DOI] [PubMed] [Google Scholar]
- Krejci A., Bernard F., Housden B. E., Collins S., Bray S. J. (2009). Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal 2, ra1 [DOI] [PubMed] [Google Scholar]
- Lai E. C. (2004). Notch signaling: control of cell communication and cell fate. Development 131, 965-973 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Orgogozo V. (2004). A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev. Biol. 269, 1-17 [DOI] [PubMed] [Google Scholar]
- Langevin J., Le Borgne R., Rosenfeld F., Gho M., Schweisguth F., Bellaiche Y. (2005). Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr. Biol. 15, 955-962 [DOI] [PubMed] [Google Scholar]
- Lecourtois M., Schweisguth F. (1995). The neurogenic suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split Complex genes triggered by Notch signaling. Genes Dev. 9, 2598-2608 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251-254 [DOI] [PubMed] [Google Scholar]
- Lehman R., Jiménez F., Dietrich U., Campos-Ortega J. A. (1983). On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Roux's Arch. Develop. Biol. 192, 62-74 [DOI] [PubMed] [Google Scholar]
- Leimeister C., Externbrink A., Klamt B., Gessler M. (1999). Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech. Dev. 85, 173-177 [DOI] [PubMed] [Google Scholar]
- Lin D. M., Auld V. J., Goodman C. S. (1995). Targeted neuronal cell ablation in the Drosophila embryo: pathfinding by follower growth cones in the absence of pioneers. Neuron 14, 707-715 [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. (2006). Notch signalling in vertebrate neural development. Nat. Rev Neurosci. 7, 93-102 [DOI] [PubMed] [Google Scholar]
- Maurange C., Gould A. P. (2005). Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci. 28, 30-36 [DOI] [PubMed] [Google Scholar]
- Menne T. V., Klambt C. (1994). The formation of commissures in the Drosophila CNS depends on the midline cells and on the Notch gene. Development 120, 123-133 [DOI] [PubMed] [Google Scholar]
- Nakao K., Campos-Ortega J. A. (1996). Persistent expression of genes of the Enhancer of split Complex suppresses neural development in Drosophila. Neuron 16, 275-286 [DOI] [PubMed] [Google Scholar]
- O'Connor-Giles K. M., Skeath J. B. (2003). Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell 5, 231-243 [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. (1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196-2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. H., Condron B. G., Zinn K. (1994). Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature 367, 429-434 [DOI] [PubMed] [Google Scholar]
- Pitsouli C., Delidakis C. (2005). The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development 132, 4041-4050 [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Reeves N. L., Posakony J. W. (2002). SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl. Acad. Sci. USA 99, 9888-9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S., White K. (1991). Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443-461 [DOI] [PubMed] [Google Scholar]
- Roegiers F., Jan L. Y., Jan Y. N. (2005). Regulation of membrane localization of Sanpodo by lethal giant larvae and neuralized in asymmetrically dividing cells of Drosophila sensory organs. Mol. Biol. Cell 16, 3480-3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M., Hirata H., Ohtsuka T., Bessho Y., Kageyama R. (2003). The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J. Biol. Chem. 278, 44808-44815 [DOI] [PubMed] [Google Scholar]
- Satow T., Bae S. K., Inoue T., Inoue C., Miyoshi G., Tomita K., Bessho Y., Hashimoto N., Kageyama R. (2001). The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J. Neurosci. 21, 1265-1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., Degnan B. M., Vervoort M. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7, 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath J. B., Doe C. Q. (1998). Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125, 1857-1865 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Thor S. (2003). Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13, 8-15 [DOI] [PubMed] [Google Scholar]
- Spana E. P., Doe C. Q. (1996). Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21-26 [DOI] [PubMed] [Google Scholar]
- Spana E. P., Kopczynski C., Goodman C. S., Doe C. Q. (1995). Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development 121, 3489-3494 [DOI] [PubMed] [Google Scholar]
- Su M. T., Fujioka M., Goto T., Bodmer R. (1999). The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development 126, 3241-3251 [DOI] [PubMed] [Google Scholar]
- Taelman V., Van Wayenbergh R., Solter M., Pichon B., Pieler T., Christophe D., Bellefroid E. J. (2004). Sequences downstream of the bHLH domain of the Xenopus hairy-related transcription factor-1 act as an extended dimerization domain that contributes to the selection of the partners. Dev. Biol. 276, 47-63 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., Singh C. M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H. L., et al. (2004). A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283-287 [DOI] [PubMed] [Google Scholar]
- Truman J. W., Schuppe H., Shepherd D., Williams D. W. (2004). Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 131, 5167-5184 [DOI] [PubMed] [Google Scholar]
- Udolph G., Rath P., Chia W. (2001). A requirement for Notch in the genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development 128, 1457-1466 [DOI] [PubMed] [Google Scholar]
- Vaessin H., Grell E., Wolff E., Bier E., Jan L. Y., Jan Y. N. (1991). prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67, 941-953 [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. (2005). Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132, 2883-2894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.