Abstract
For an enzyme functioning predominantly in a seemingly housekeeping role of 5′ tRNA maturation, RNase P displays a remarkable diversity in subunit make-up across the three domains of life. Despite the protein complexity of this ribonucleoprotein enzyme increasing dramatically from bacteria to eukarya, the catalytic function rests with the RNA subunit during evolution. However, the recent demonstration of a protein-only human mitochondrial RNase P has added further intrigue to the compositional variability of this enzyme. In this review, we discuss some possible reasons underlying the structural diversity of the active sites, and use them as thematic bases for elaborating new directions to understand how functional variations might have contributed to the complex evolution of RNase P.
Keywords: RNase P, precursor tRNA, diversity, evolution, organellar
1. INTRODUCTION
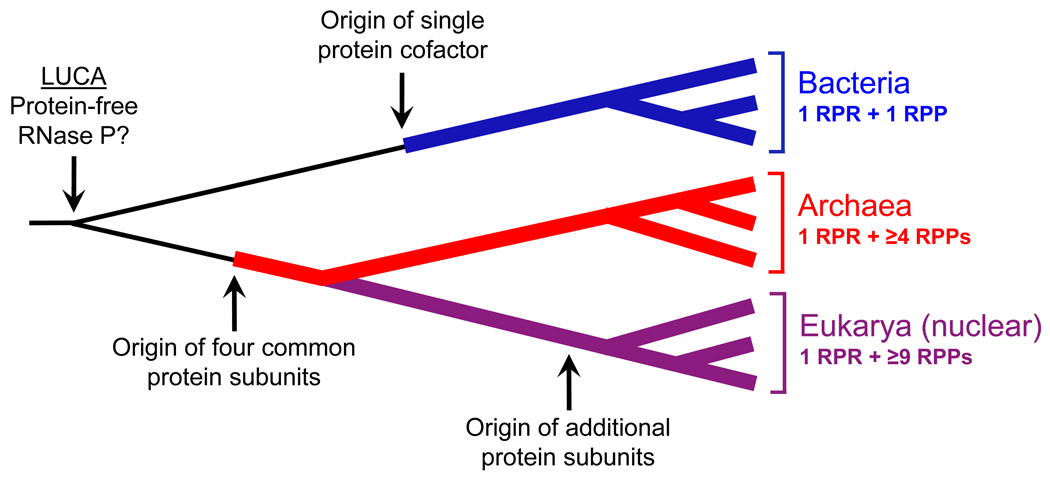
What began four decades ago as a quest to uncover the catalyst responsible for removing the 5′ leaders of precursor tRNAs (pre-tRNAs) during tRNA biogenesis resulted not only in the discovery of the first true RNA enzyme [1], but has also revealed the surprisingly diverse guises adopted by this enzyme in different life forms. This endonucleolytic activity termed RNase P functions predominantly as a ribonucleoprotein (RNP), albeit with different subunit make-up depending on the source: a single RNase P RNA (RPR) is associated with one, (at least) four and nine RNase P protein (RPP) subunits in Bacteria, Archaea and Eukarya (nucleus), respectively (Fig. 1) [2–4]. Although catalysis is associated with the RNA [1,5,6], the proteins play vital supporting roles, highlighting the intimate functional coordination among the subunits. The higher protein:RNA mass ratio in archaeal and eukaryal RNase P than in their bacterial counterpart offers a paradigm to understand the possible reassignment of structural and functional attributes of RNAs to protein cofactors that were recruited during evolution. The finding that some organellar isoforms of RNase P are protein enzymes [7] invites the question why cells, now relying almost exclusively on proteins for catalytic and structural roles, have retained RNA-mediated catalysis at all in RNase P. Such questions spawned by the remarkable diversity of RNase P form the mainstay of this review. While we take a closer look at the likely bases for the variability in RNase P and discuss some future research directions in this regard, we refer the reader to a new book on RNase P [8] and some recent comprehensive reviews [9–12] for detailed information on various aspects not elaborated here.
Figure 1.
A cladogram depicting a possible scheme for evolution of RNA-based RNase P (see sections 1 and 4 for details). Scales are arbitrary. LUCA, last universal common ancestor; RPR, RNase P RNA; RPP, RNase P protein.
2. VARIABLE COMPOSITION OF RNase P
Although there are some thematic variations, we highlight the most common make-up of the RNase P holoenzyme from each domain of life. We describe the organellar variants in some detail as they exemplify the remarkable variability of RNase P.
2.1. Bacterial RNase P
From early observations that mutants thermosensitive for RNase P activity map to two distinct loci in the Escherichia coli genome [13,14], two subunits were expected in the holoenzyme. However, RNase P was not anticipated then to be an RNP. Of historical significance were biochemical reconstitution and genetic studies revealing the RNA subunit (encoded by rnpB) to be the catalytic moiety and the protein subunit (encoded by rnpA) a cofactor [1,15–18]; both subunits are essential in vivo. The RPR is roughly 120 kDa and the protein 15 kDa in most bacteria, and the tertiary structures of each subunit has been solved [19–23]. The single bacterial RPP has multiple roles [24–32]: (i) promoting direct interactions with the 5′ leader sequence to increase the affinity for the pre-tRNA (substrate) over mature tRNA (product), (ii) enhancing the RPR’s rate of cleavage, and (iii) enabling the RPR to function at physiological Mg2+ concentrations by increasing the affinity for active-site Mg2+ ions. In a manner reminiscent of EF-Tu [33], bacterial RPP appears to compensate for differences in various pre-tRNA structures by altering its energetic contributions to leader binding and enhancing the rate of RPR-mediated cleavage by 3- to 1,000-fold to ensure that processing of different substrates by the holoenzyme occurs at an almost invariable rate [30].
2.2. Eukaryal (nuclear) RNase P
Isolation and characterization of yeast and human native nuclear RNase P revealed that they have an RPR plus nine and ten RPPs, respectively [4,34–37]; genetic depletion in yeast revealed that all subunits are essential for RNase P activity and cell viability [34]. While the RPR is typically ~100 kDa, the proteins range from ~15 to 100 kDa. The human RPPs are termed RPP14, RPP20, RPP21, RPP25, RPP29, RPP30, RPP38, RPP40, POP5 and POP1, with RPP40 being the only protein with no homolog in yeast RNase P. Although in vitro pull-down/crosslinking and yeast two-/three-hybrid studies [38–41] have uncovered some RPP-RPP and RPP-RPR interactions in these large RNPs (~400 kDa), the functional roles of individual RPPs are lacking in the absence of in vitro reconstitution and tertiary structures. The isolation of conditional defective mutants has provided some clues into role of RPPs [42], although their precise molecular defects remain to be discerned.
Sequence homology-based inventories of eukaryal RPP homologs reveal their absence in many organisms [43]; it is unclear if this is due to sequence divergence or the real absence of RPPs. Also, although there are some animal RPP homologs in plants, no RPR has been identified to date, despite characterization of the activity indicating presence of an RNA component [44,45].
2.3. Archaeal RNase P
Using polyclonal antisera raised individually against the four Methanothermobacter thermautotrophicus (Mth) polypeptides (RPP21, RPP29, RPP30 and POP5) with sequence homology to the corresponding yeast/human RPPs, RNase P activity was immunoprecipitated from a partially purified Mth preparation [46]. The archaeal RPPs, whose tertiary structures have been solved [47–56], range from 10 to 30 kDa; the RPR is roughly 100 kDa. The holoenzyme from different archaea has now been reconstituted in vitro from recombinant subunits [48,57–59]. Such assays revealed that the four archaeal RPPs function as two binary complexes (RPP21–RPP29 and RPP30-POP5) [58,59], consistent with results from yeast two-hybrid and structural studies [47,49,51,55,60–62]. Although each binary complex aids RPR catalysis, kinetic analyses suggest that they fulfill different roles: RPP21–RPP29 increases affinity of the RPR for the pre-tRNA substrate (W-Y Chen and Gopalan, unpublished results), while POP5-RPP30 enhances the rate of chemical cleavage nearly 100-fold [58]. The functional parallel of POP5-RPP30 to the bacterial RPP [30,58] is mirrored by the structural similarity between POP5 and the bacterial RPP [22,55], despite their weak primary sequence homology, perhaps illustrating a case of convergent evolution.
Recently, we have obtained evidence that the ribosomal protein L7Ae, homologous to human RPP38, co-purifies with archaeal RNase P activity (I-M Cho and Gopalan, unpublished results). This protein was previously suspected to be a putative RPP based on its ability to influence the thermal stability and kinetic properties of an in vitro reconstituted archaeal RNase P [63]. Detailed investigations into the mechanism of action of L7Ae are ongoing.
2.4. Organellar RNase P
Except for the recent description of the protein-only RNase P from human mitochondria [7], the make-up of RNase P from protein-synthesizing organelles (mitochondria and chloroplasts) has remained elusive despite many years of intensive effort, even in favored model organisms such as yeast. Organelles are derived from bacterial endosymbionts, and therefore a bacterial-like RNase P structure would be a reasonable expectation for organellar RNase P. Surprisingly, this is more the exception than the rule.
RPR
Homologs of the bacterial RPR gene (rnpB) are found in a few sequenced mitochondrial genomes of basal lineages, such as those with either a genomic organization similar to that of the ancestral endosymbiont (Reclinomonas) [64] or a high number of retained genes (prasinophyte algae) [65]. In cases like Ascomycete fungi, the mitochondrial RPR is highly degenerate and difficult to recognize [66]. In others such as Zygomycete fungi, the RPR sequence is highly variable in length, ranging from 188 to 982 nts [67]. To date, no mitochondrial RPR has been shown to be active in vitro.
Based on the available sequences of chloroplast genomes, rnpB homologs are present in some of the chloroplasts derived from primary endosymbiosis: the glaucophyte alga Cyanophora paradoxa [68], red algae [66,69–71], and several green algae (prasinophytes) [72,73], but not in other green algae or higher plants. It is also absent in chloroplasts derived from secondary endosymbiosis (e.g., Cryptophyte, Apicomplexa, Euglenozoa, Haptophyte, Chlorarachniophyceae, and Stramenopiles). The predicted secondary structure of the chloroplastic RPRs fits the bacterial type A consensus, although some of the tertiary contacts are typically missing [74]. These RPRs failed to show activity in vitro [74], except for that from C. paradoxa [75], which is weakly active by itself and could be reconstituted with a bacterial RPP [75,76]. Although this bacterial-like RPR is enriched in purified fractions of C. paradoxa plastid RNase P [77], nothing is known about its protein subunit(s). In higher plants, biochemical data suggest that the chloroplast enzyme lacks RNA, but its components have not been identified so far (see 5.4) [78,79].
RPP(s)
No protein subunit homologous to bacterial RPP has been identified in organellar RNase P (see below). Only in yeast and human mitochondria, is there any information on the protein composition of RNase P. In yeast, only the nuclear-encoded RPM2 is known and shown genetically to be required for mitochondrial RNase P activity [80]. It is a 100-kDa protein completely unrelated to bacterial, archaeal or eukaryal nuclear RPPs and has no homologs beyond Saccharomycetales.
The human mitochondrial (mt) RNase P is composed of three proteins (MRPP1, MRPP2, and MRPP3), all unrelated to any known RPP [7]. MRPP1 is a tRNA m1G methyltransferase (Trm10) involved in methylation of G9, MRPP2 is a member of the short-chain dehydrogenase/reductase family, and MRPP3 is a metallonuclease with two pentatricopeptide repeats and is postulated to house the catalytic site of the enzyme. Therefore, the ancestral RNA-containing RNase P has been replaced by a patchwork of three proteins in human mtRNase P, two of which have no known function involving tRNAs.
3. CATALYSIS
Cleavage of the scissile phosphodiester bond in pre-tRNAs (and other substrates) by either RNP- or protein-based RNase P results in a 5' RNA with a 3'-OH and a 3' RNA with a 5'-phosphate. Despite plurality in the nature of the enzyme (E) and its substrate (S), rearrangements in the ES complex must somehow lead to a similar transition state and site-specific phosphodiester hydrolysis. Although how the enzyme accomplishes this reaction is unclear, some aspects of the mechanism are well characterized in studies on bacterial RNase P discussed below.
3.1. Substrate recognition
Pre-tRNAs, the primary substrates for RNase P, are cleaved between N−1 and N+1 (Fig. 2), except for bacterial and some organellar pre-tRNAHis wherein cleavage occurs between N−2 and N−1 [11,81]. Cleavage involves (i) docking the pre-tRNA in the active site, (ii) positioning the catalytic metal ions that promote the chemical cleavage, and (iii) preventing a nucleophilic attack by the neighboring 2'-OH at N−1 that would generate cleavage products with incorrect ends (5'-OH and 2',3'-cyclic phosphate) [11].
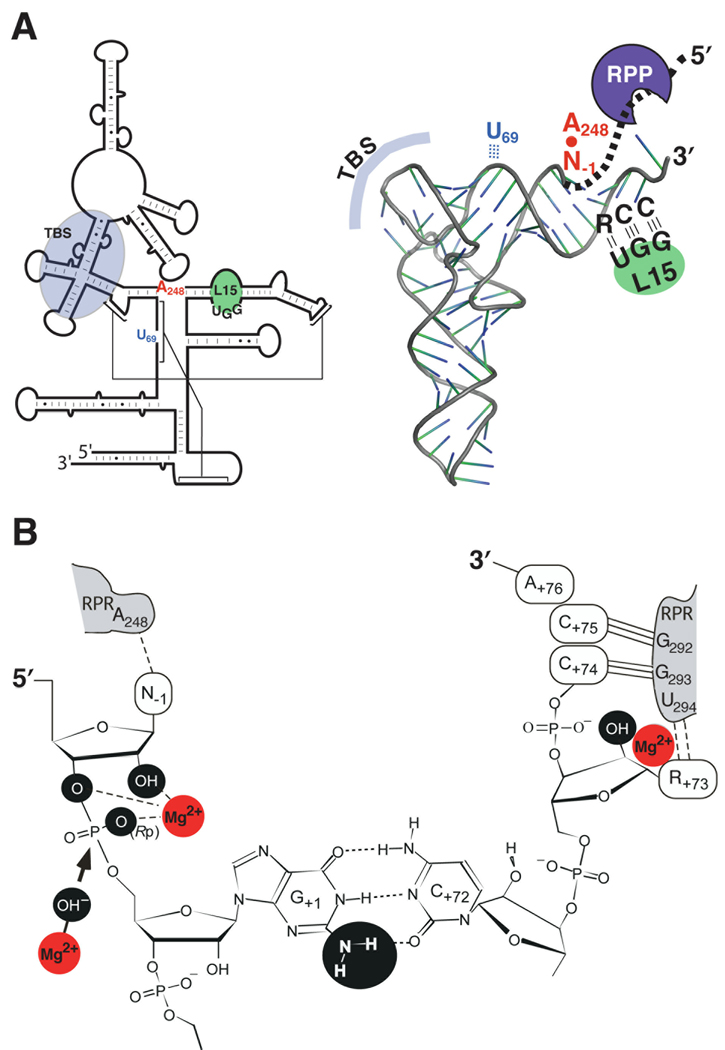
Figure 2.
Substrate recognition and catalysis by bacterial RNase P (see section 3 for details). (A) Secondary structures of the E. coli RPR (left) and a pre-tRNA (right) illustrating the domains required for interactions during RNase P catalysis. The interaction of the leader sequence in the pre-tRNA (bold, dashed line) with the RPP is also indicated. TBS, T stem-loop-binding site. (B) A close-up view showing one model of the canonical RNase P cleavage site. Chemical groups (black spheres) and Mg2+ (red spheres) suggested to contribute to catalysis are marked. Also depicted is a Mg2+-activated hydroxide nucleophile (arrow), which attacks the phosphorous atom in the scissile bond.
We do not have a crystal structure of RNase P bound to its substrate. However, based on biochemical and genetic data, several interactions between bacterial RNase P and its substrate have been well characterized (Fig. 2A). These interactions are (i) the T-stem-loop region (TSL) in the pre-tRNA and the P7–P11 region in the RPR, referred to as the TSL-binding site (TBS) [11,82–84], (ii) the 3'-RCC sequence of the pre-tRNA which pairs with a conserved GGU sequence in the L15-loop of RPR [85], (iii) the base at N−1 in the pre-tRNA interacts with A248 in the RPR (E. coli numbering) [86,87], (iv) the acceptor stem of the pre-tRNA and U69 in the RPR [88], (v) N−2 and N−4-N−7 in the 5' leader of the pre-tRNA and the bacterial RPP [25,28,89]. Many of these interactions influence the positioning of catalytic metal ions at and proximal to the cleavage site [26,88,90]. Interestingly, most pre-tRNAs carry a G+1, which has been suggested to act as a guiding nucleotide during formation of the RNase P-substrate complex [91,92]. Mutant bacterial RPRs or substrates (e.g., pre-tRNAs or model substrates) in which these contacts were individually disrupted have revealed some redundancy in the determinants that specify the cleavage site and align the scissile bond for efficient cleavage (for a recent review, see [11]).
The TBS structures in archaeal/eukaryal and bacterial RPRs are quite different, and the L15-loop, which in bacterial RPR constitutes the binding site for the 3' end of the substrate, is missing in eukaryal (and some archaeal) RPRs [9,93]. Our recent observations suggesting that the four archaeal RPPs influence substrate recognition and cleavage-site selection is therefore not unexpected (S. Sinapah et al., unpublished results). It remains to be determined, however, if the archaeal/eukaryal RPPs contribute directly to catalysis, or if they merely serve as a scaffold for their cognate RPRs and bring into register various residues and chemical groups required for catalysis.
If RPRs base pair with pre-tRNAs, then how does the protein-based RNase P recognize pre-tRNAs? In human mtRNase P, the Trm10 homolog MRPP1, responsible for G9 methylation, has been proposed to be vital for substrate recognition [7]. But only five of the 22 human mitochondrial tRNAs contain a G9. Activity of in vitro reconstituted human mtRNase P has been reported with only two pre-tRNAs, both containing a G9. Might a different RNase P enzyme be involved in processing the non-G9-containing pre-tRNAs? This question is pertinent as there has been controversy over the nature of human mtRNase P, specifically if it is protein- or RNP-based [94]. However, RNAi-mediated silencing of each of the three MRPPs induces accumulation of A9-containing pre-tRNAVal, indicating that the pre-tRNA processing function of MRPP1 is not restricted to G9-containing pre-tRNAs. Nevertheless, it is important to analyze the ability of protein-only human mtRNase P to recognize a broader range of pre-tRNAs (see 5.4).
3.2. Cleavage
Optimal cleavage by RNase P requires Mg2+. With bacterial RPRs, Mg2+ can be replaced by other divalent metal ions (e.g., Mn2+, Ca2+) albeit with decreased activity and, in some cases, fidelity [95–100]. It has been suggested that at least two Mg2+ ions participate directly in the chemistry of cleavage, which is believed to proceed via an SN2 mechanism: one hydrated Mg2+ ion generates the hydroxide nucleophile, and the other coordinates and stabilizes the oxyanion leaving group [101–103]. This mechanism was first observed in protein enzymes performing phosphoryl transfer (e.g., alkaline phosphatase) [104,105], and several studies have supported such a scheme in RNase P catalysis [90,91,96,98,99,101–103,106–111]. Hence, cleavage requires that the catalytic Mg2+ ions be positioned such that the correct phosphorous is attacked. Specific chemical groups at and proximal to the cleavage site have been identified to contribute to Mg2+ binding and bacterial RNase P catalysis: the 2'-OH at N−2, N−1 and R+73; N7 at R+73; the (pro)-Rp-oxygen; the bridging 3' oxygen (leaving group) and the exocyclic amine of G+1 [84,86,90,91,100,102,103,107–110,112–119]. In addition, replacing the 2'-OH at G+1 with hydrogen and the (pro)-Sp-oxygen at the scissile linkage with sulfur alters the cleavage-site [91,102,103,110,114], but whether these functional groups affect how catalytic Mg2+ ions are positioned or directly interact with RPR is not yet understood.
There appear to be negative and positive determinants for correct cleavage. The neighboring 2'-OH at N−1 in the substrate coordinates Mg2+ either by acting as an outer or inner sphere ligand [87,90,108]. Potentially, this 2'-OH could nucleophilically attack the phosphorous atom giving products with incorrect ends: 5'-OH and 2',3'-cyclic phosphate. Hence, the 2'-OH at N−1 has to be positioned in the RNase P-substrate (ES) complex such that this outcome is avoided and so this 2'-OH can be considered as a negative determinant. Among various possibilities for how this incorrect cleavage is prevented, one suggests that the 2'-OH at N−1 hydrogen bonds with the RPR and, together with the (pro)-Rp-oxygen, coordinates the Mg2+ which generates the attacking nucleophile for correct cleavage [108]. Since coordination of Mg2+ to the (pro)-Rp-oxygen would make the phosphorous more susceptible for a nucleophilic attack, the Rp-oxygen could be viewed as a positive determinant.
We have analyzed three-dimensional structures of short RNA helices to obtain information about the structural topography of the cleavage site in the absence of RNase P [120]. Such a study suggests that a fully hexahydrated Mg2+ could be positioned in the deep groove close (≈7–8 Å) to the cleavage site. Although the geometry is not ideal for in-line attack, in the presence of the RPR, this critical metal ion might be repositioned in the ES complex to activate a water molecule leading to nucleophilic attack on the scissile bond. Such a nucleophile-generating metal ion could be distinct from the metal ion that coordinates the (pro)-Rp-oxygen at the scissile bond and the 2'-OH at N−1 (Fig. 2B). Despite having a crystal structure for two bacterial RPRs in which some metal ion-binding sites have been identified [20,23,121], we still lack a clear picture of where the catalytically relevant metal ions are positioned.
Although the make-up of bacterial and eukaryal RNase P is different, the mechanism of cleavage in these disparate RNPs appears to have been preserved during evolution. Reminiscent of bacterial RNase P are the findings in the eukaryotic RNase P-catalyzed reaction that (i) sulfur-substitution of the (pro)-Sp-oxygen at the scissile bond shifts the cleavage site [116], and (ii) both the 2'-OH of N−1 and the (pro)-Rp-oxygen at the cleavage site contribute to catalysis, likely by coordinating Mg2+ [113,116]. In striking contrast to these RNPs, spinach chloroplast RNase P, suspected to be a protein-only enzyme, was largely insensitive to substitution of the (pro)-Rp-oxygen, indicating a different mechanism, yet to be deciphered [117].
4. BASIS FOR DIVERSITY OF RNase P?
4.1. Non-organellar RNase P
RPRs from all three domains of life possess the structural elements crucial for pre-tRNA processing; this common attribute of RPRs was anticipated from their shared ancestry attested by sequence and structural similarity of their putative catalytic core. None of the protein cofactors associated with the RNP forms of RNase P, either alone or in combination, displays any pre-tRNA processing activity. These observations could be integrated into an evolutionary scenario wherein a progenitor RNA, a possible remnant from the RNA world, recruited/associated itself with different protein cofactors; dynamic co-evolution of the RPR, concomitant with remodeling of the RNP to fit the needs of the respective cells, could account for the extant RPR variants (Fig. 1). However, it is intriguing why archaeal and eukaryal RNase P are associated with four and nine RPPs, respectively, especially when a single, small bacterial RPP (a mere one-tenth the size of the cognate RPR) renders the RNA catalyst more versatile and efficient under near-physiological conditions.
Albeit appealing, the notion that increased protein complexity helps RNase P fulfill additional functions or enables it to be finely regulated remains to be proven (see 5.2) [122,123]. Interestingly, the catalytic efficiency (at 37°C) of RNase P holoenzymes from the three domains of life are similar: kcat/Km ~107 M−1s−1 (caveat emptor - there are differences in assay conditions, substrates used, etc. in these different studies) [32,42,124]. Moreover, a recent study with affinity-purified yeast native RNase P showed that its turnover at steady state was limited by product release, akin to the rate-limiting step of the bacterial relative, thus indicating a similar mechanism for these two disparate holoenzymes [125].
In light of this uniformity amid diversity, what then might account for the additional archaeal/eukaryal RPPs? Some RPPs may be required to merely enhance the half-life (t1/2) of the RPR, which when naked may be subject to nucleases. Even with bacterial RNase P, the mature RPR has a t1/2 of 10 min, while that of the holoenzyme (with the single RPP) is 60 min [126]. The crystal structure of the bacterial RPR reveals that its catalytic and substrate-specificity domains are held together by intra-molecular braces formed by docking of tetraloops on helices [20,23]. While the absence of these RPR elements likely eliminated such struts in archaeal/eukaryal RPRs, at least some archaeal/eukaryal RPPs may have taken over this structural role to enhance overall stability [123].
It is conceivable that the additional RPPs aid the RPR’s fidelity of processing by virtue of proof-reading; in fact, one of the human RPPs has been shown to have weak ATPase activity [127]. Alternatively, they might make the archaeal/eukaryal RNase P holoenzyme more versatile by enabling processing of a wider array of substrates (both non-coding RNAs and mRNAs); for example, Saccharomyces cerevisiae RNase P is implicated in the maturation of some intron-encoded box C/D snoRNAs [128]. However, such a claim related to versatility has to be tempered by the fact that there is a growing number of non-tRNA substrates for even bacterial RNase P (e.g., polycistronic mRNAs) [129,130].
With emerging evidence for functional coupling of RNase P with other machineries, it is conceivable that this crosstalk requires RPPs for specialized protein-protein interactions. In fact, inactivation of human RNase P results in decreased transcription of several non-coding RNAs in a cell cycle-dependent fashion; it has been speculated that RNase P (not necessarily with its full suite of RPPs) might act as a scaffold for the pol I and pol III transcription machineries or even facilitate chromatin remodeling [131,132]. Moreover, there is recent support for linkage between RNase P expression/assembly and mitochondrial fatty acid biosynthesis in yeast and vertebrates, although how and why these pathways intersect is unclear [133].
While the bacterial RNase P holoenzyme might be able to process in vitro the newly identified substrates of eukaryal RNase P, the additional RPPs in the latter might be critical for sub-cellular localization, regulation and functional coordination with other cellular assemblages. Focusing on the catalytic capabilities alone of the different RNase P variants might then undervalue the roles of archaeal/eukaryal RPPs in their respective milieu.
4.2. Organellar RNase P
Despite lacking in-depth knowledge about organellar RNase P, available information attests to its rapid evolution as borne out by its diverse structure and composition (see 2.4). In the protist Reclinomonas, mtRNase P contains a bacterial-like RPR similar to those of the α-proteobacterial ancestor, while in human mitochondria it is devoid of RNA [7,64]. A similar trend is observed in chloroplasts: a glaucophyte, red algae and prasinophytes contain a bacterial-like RPR similar to those of their cyanobacterial ancestor, while in higher plants the enzyme appears to be protein only [68–73,79]. What is the basis underlying this evolutionary change? Although it is attributable to the high evolutionary rate of organelles and the plasticity of their genomes, reasons based on RNase P function also merit consideration. RNase P has many substrates besides pre-tRNAs in bacteria and the same might be true in the eukaryotic nucleus. Evolution of nuclear RNase P is therefore constrained by the need to recognize these different substrates efficiently and cleave them with high fidelity. Due to reduced and streamlined genomes in organelles, many of the non-tRNA substrates known in bacteria are absent, perhaps then allowing the organellar enzyme to diverge more freely albeit at the cost of a narrower substrate specificity. Alternatively, the idiosyncratic nature of organellar tRNAs and mRNAs could be the driving force behind the evolution of the enzyme. Clearly, importing and assembling a large RNP (at least in some cases) was disfavored over that of a collage of pre-existing organellar proteins [7].
5. REMAINING CHALLENGES
We enumerate below some directions that might facilitate efforts to unravel the bases for the striking variations in RNase P.
5.1. Is the protein-rich subunit composition of RNase P in higher organisms a necessity for its temporal and spatial control?
The subunit make-up of eukaryotic RNase P need not be invariable. It is possible that core components are required for RNase P catalysis, and additional RPPs are recruited to promote assembly and sub-cellular localization in response to temporal and spatial cues. Support for such a scenario comes from various findings. First, in vitro reconstituted archaeal RNase P, made up of the RPR and five RPPs (including L7Ae), displays a kcat/Km comparable to that of yeast native RNase P (I-M Cho and Gopalan, unpublished results) [42]. Second, a native precursor form of yeast RNase P (RPR + 7 RPPs, without POP3/RPP38 and Rpr2p/RPP21) was found to display an identical steady-state rate as the mature form (RPR + 9 RPPs) [134]. Third, animal RPP20 associates with the SMN complex that is required for assembly of large RNPs [135]. Therefore, spatially- and temporally-resolved proteomic inventories are needed to understand these possible dynamic changes in RNase P composition that might be choreographed by development, differentiation and/or environmental factors. Such changes in the make-up of the holoenzyme might be essential for altering substrate specificity, influencing localization or conferring regulation. Appropriate choice of model organisms (e.g., Caenorhabditis elegans) will be critical to generate a spatio-temporal compositional map of RNase P.
Our knowledge of a possible “catalytic core” could also be shaped by a systematic computational analysis of genomes of simpler eukaryotes (e.g., Trichoplax adhaerens, Monosiga brevicollis). Of course, the presence of such a eukaryal RNase P with fewer RPPs compared to yeast/human RNase P will need to be experimentally validated.
An examination of the 3' UTRs of all eukaryal RPP mRNAs for possible micro-RNA (miRNA)-binding sites might help define the core RPPs and those that are regulatory or transiently associated. A recent study has suggested that housekeeping genes escape miRNA regulation by having shorter 3' UTRs compared to highly regulated genes which are subject to stringent control by miRNAs [136]. Alternative splicing, such as that described for human RPP21, could be another mechanism to regulate eukaryotic RNase P subunit composition and attendant cellular processes [36,137]. The finding of three additional mouse genes encoding shorter RPRs led to the idea, untested so far, that these 3'-truncated forms might soak up RPPs and thereby negatively regulate mouse RNase P activity in a cell-specific manner [138].
In vitro reconstitution of eukaryotic RNase P, albeit intractable to date, is essential to delineate the subunits minimally required for catalysis. Although the human RPR is weakly active without RPPs [5], and some activity has been reported when reconstituted with RPP21 and RPP29 [139], a sequential assembly map with all 10 RPPs, together with a kinetic and thermodynamic analysis of each partially reconstituted complex, will help delineate the functional contribution of individual RPPs. Such studies with archaeal RNase P have proven instructive [58,59] and might even provide a framework for studies on the more complex eukaryotic counterpart.
5.2. What is the full spectrum of substrates of RNase P?
Coupling DNA microarrays with conditional mutants of RNase P in bacteria and yeast is a powerful approach to conduct genome-wide searches and identify new substrates (e.g., polycistronic mRNAs, snoRNAs) [128,130,140]. Rapid affinity purification to obtain substrate-bound yeast native RNase P and subsequent deep sequencing of bound substrates have also proven fruitful [128]; the caveat exists that transiently-associated substrates might not be identified. Similar approaches in other unicellular systems might expand the repertoire of RNase P substrates. For multi-cellular organisms, the availability of inducible RNAi constructs [141,142], critical in dissecting the role of tissue-specific proteins in developmental processes (in flies and worms), opens up the possibility of similarly exploring the specialized role(s) of “accessory RPPs”. Once an RPP is down-regulated using transgenic RNAi, RNAs in tissues of interest could be examined using either deep sequencing or tiling arrays to identify novel RNase P substrates whose processing likely required the knocked-down RPP. Correlating molecular and phenotypic changes will add to our understanding of the RPP’s role in an organismal context.
The ability of bacterial and eukaryal RNase P to cleave a large number of structurally different substrates begs the question of what features are essential for recognition in vivo. A careful compilation of non-tRNA substrates and analyzing their structures (experimental probing or in silico approaches) might aid future computational prediction of RNase P substrates.
5.3. Is there coordinate control of expression of RPR and RPPs?
RNase P, like other RNPs, presents a challenge to the cell for balanced synthesis of its RNA and protein components. In bacteria, the RPR has a significantly shorter half-life in vivo without its cognate protein [126]; therefore, economic considerations alone would suggest that there be mechanisms (currently unknown) for synchronized expression of the RNA and protein subunits. In addition to the possible involvement of post-transcriptional processes to ensure stoichiometric expression of RPR and RPPs in both bacteria and eukarya, coordinated transcription of the eukaryotic RPR (by pol III) and nine RPP mRNAs (by pol II) faces the problem of establishing crosstalk between different polymerases. In fact, in two independent studies [143,144], down-regulation of a single RPP in cultured human cells resulted in a concomitant decrease of up to four other RPPs (but not the RPR), likely due to transcriptional repression. Transcriptome data pursuant to either transcription factor knock-down or overexpression could be used to uncover mechanisms that permit coordinate control. Also, recent advances using destabilizing domains in a protein of interest and a synthetic, cell-permeable organic molecule to rapidly and reversibly manipulate its stability/function [145] are worth exploiting to examine how altering a single RPP affects other RNase P holoenzyme components.
Superimposed on this coordinated RPR and RPP expression is the need for functional coupling of RNase P with other members of the translational apparatus. In bacteria and archaea, one RPP gene is in the same operon as a ribosomal protein gene [15,146]; how the crosstalk is accomplished in eukaryotes is unclear. Recently, two studies have discovered that yeast RNase P might process antisense RNAs from genes encoding ribosomal proteins [128,140]; whether this is a mechanism for correlating RNase P activity with expression of ribosomal proteins remains to be proven.
5.4. Is the protein-only RNase P recently found in human mitochondria unique to metazoa?
The finding of a proteinaceous human mtRNase P [7], along with the proposal of the same in plant choroplasts [79], suggests that an organellar protein-only RNase P might be more common. Of the three protein subunits of human mtRNase P, only the putative catalytic MRPP3 has a widespread distribution in eukarya; both MRPP1 and MRPP2 seem to be restricted to the animal kingdom [7]. Such non-overlapping presence of the three subunits implies that alternative organellar solution(s) might be used in other eukaryotes. It is possible that MRPP3 from a non-metazoan lineage can support RNase P activity either alone or with additional lineage-specific proteins. The finding that pentatricopeptide repeat (PPR) proteins support translation in organelles by binding to mRNAs and tRNAs [147] suggests that an MRPP3 homolog might be able to use (i) its PPR domain to bind pre-tRNAs, and (ii) its metallonuclease domain to cleave the 5' leader. Moreover, given the combinatorial capabilities afforded by exon shuffling, it would not be surprising to find hybrid versions where two or three RPPs of human mtRNase P are fused together. Database mining and validating such variants will be fruitful.
Nuclear RNase P has been found to date to be RNP-based, but there might well be exceptions. Although the sequenced nuclear genomes of kinetoplastids encode no identifiable eukaryal nuclear RPR or RPPs [43], MRPP3 homologs are found in Leishmania species and Trypanosoma brucei [7], and these merit further investigation.
5.5. Organellar RNase P variants - surprises in store?
Biochemical purification of organellar RNase P, albeit arduous, was the key to determining the protein-only make-up of human mtRNase P [7]. However, this procedure requires the availability of high-quality purified organelles in sufficient amount from an organism with its nuclear and organellar genomes sequenced. That is not the case for many of the organisms in which the organellar genome encodes an rnpB. For instance, it is currently difficult to isolate from red algae chloroplasts that are free of contaminating nuclei and cytoplasm. Other algae like Ostreococcus tauri are difficult to cultivate to high levels as axenic cultures. Biochemical advances will therefore depend on identifying new model organisms that have complete genome information, are amenable to genetic manipulation, and permit isolation of large amounts of organelles.
Although currently available organellar genomic sequences have not provided clues on possible RPPs, several prasinophyte nuclear genomes encode proteins containing domains homologous to bacterial RPP [148–150]. The function of these proteins is unknown as is their localization to organelles, where an rnpB is identified in some cases. This is nevertheless an interesting scenario wherein the RNA component of the bacterial RNase P predecessor was retained in the organellar genome while the protein was transferred to the nucleus.
5.6. Is the protein-only RNase P functionally equivalent to the RNP version?
Since genetic complementation assays are available in bacteria and yeast, it would be interesting to examine if the three-protein human mtRNase P would suffice in heterologous settings where an RNP is typically employed; of course, such experiments are predicated on successful expression and reconstitution of the MRPPs in a non-native environment. It is also essential to determine in vitro the kinetic properties of the RNP and protein variants of RNase P (at physiological Mg2+ and salt concentrations) to appreciate possible limitations of the latter that might have precluded its widespread adoption in lieu of the former.
5.7. Coordination with other machineries
To fully understand the basis for the increased protein complexity in archaeal and eukaryal RNase P, it is vital to map their possible linkage with other cellular machineries. Since affinity purification of human and yeast RNase P is possible, these purifications should be performed under conditions that preserve weak macromolecular interactions, allowing subsequent proteomic studies to identify interactors. Immunoprecipitation (IP) with an RPP-specific antibody under mild conditions followed by examining the IP contents on a comprehensive antibody micro-array (when available) could also provide useful leads.
6. SUMMARY
For a seemingly simple housekeeping-type enzyme, RNase P continues to provide surprises in terms of the structural diversity of its active sites and the complex evolution underpinning this variability. Perhaps, it merely attests the idea, now appreciated in protein enzymes, that there are many ways to hydrolyze phosphodiester bonds in nucleic acids. High-resolution structures of RNase P-substrate complexes from the three domains of life (including organellar variants) should inform us if a uniform active-site architecture could result from RNA- and protein-based RNase P. Meanwhile, the daunting challenge will be to figure out the possible bases for this remarkable plasticity of RNase P.
ACKNOWLEDGMENTS
We are deeply indebted to Professor Sidney Altman (Yale University) for his continued support as a mentor and a valuable colleague. We are grateful for support from Ministerio de Ciencia y Tecnología, Spain (BFU2007-60651, to AV) and Junta de Andalucía, Spain (P06-CVI-01692, to AV); the Swedish Research Council (to LAK), Linné support from the Swedish Research Council to Uppsala RNA Research Center and the Swedish Foundation for Strategic Research (to LAK); and the NSF (MCB-0843543, to VG) and the NIH (GM067807, to Mark Foster and VG; AI082242, to Dan Schoenberg and VG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 2.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Gopalan V, Altman S. Ribonuclease P: structure and catalysis. In: Gesteland RF, Cech TR, Atlkins JF, editors. The RNA World. New York: Cold Spring Harbor Laboratory Press; 2006. only online at http://rna.cshl.edu. [Google Scholar]
- 4.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl. Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Altman S. Ribonuclease P. New York: Springer-Verlag; 2009. [Google Scholar]
- 9.Ellis JC, Brown JW. The RNase P family. RNA Biol. 2009;6:362–369. doi: 10.4161/rna.6.4.9241. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Prog. Mol. Biol. Transl. Sci. 2009;85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 11.Kirsebom LA, Trobro S. RNase P RNA-mediated cleavage. IUBMB Life. 2009;61:189–200. doi: 10.1002/iub.160. [DOI] [PubMed] [Google Scholar]
- 12.Smith JK, Hsieh J, Fierke CA. Importance of RNA-protein interactions in bacterial ribonuclease P structure and catalysis. Biopolymers. 2007;87:329–338. doi: 10.1002/bip.20846. [DOI] [PubMed] [Google Scholar]
- 13.Ozeki H, Sakano H, Yamada S, Ikemura T, Shimura Y. Temperature-sensitive mutants of Escherichia coli defective in tRNA biosynthesis. Brookhaven. Symp. Biol. 1975;26:89–105. [PubMed] [Google Scholar]
- 14.Schedl P, Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl. Acad. Sci. USA. 1973;70:2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen FG, Hansen EB, Atlung T. Physical mapping and nucleotide sequence of the rnpA gene that encodes the protein component of ribonuclease P in Escherichia coli. Gene. 1985;38:85–93. doi: 10.1016/0378-1119(85)90206-9. [DOI] [PubMed] [Google Scholar]
- 16.Kole R, Altman S. Reconstitution of RNase P activity from inactive RNA and protein. Proc. Natl. Acad. Sci. USA. 1979;76:3795–3799. doi: 10.1073/pnas.76.8.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kole R, Baer MF, Stark BC, Altman S. E. coli RNAase P has a required RNA component. Cell. 1980;19:881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- 18.Stark BC, Kole R, Bowman EJ, Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc. Natl. Acad. Sci. USA. 1978;75:3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazantsev AV, Krivenko AA, Harrington DJ, Carter RJ, Holbrook SR, Adams PD, Pace NR. High-resolution structure of RNase P protein from Thermotoga maritima. Proc. Natl. Acad. Sci. USA. 2003;100:7497–7502. doi: 10.1073/pnas.0932597100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl. Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spitzfaden C, Nicholson N, Jones JJ, Guth S, Lehr R, Prescott CD, Hegg LA, Eggleston DS. The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J. Mol. Biol. 2000;295:105–115. doi: 10.1006/jmbi.1999.3341. [DOI] [PubMed] [Google Scholar]
- 22.Stams T, Niranjanakumari S, Fierke CA, Christianson DW. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- 23.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 24.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crary SM, Niranjanakumari S, Fierke CA. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5' leader sequence of pre-tRNAAsp. Biochemistry. 1998;37:9409–9416. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- 26.Kurz JC, Fierke CA. The affinity of magnesium binding sites in the Bacillus subtilis RNase P●pre-tRNA complex is enhanced by the protein subunit. Biochemistry. 2002;41:9545–9558. doi: 10.1021/bi025553w. [DOI] [PubMed] [Google Scholar]
- 27.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 28.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl. Acad. Sci. USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reich C, Olsen GJ, Pace B, Pace NR. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988;239:178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L, Harris ME. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007;13:1505–1515. doi: 10.1261/rna.571007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tallsjo A, Kirsebom LA. Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993;21:51–57. doi: 10.1093/nar/21.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 34.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eder PS, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl. Acad. Sci. USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarrous N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Eenennaam H, Jarrous N, van Venrooij WJ, Pruijn GJ. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life. 2000;49:265–272. doi: 10.1080/15216540050033113. [DOI] [PubMed] [Google Scholar]
- 38.Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl. Acad. Sci. USA. 2002;99:2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang T, Altman S. Protein-protein interactions with subunits of human nuclear RNase P. Proc. Natl. Acad. Sci. USA. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang T, Guerrier-Takada C, Altman S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao S, Hsieh J, Nugent RL, Coughlin DJ, Fierke CA, Engelke DR. Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP. RNA. 2006;12:1023–1037. doi: 10.1261/rna.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenblad MA, Lopez MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franklin SE, Zwick MG, Johnson JD. Characterization and partial purification of two pre-tRNA 5'-processing activities from Daucus carrota (carrot) suspension cells. Plant J. 1995;7:553–563. doi: 10.1046/j.1365-313x.1995.7040553.x. [DOI] [PubMed] [Google Scholar]
- 45.Pulukkunat DK. M.S. thesis. Columbus, OH: The Ohio State University; 2002. [Google Scholar]
- 46.Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8:296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amero CD, Boomershine WP, Xu Y, Foster M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry. 2008;47:11704–11710. doi: 10.1021/bi8015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl. Acad. Sci. USA. 2003;100:15398–15403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. Structure of an archaeal homolog of the human protein complex Rpp21–Rpp29 that is a key core component for the assembly of active ribonuclease P. J. Mol. Biol. 2008;384:652–662. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 50.Kakuta Y, Ishimatsu I, Numata T, Kimura K, Yao M, Tanaka I, Kimura M. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: an archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry. 2005;44:12086–12093. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- 51.Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme. J. Mol. Biol. 2006;357:583–591. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 52.Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10:1423–1432. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sidote DJ, Heideker J, Hoffman DW. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry. 2004;43:14128–14138. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- 54.Sidote DJ, Hoffman DW. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42:13541–13550. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- 55.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc. Natl. Acad. Sci. USA. 2006;103:873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takagi H, Watanabe M, Kakuta Y, Kamachi R, Numata T, Tanaka I, Kimura M. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2004;319:787–794. doi: 10.1016/j.bbrc.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 57.Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 58.Pulukkunat DK, Gopalan V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008;36:4172–4180. doi: 10.1093/nar/gkn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl. Acad. Sci. USA. 2006;103:16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall TA, Brown JW. Interactions between RNase P protein subunits in archaea. Archaea. 2004;1:247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kifusa M, Fukuhara H, Hayashi T, Kimura M. Protein-protein interactions in the subunits of ribonuclease P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci. Biotechnol. Biochem. 2005;69:1209–1212. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal RNase P binary protein complex: Formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009;393:1043–1055. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukuhara H, Kifusa M, Watanabe M, Terada A, Honda T, Numata T, Kakuta Y, Kimura M. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006;343:956–964. doi: 10.1016/j.bbrc.2006.02.192. [DOI] [PubMed] [Google Scholar]
- 64.Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 65.Turmel M, Lemieux C, Burger G, Lang BF, Otis C, Plante I, Gray MW. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1730. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seif ER, Forget L, Martin NC, Lang BF. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA. 2003;9:1073–1083. doi: 10.1261/rna.5880403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seif E, Leigh J, Liu Y, Roewer I, Forget L, Lang BF. Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 2005;33:734–744. doi: 10.1093/nar/gki199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shevelev EL, Bryant DA, Loffelhardt W, Bohnert HJ. Ribonuclease-P RNA gene of the plastid chromosome from Cyanophora paradoxa. DNA Res. 1995;2:231–234. doi: 10.1093/dnares/2.5.231. [DOI] [PubMed] [Google Scholar]
- 69.Hagopian JC, Reis M, Kitajima JP, Bhattacharya D, de Oliveira MC. Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastids. J. Mol. Evol. 2004;59:464–477. doi: 10.1007/s00239-004-2638-3. [DOI] [PubMed] [Google Scholar]
- 70.Ohta N, et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2003;10:67–77. doi: 10.1093/dnares/10.2.67. [DOI] [PubMed] [Google Scholar]
- 71.Reith M, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 1995;13:333–335. [Google Scholar]
- 72.Turmel M, Gagnon MC, O'Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol. Biol. Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- 73.Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl. Acad. Sci. USA. 1999;96:10248–10253. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De la Cruz J, Vioque A. A structural and functional study of plastid RNAs homologous to catalytic bacterial RNase P RNA. Gene. 2003;321:47–56. doi: 10.1016/s0378-1119(03)00831-x. [DOI] [PubMed] [Google Scholar]
- 75.Li D, Willkomm DK, Schon A, Hartmann RK. RNase P of the Cyanophora paradoxa cyanelle: A plastid ribozyme. Biochimie. 2007;89:1528–1538. doi: 10.1016/j.biochi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 76.Pascual A, Vioque A. Functional reconstitution of RNase P activity from a plastid RNA subunit and a cyanobacterial protein subunit. FEBS Lett. 1999;442:7–10. doi: 10.1016/s0014-5793(98)01621-4. [DOI] [PubMed] [Google Scholar]
- 77.Baum M, Cordier A, Schon A. RNase P from a photosynthetic organelle contains an RNA homologous to the cyanobacterial counterpart. J. Mol. Biol. 1996;257:43–52. doi: 10.1006/jmbi.1996.0145. [DOI] [PubMed] [Google Scholar]
- 78.Thomas BC, Li X, Gegenheimer P. Chloroplast ribonuclease P does not utilize the ribozyme-type pre-tRNA cleavage mechanism. RNA. 2000;6:545–553. doi: 10.1017/s1355838200991465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang MJ, Davis NW, Gegenheimer P. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J. 1988;7:1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morales MJ, Dang YL, Lou YC, Sulo P, Martin NC. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc. Natl. Acad. Sci. USA. 1992;89:9875–9879. doi: 10.1073/pnas.89.20.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orellana O, Cooley L, Soll D. The additional guanylate at the 5' terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol. Cell. Biol. 1986;6:525–529. doi: 10.1128/mcb.6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brannvall M, Kikovska E, Wu S, Kirsebom LA. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J. Mol. Biol. 2007;372:1149–1164. doi: 10.1016/j.jmb.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 83.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2'-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl. Acad. Sci. USA. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 85.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zahler NH, Christian EL, Harris ME. Recognition of the 5' leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zahler NH, Sun L, Christian EL, Harris ME. The pre-tRNA nucleotide base and 2'-hydroxyl at N(−1) contribute to fidelity in tRNA processing by RNase P. J. Mol. Biol. 2005;345:969–985. doi: 10.1016/j.jmb.2004.10.080. [DOI] [PubMed] [Google Scholar]
- 88.Christian EL, Smith KM, Perera N, Harris ME. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA. 2006;12:1463–1467. doi: 10.1261/rna.158606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rueda D, Hsieh J, Day-Storms JJ, Fierke CA, Walter NG. The 5' leader of precursor tRNAAsp bound to the Bacillus subtilis RNase P holoenzyme has an extended conformation. Biochemistry. 2005;44:16130–16139. doi: 10.1021/bi0519093. [DOI] [PubMed] [Google Scholar]
- 90.Brannvall M, Kikovska E, Kirsebom LA. Cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage. Nucleic Acids Res. 2004;32:5418–5429. doi: 10.1093/nar/gkh883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perreault JP, Altman S. Important 2'-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J. Mol. Biol. 1992;226:399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- 92.Svard SG, Kirsebom LA. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J. Mol. Biol. 1992;227:1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]
- 93.Brown JW. The ribonuclease P database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brannvall M, Kirsebom LA. Manganese ions induce miscleavage in the Escherichia coli RNase P RNA-catalyzed reaction. J. Mol. Biol. 1999;292:53–63. doi: 10.1006/jmbi.1999.3048. [DOI] [PubMed] [Google Scholar]
- 96.Brannvall M, Kirsebom LA. Metal ion cooperativity in ribozyme cleavage of RNA. Proc. Natl. Acad. Sci. USA. 2001;98:12943–12947. doi: 10.1073/pnas.221456598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gardiner KJ, Marsh TL, Pace NR. Ion dependence of the Bacillus subtilis RNase P reaction. J. Biol. Chem. 1985;260:5415–5419. [PubMed] [Google Scholar]
- 98.Guerrier-Takada C, Haydock K, Allen L, Altman S. Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli. Biochemistry. 1986;25:1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- 99.Kazakov S, Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1991;88:9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smith D, Burgin AB, Haas ES, Pace NR. Influence of metal ions on the ribonuclease P reaction. Distinguishing substrate binding from catalysis. J. Biol. Chem. 1992;267:2429–2436. [PubMed] [Google Scholar]
- 101.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–10559. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 102.Chen Y, Li X, Gegenheimer P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-RP oxygen of the scissile bond. Biochemistry. 1997;36:2425–2438. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- 103.Warnecke JM, Furste JP, Hardt WD, Erdmann VA, Hartmann RK. Ribonuclease P (RNase P) RNA is converted to a Cd2+-ribozyme by a single Rpphosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl. Acad. Sci. USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim EE, Wyckoff HW. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J. Mol. Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 105.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cuzic S, Hartmann RK. Studies on Escherichia coli RNase P RNA with Zn2+ as the catalytic cofactor. Nucleic Acids Res. 2005;33:2464–2474. doi: 10.1093/nar/gki540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perreault JP, Altman S. Pathway of activation by magnesium ions of substrates for the catalytic subunit of RNase P from Escherichia coli. J. Mol. Biol. 1993;230:750–756. doi: 10.1006/jmbi.1993.1197. [DOI] [PubMed] [Google Scholar]
- 108.Persson T, Cuzic S, Hartmann RK. Catalysis by RNase P RNA: unique features and unprecedented active site plasticity. J. Biol. Chem. 2003;278:43394–43401. doi: 10.1074/jbc.M305939200. [DOI] [PubMed] [Google Scholar]
- 109.Smith D, Pace NR. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 110.Warnecke JM, Held R, Busch S, Hartmann RK. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 111.Zuleeg T, Hartmann RK, Kreutzer R, Limmer S. NMR spectroscopic evidence for Mn2+(Mg2+) binding to a precursor-tRNA microhelix near the potential RNase P cleavage site. J. Mol. Biol. 2001;305:181–189. doi: 10.1006/jmbi.2000.4299. [DOI] [PubMed] [Google Scholar]
- 112.Brannvall M, Kirsebom LA. Complexity in orchestration of chemical groups near different cleavage sites in RNase P RNA mediated cleavage. J. Mol. Biol. 2005;351:251–257. doi: 10.1016/j.jmb.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 113.Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 114.Kikovska E, Brannvall M, Kirsebom LA. The exocyclic amine at the RNase P cleavage site contributes to substrate binding and catalysis. J. Mol. Biol. 2006;359:572–584. doi: 10.1016/j.jmb.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 115.Kleineidam RG, Pitulle C, Sproat B, Krupp G. Efficient cleavage of pre-tRNAs by E. coli RNAse P RNA requires the 2'-hydroxyl of the ribose at the cleavage site. Nucleic Acids Res. 1993;21:1097–1101. doi: 10.1093/nar/21.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pfeiffer T, Tekos A, Warnecke JM, Drainas D, Engelke DR, Seraphin B, Hartmann RK. Effects of phosphorothioate modifications on precursor tRNA processing by eukaryotic RNase P enzymes. J. Mol. Biol. 2000;298:559–565. doi: 10.1006/jmbi.2000.3655. [DOI] [PubMed] [Google Scholar]
- 117.Thomas BC, Li X, Gegenheimer P. Chloroplast ribonuclease P does not utilize the ribozyme-type pre-tRNA cleavage mechanism. RNA. 2000;6:545–553. doi: 10.1017/s1355838200991465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Warnecke JM, Sontheimer EJ, Piccirilli JA, Hartmann RK. Active site constraints in the hydrolysis reaction catalyzed by bacterial RNase P: analysis of precursor tRNAs with a single 3'-S-phosphorothiolate internucleotide linkage. Nucleic Acids Res. 2000;28:720–727. doi: 10.1093/nar/28.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kikovska E, Brannvall M, Kufel J, Kirsebom LA. Substrate discrimination in RNase P RNA-mediated cleavage: importance of the structural environment of the RNase P cleavage site. Nucleic Acids Res. 2005;33:2012–2021. doi: 10.1093/nar/gki344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kikovska E, Mikkelsen NE, Kirsebom LA. The naturally trans-acting ribozyme RNase P RNA has leadzyme properties. Nucleic Acids Res. 2005;33:6920–6930. doi: 10.1093/nar/gki993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kazantsev AV, Krivenko AA, Pace NR. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA. 2008;15:266–276. doi: 10.1261/rna.1331809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marvin MC, Engelke DR. RNase P: increased versatility through protein complexity? RNA Biol. 2009;6:40–42. doi: 10.4161/rna.6.1.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gopalan V. Uniformity amid diversity in RNase P. Proc. Natl. Acad. Sci. USA. 2007;104:2031–2032. doi: 10.1073/pnas.0611193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Andrews AJ, Hall TA, Brown JW. Characterization of RNase P holoenzymes from Methanococcus jannaschii and Methanothermobacter thermoautotrophicus. Biol. Chem. 2001;382:1171–1177. doi: 10.1515/BC.2001.147. [DOI] [PubMed] [Google Scholar]
- 125.Hsieh J, Walker SC, Fierke CA, Engelke DR. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA. 2009;15:224–234. doi: 10.1261/rna.1309409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim Y, Lee Y. Novel function of C5 protein as a metabolic stabilizer of M1 RNA. FEBS Lett. 2009;583:419–424. doi: 10.1016/j.febslet.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 127.Li Y, Altman S. A subunit of human nuclear RNase P has ATPase activity. Proc. Natl. Acad. Sci. USA. 2001;98:441–444. doi: 10.1073/pnas.021555498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coughlin DJ, Pleiss JA, Walker SC, Whitworth GB, Engelke DR. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc. Natl. Acad. Sci. USA. 2008;105:12218–12223. doi: 10.1073/pnas.0801906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl. Acad. Sci. USA. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Y, Cole K, Altman S. The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA. 2003;9:518–532. doi: 10.1261/rna.2198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 2006;20:1621–1635. doi: 10.1101/gad.386706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS One. 2008;3:e4072. doi: 10.1371/journal.pone.0004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hiltunen JK, Schonauer MS, Autio KJ, Mittelmeier TM, Kastaniotis AJ, Dieckmann CL. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 2009;284:9011–9015. doi: 10.1074/jbc.R800068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Srisawat C, Houser-Scott F, Bertrand E, Xiao S, Singer RH, Engelke DR. An active precursor in assembly of yeast nuclear ribonuclease P. RNA. 2002;8:1348–1360. doi: 10.1017/s1355838202027048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hua Y, Zhou J. Rpp20 interacts with SMN and is re-distributed into SMN granules in response to stress. Biochem. Biophys. Res. Commun. 2004;314:268–276. doi: 10.1016/j.bbrc.2003.12.084. [DOI] [PubMed] [Google Scholar]
- 136.Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10:431. doi: 10.1186/1471-2164-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7:1153–1164. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li K, Williams RS. Cloning and characterization of three new murine genes encoding short homologues of RNase P RNA. J. Biol. Chem. 1995;270:25281–25285. doi: 10.1074/jbc.270.42.25281. [DOI] [PubMed] [Google Scholar]
- 139.Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol. Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 140.Samanta MP, Tongprasit W, Sethi H, Chin CS, Stolc V. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc Natl Acad Sci USA. 2006;103:4192–4197. doi: 10.1073/pnas.0507669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Briese M, Esmaeili B, Johnson NM, Sattelle DB. pWormgatePro enables promoter-driven knockdown by hairpin RNA interference of muscle and neuronal gene products in Caenorhabditis elegans. Invert. Neurosci. 2006;6:5–12. doi: 10.1007/s10158-005-0011-x. [DOI] [PubMed] [Google Scholar]
- 142.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cohen A, Reiner R, Jarrous N. Alterations in the intracellular level of a protein subunit of human RNase P affect processing of tRNA precursors. Nucleic Acids Res. 2003;31:4836–4846. doi: 10.1093/nar/gkg691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kovrigina E, Wesolowski D, Altman S. Coordinate inhibition of expression of several genes for protein subunits of human nuclear RNase P. Proc. Natl. Acad. Sci. USA. 2003;100:1598–1602. doi: 10.1073/pnas.0337661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 147.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 2008;28:5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Derelle E, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]