Abstract
Inhibitors and activators of protein-protein interactions are valuable as biological probes and medicinal agents, but are often difficult to identify. Herein we describe a high-throughput assay, based upon photonic crystal (PC) biosensors, for the identification of modulators of protein—protein interactions. Through the use of a D-biotin-tris-NTA (BTN) hybrid compound, any His6-tagged protein can be immobilized on the surface of a PC biosensor. Binding of the bound protein to its cognate partner is detected via a shift in the peak wavelength value. We demonstrate this assay with three protein-protein pairs (caspase-9—XIAP, caspase-7—XIAP, FKBP12-FRB) and their small molecule modulators.
While the value of small molecules that inhibit or enhance protein—protein interactions is widely recognized,1 the identification of such compounds remains problematic.2 This is partly due to the wide and shallow nature of many protein—protein interfaces, but also because of the inherent difficulties associated with detecting protein—protein binding en masse. Assays utilized for the detection of protein—protein interactions include enzyme-linked immunosorbent assays (ELISAs),3 fluorescence polarization (FP),4 and surface plasmon resonance (SPR).5 ELISAs and FP have been applied in a high-throughput screening (HTS) mode for the identification of small molecule inhibitors of protein—protein interactions,1b,6 while SPR can be multiplexed for low-to-medium throughput applications.7 To develop an assay for the high-throughput identification of modulators of protein—protein interactions we have devised a strategy that requires only a His6-tagged protein and an alternately tagged or untagged version of the cognate partner. This assay, based on photonic crystal (PC) biosensor technology, is able to measure protein—protein interactions in real time, and is independent of antibodies or fluorescence measurements.
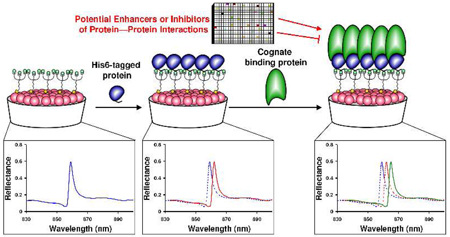
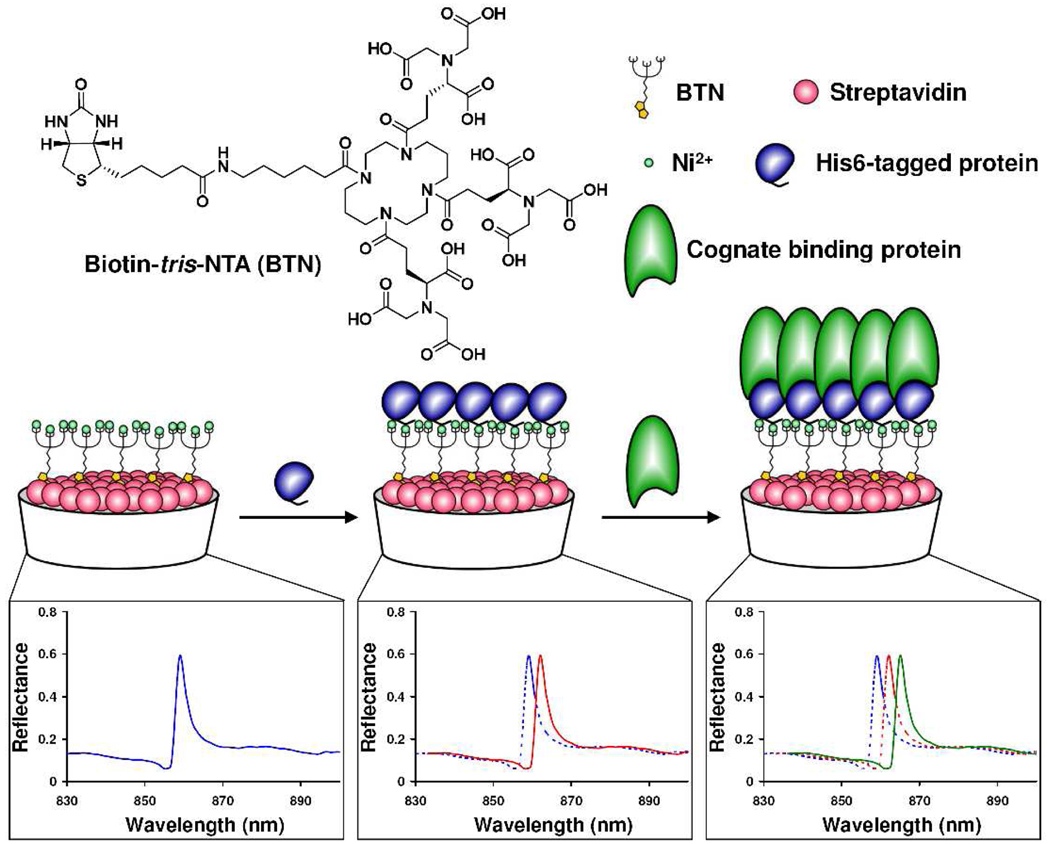
PC biosensors are surface structures comprised of a subwavelength polymer grating coated with a high refractive index material (TiO2).8 The PC surface resonantly reflects a single wavelength, that is modulated by the adsorption of biomaterial to the sensor surface.8 Sheets of PC biosensors can be attached to bottomless 384-well plates for HTS, and coated with streptavidin (SA). As demonstrated previously,9 PC biosensor technology can be used for measuring protein—DNA interactions and HTS. As modulators of protein—protein interactions are valuable as medicinal agents and biological probes, but difficult to identify, we sought to adapt PC biosensor technology to the detection of protein—protein interactions. As diagrammed in Figure 1, we envisioned a D-biotin-tris-NTA hybrid compound (BTN) as the linkage between the biosensor surface and a His6-tagged protein of interest. As demonstrated herein, after charging the sensor with Ni2+ a His6-tagged protein can bind to the functionalized sensor, and the interaction between two proteins of interest can readily be detected by a peak wavelength value (PWV) shift, a change in the peak reflectance of the sensor.
Figure 1.
Structure of D-Biotin-tris-NTA (BTN) and a schematic of a His6-tagged protein binding to the BTN-coupled PC biosensors, followed by cognate protein binding. Each binding event results in a shift in the peak wavelength value of the sensor.
For these experiments, BTN was synthesized in a manner similar to analogous compounds (see Supporting Information).10 After incubating individual wells of the SA-coated 384-well biosensor plate with BTN and then charging with 500 µM NiCl2, four different His-tagged proteins (caspase-3,-7,-9, and FKBP12, ranging in size from 12 to 37 kDa) were shown to bind the BTN-functionalized PC biosensor surface in a dose-dependent manner as indicated by PWV shifts; this binding was not observed in the absence of NiCl2 (See Supporting Figures S1, S2).
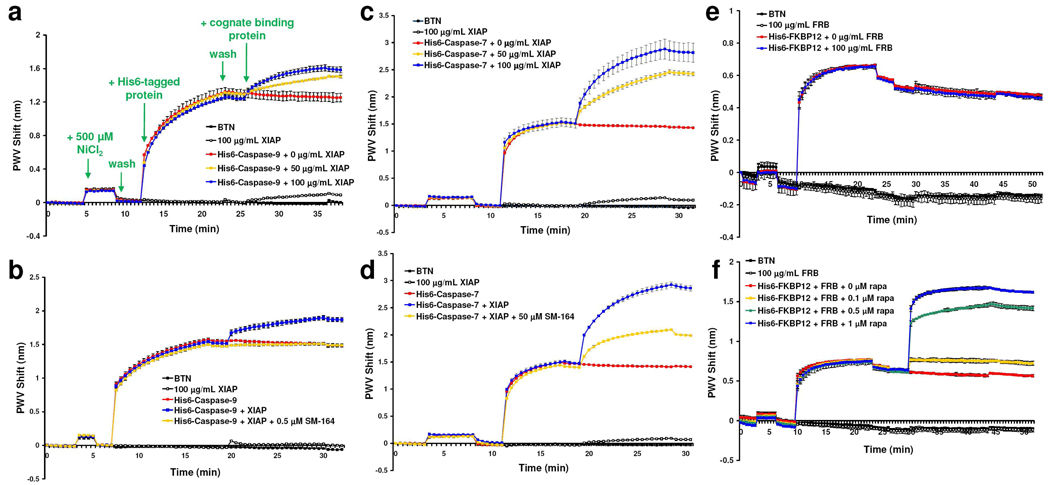
Next, three well-studied protein—protein pairs with known small molecule modulators were chosen for proof-of-principle experiments: XIAP—caspase-9,11 XIAP—caspase-7,11 and FKBP12—mTOR (FRB domain).12 XIAP binds to caspase-7 (KD = 0.5 nM)13a and caspase-913b (KD = 0.5 µM) with KD values in the range typical of many protein-protein interactions. As shown, immobilized His6-caspase-9 (Fig. 2a) and His6-caspase-7 (Fig. 2c) bind to their cognate protein (GST-XIAP), while His6-FKBP12 does not bind GST-FRB in the absence of rapamycin (Fig. 2e). The binding of GST-XIAP to His6-caspase-3 was also readily detected using this technology (Supporting Figure S3).
Figure 2.
Protein—protein interactions and their inhibition or enhancement by small molecules. (a) The association of GST-XIAP and His6-caspase-9, and (b) their inhibition by SM-164. (c) The association of GST-XIAP and His6-caspase-7, and (d) their inhibition by SM-164. (e) The inability of GST-FRB to bind to His6-FKBP12, and (f) the enhancement of this interaction by rapamycin. All experiments were conducted at 25 °C, and preincubation times and other experimental details are described in the Supporting Information. All error bars represent the range (n = 2).
The XIAP—caspase-9 and XIAP—caspase-7 interactions can be inhibited by SM-164,11 while FKBP12 will bind to FRB only in the presence of rapamycin. The inhibitory properties of SM-164 were readily apparent using the BTN-conjugated biosensor. As shown in Fig. 2b & d, preincubation of XIAP with SM-164 resulted in a dramatic reduction in protein—protein binding. The greater potency of SM-164 in the caspase-9 system is supported by the differential binding affinity of XIAP for members of the caspase family and the synthetic design of SM-164.13 In an analogous fashion, enhancement of protein—protein interactions was demonstrated by the binding of GST-FRB to His6-FKBP12 in the presence of rapamycin (Fig. 2f). Disruption of the onsensor His6-caspase-9—GST-XIAP complex through addition of SM-164 could also readily be detected (Supporting Fig. S4).
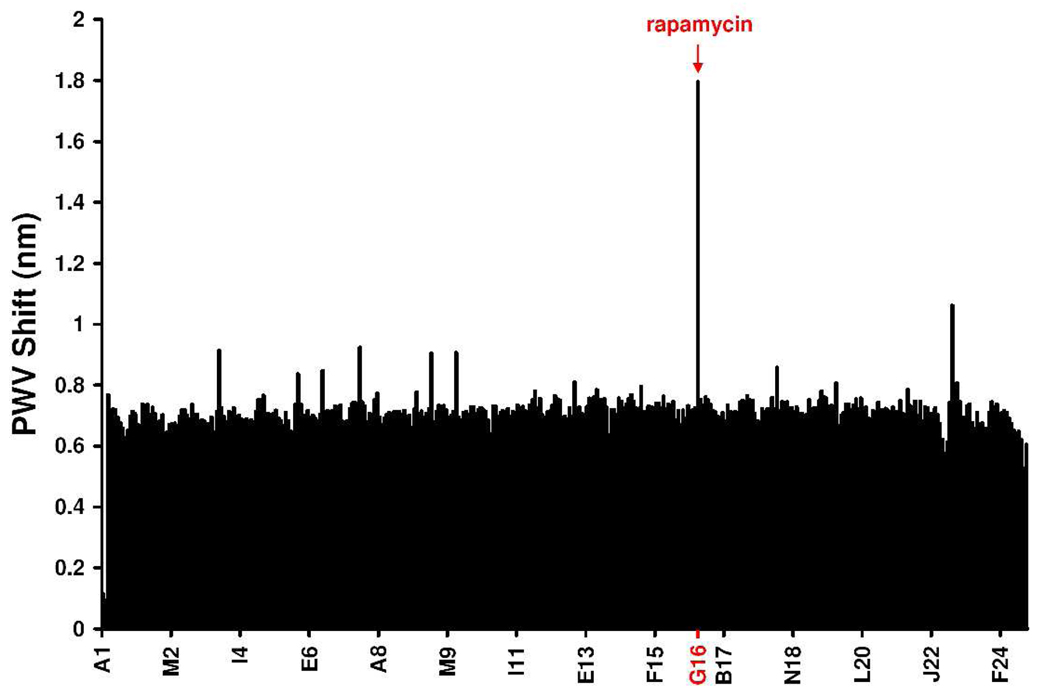
Finally, to demonstrate the HTS capability of the PC biosensor assay, we performed a “needle-in-a-haystack” experiment with His6-FKBP12 and GST-FRB (Figure 3). 320 compounds from an in-house screening library were added to different wells of a 384-well SA biosensor plate that had been charged with BTN, NiCl2, and His6-FKBP12. Rapamycin was then spiked into a well; the final concentration of screening compounds was 50 µM, whereas rapamycin was at 1 µM. As shown in Figure 3, after addition of GST-FRB the rapamycin-containing well is easily identified amongst the screening compounds, demonstrating that this assay is capable of being performed in a HTS mode for identifying modulators of protein-protein interactions. In an analogous HTS experiment, SM-164 was readily detected as an inhibitor of caspase-9—XIAP binding (Supporting Figure S5).
Figure 3.
Enhancement of GST-FRB binding to His6-FKBP12 is detectable in an HTS. A1 and B1 contain no His6-FKBP12 (only DMSO & GST-FRB), and G16 contains rapamycin (1 µM). All other wells contain His6-FKBP12, GST-FRB, and screening compounds (at 50 µM).
As a reusable biosensor surface would substantially reduce the cost of a PC biosensor screen, we explored the possibility of regenerating and reusing the BTN-conjugated PC biosensor plates. Treatment of the wells with 350 mM EDTA, followed by washing, regenerated the sensor surface with minimal loss of binding capacity, even when binding and stripping procedures were repeated five times (Supporting Figure S6). In addition to the cost reduction, if the BTN-conjugated biosensor plates are reused five times, then screening 200,000 compounds (at one compound per well in 384-well plates) would require less than 20 mg of the BTN reagent. Finally, although the major advantage of this technology is its use in HTS, it is worth noting that IC50 values for inhibitors can be calculated from dose response curves. For example, we find that SM-164 inhibits the XIAP-caspase-9 interaction with an IC50 = 0.39 µM, whereas SM-122, a compound that binds more weakly to XIAP than SM-164,11 has an IC50 = 2.72 µM (Supporting Figure S7).
Small molecule modulators of protein—protein interactions have tremendous potential in biology and medicine. The system described herein for their identification is general (interfacing with widely-utilized His6 fusion proteins), does not require antibodies or fluorescent tags, and utilizes commercially available biosensor readers and plates. Applications of this technique for the high-throughput identification of novel protein—protein modulators are on-going and will be reported in due course.
Supplementary Material
Acknowledgment
We thank Prof. Colin Duckett (U. Michigan) for the pGEX-XIAP, Prof. Shaomeng Wang (U. Michigan) for SM-164 and SM-122, Prof Qian Tin (Cornell University) for pET28a-caspase-9, and Prof. Jie Chen (U. Illinois) for pGEX-FKBP12 and pGEX-FRB. This work was supported by the NIH (R01CA118562) and the Korea Research Foundation Grant (KRF-2008-357-E00064 to S.-H. K.).
Footnotes
Supporting Information Available: Materials and Methods, supporting figures, and NMR spectra. This information is available free of charge via the internet at http://pubs.acs.org/.
References
- 1.(a) Berg T. Curr. Opin. Drug. Discov. Devel. 2008;11:666. [PubMed] [Google Scholar]; (b) Fry DC. Biopolymers. 2006;84:535. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 2.(a) Domling A. Curr. Opin. Chem. Biol. 2008;12:281. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]; (b) Whitty A, Kumaravel G. Nat. Chem. Biol. 2006;2:112. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 3.Taipa MA. Comb. Chem. High Throughput Screen. 2008;11:325. doi: 10.2174/138620708784246031. [DOI] [PubMed] [Google Scholar]
- 4.Eggeling C, Brand L, Ullmann D, Jager S. Drug Discov. Today. 2003;8:632. doi: 10.1016/s1359-6446(03)02752-1. [DOI] [PubMed] [Google Scholar]
- 5.Berggard T, Linse S, James P. Proteomics. 2007;7:2833. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 6.(a) Fletcher S, Hamilton AD. Curr. Top. Med. Chem. 2007;7:922. doi: 10.2174/156802607780906735. [DOI] [PubMed] [Google Scholar]; (b) Vicent MJ, Perez-Paya E, Orzaez M. Curr. Top. Med. Chem. 2007;7:83. doi: 10.2174/156802607779318307. [DOI] [PubMed] [Google Scholar]
- 7.Boozer C, Kim G, Cong S, Guan H, Londergan T. Curr. Opin. Biotechnol. 2006;17:400. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 8.(a) Cunningham BT, Laing L. Expert Rev. Proteomics. 2006;3:271. doi: 10.1586/14789450.3.3.271. [DOI] [PubMed] [Google Scholar]; (b) Cunningham BT, Li P, Schulz S, Lin B, Baird C, Gerstenmaier J, Genick C, Wang F, Fine E, Laing L. J. Biomol. Screen. 2006;6:481. doi: 10.1177/1087057104267604. [DOI] [PubMed] [Google Scholar]
- 9.Chan LL, Pineda M, Heeres JT, Hergenrother PJ, Cunningham BT. ACS Chem. Biol. 2008;3:437. doi: 10.1021/cb800057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Lata S, Gavutis M, Tampe R, Piehler J. J. Am. Chem. Soc. 2006;128:2365. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]; (b) Lata S, Reichel A, Brock R, Tampe R, Piehler J. J. Am. Chem. Soc. 2005;127:10205. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- 11.Sun H, Nikolovska-Coleska Z, Lu J, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller PP, Stuckey JA, Wang S. J. Am. Chem. Soc. 2007;129:15279. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi J, Chen J, Schreiber SL, Clardy J. Science. 1996;273:239. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 13.(a) Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Cell. 2001;104:781. [PubMed] [Google Scholar]; (b) Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. Nature. 2001;410:112. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]; (c) Sun H, Nikolovska-Coleska Z, Lu J, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller PP, Stuckey JA, Wang S. J. Am. Chem. Soc. 2007;129:15279. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.