Abstract
Satellite cells are skeletal muscle stem cells that provide myogenic progeny for myofiber growth and repair. Temporal expression of muscle regulatory factors (MRFs) and the paired box transcription factor Pax7 defines characteristic phases of proliferation (Pax7+/MyoD+/myogenin−) and differentiation (Pax7−/MyoD+/myogenin+) during myogenesis of satellite cells. Here, using bromodeoxyuridine (BrdU) labeling and triple immunodetection, we analyzed expression patterns of Pax7 and the MRFs MyoD, Myf5, or myogenin within S phase myoblasts prepared from posthatch chicken muscle. Essentially all BrdU incorporation was restricted to Pax7+ cells, of which the majority also expressed MyoD. The presence of a minor BrdU+/Pax7+/myogenin+ population in proliferation stage cultures suggests that myogenin upregulation is alone insufficient for terminal differentiation. Myf5 was detected strictly within Pax7+ cells and decreased during S phase while MyoD presence persisted in cycling cells. This study provides novel data in support of a unique role for Myf5 during posthatch myogenesis.
INTRODUCTION
Skeletal muscle myofibers are formed during embryogenesis and continue to enlarge postnatally until their mature size has been reached. This postnatal myofiber growth entails an increase in myofiber protein accretion and in the number of myofiber nuclei (Allen et al., 1979; Edgerton and Roy, 1991; Mozdziak et al., 1997). The primary source of these additional myofiber nuclei are the satellite cells, myogenic stem cells situated on the surface of the myofiber between the myofiber plasmalemma and its overlying basal lamina (Mauro, 1961; Zammit et al., 2006; Allouh et al., 2008; Yablonka-Reuveni et al., 2008). Satellite cells emerge in late fetal stage as a distinct myogenic population, replacing the myoblasts that contribute to muscle development during embryogenesis (Cossu and Molinaro 1987; Stockdale 1992; Yablonka–Reuveni 1995). During postnatal growth, satellite cells proliferate and then fuse with the enlarging myofibers (Moss and Leblond, 1971; Halevy et al., 2004; Schultz et al., 2006). In mature muscles, the satellite cells are typically quiescent, but are activated in response to muscle damage and provide progeny for myofiber repair (Grounds and Yablonka-Reuveni, 1993; Hawke and Garry, 2001).
Satellite cells and their progeny are characterized by temporal expression of transcription factors associated with different stages of myogenic progression. These factors include the paired box transcription factor Pax7, and the myogenic regulatory factors (MRFs), MyoD and myogenin (Smith et al., 1993; Smith et al., 1994; Yablonka-Reuveni-Reuveni and Rivera, 1994; Seale et al., 2000; Zammit et al., 2006; Yablonka-Reuveni et al., 2008). Based on characteristic temporal expression of Pax7, MyoD, and myogenin during posthatch myogenesis in the chicken, we proposed a model of satellite cell proliferation (Pax7+/MyoD+/myogenin−), differentiation (Pax7−/MyoD+/myogenin+), and reservation, also referred to as renewal (Pax7+/MyoD−/myogenin−) (Halevy et al., 2004). We also detected a minority population of Pax7+/myogenin+ cells that may represent a transitory compartment at the onset of differentiation (Halevy et al., 2004). A similar model of proliferation, differentiation, and reservation/renewal was based on combinatorial expression of Pax7, MyoD and myogenin during myogenesis initiated by mouse satellite cells (Zammit et al., 2004; Shefer et al., 2006; Day et al., 2007).
Another myogenic regulatory factor, Myf5, was shown to be expressed by satellite cells and their progeny during rodent postnatal myogenesis (Smith et al., 1993; Smith et al., 1994). However, most of the studies have been based on Myf5-driven reporter activity and Myf5 mRNA expression (Smith et al., 1994; Beauchamp et al., 2000; Zammit et al., 2006; Day et al., 2007), whereas the presence of Myf5 protein is not well characterized. Increased Myf5 protein levels in cultures of satellite cells from MyoD null mice suggested a compensatory role for Myf5 in the absence of MyoD (Yablonka-Reuveni et al., 1999). Although adult myogenesis can proceed in the absence of Myf5 or MyoD, satellite cells from Pax7-deficient mice are rapidly depleted during postnatal growth (Rudnicki et al., 1992; Grounds, 1999; Seale et al., 2000; Oustanina et al., 2004; Gayraud-Morel et al., 2007; Ustanina et al., 2007). These studies indicate that Myf5 and MyoD may have overlapping roles. However, analyses of primary myoblasts and myogenic cell lines demonstrated that protein levels of Myf5 and MyoD are regulated at different stages of the cell cycle and advocate non-redundant functions (Kitzmann et al., 1998; Lindon et al., 1998).
The present study follows our previous reports that characterized MyoD, myogenin, and Pax7 expression throughout the phase of posthatch chicken myogenesis during which satellite cells are proliferative and enable rapid myofiber growth (Yablonka-Reuveni and Paterson, 2001; Halevy et al., 2004). Herein, we analyzed Myf5 protein expression, and determined the status of Myf5 compared to MyoD and myogenin as cells transitioned through S phase. Proliferating cells were monitored by bromodeoxyuridine (BrdU) incorporation in combination with transcription factor detection using triple immunostaining for BrdU, Pax7 and each MRF (Myf5, MyoD, or myogenin). Myf5 was detected strictly within Pax7+ cells, which is distinct from MyoD detection in both Pax7+ and Pax7− cells. Our data indicates a wider range of MyoD presence through the cell cycle while Myf5 is detected during a narrower phase within Pax7+ cells. A small number of myogenin+ cells were also detected within the BrdU+/Pax7+ population. We propose a refined model of posthatch myogenesis where the dynamics of MyoD, Myf5, and myogenin detection in Pax7+ proliferating cells suggest unique roles for each MRF that may drive cycling cells toward differentiation, reservation, or continuation in the cell cycle.
RESULTS
Myogenic regulatory factor detection in early and late stage myogenic cultures
To determine the patterns of MRF expression within myogenic cells at S phase, chicken primary cultures were incubated with BrdU and subsequently immunostained for Pax7 and BrdU, and each MRF. To follow changes in MRF detection in cycling cells, early stage (day 4) and late stage (day 10) myogenic cultures were incubated with BrdU for a short (2 hr) and long (6 hr) pulse to ensure maximal labeling of proliferating cells regardless of possible differences in the length of S phase. DAPI counter-staining for individual nuclei was not possible as blue fluorescence was dedicated to detection of BrdU incorporation. Cells positive for Pax7 and/or MRF were considered myogenic cells, and cells negative for both Pax7 and MRF were excluded from the analysis. As previously published, our cell isolation and culture conditions yielded highly enriched myogenic cultures; the contribution of nonmyogenic cells was no more than 5% of total cells.
In early stage cultures, 95-99% of all myogenic cells were Pax7+ and virtually all nuclei that incorporated BrdU were Pax7+ (Fig. 1, Table 1). For each MRF examined, less than 5% of the cells expressed MRF alone (Pax7− cells), and did not incorporate BrdU. Approximately 48-70% of total myogenic nuclei at early stage were labeled with BrdU (percentage depended on the MRF analyzed and BrdU pulse duration). The highest percentage of the BrdU+/MRF+ cell populations out of total myogenic cells was represented by Pax7+/MyoD+ cells, followed by Pax7+/Myf5+ cells, and the lowest percentage was the Pax7+/myogenin+ cells. The respective percentages of BrdU+/Pax7+/MRF+ cells for 2 and 6 hour BrdU pulse were: ~42 and 52 (MyoD+), ~38 and 26 (Myf5+), ~14 and 8 (myogenin+).
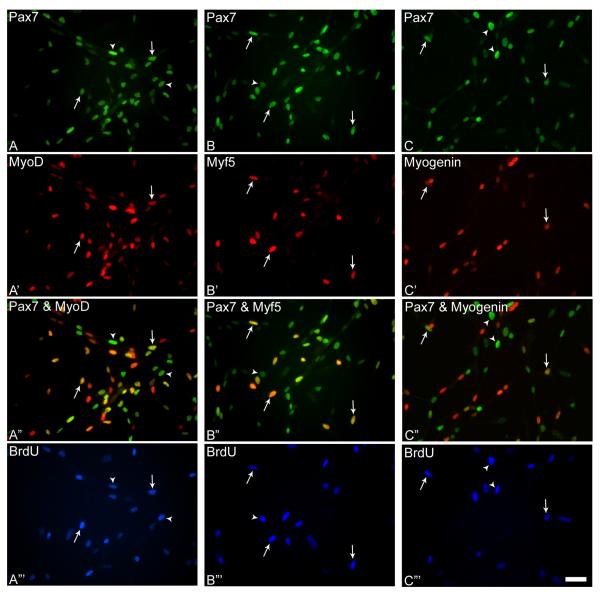
Figure 1.
Representative photomicrographs of day 4 cultures incubated with BrdU and triple-stained with antibodies against Pax7 and MyoD (A, A’), Pax7 and Myf5 (B, B’), Pax7 and myogenin (C, C’), and BrdU (A”’- C”’). Merged images of Pax7 and MRF immunostained cultures are depicted in A”- C”. Arrows indicate triple-stained nuclei in parallel images, and arrowheads indicate nuclei double-stained for Pax7 and BrdU. Scale bar, 15μM.
Table 1.
Representative distribution (% ± SEM) of cells labeled with BrdU, Pax7 and MRF in early and late stage cultures§
| BrdU pulse (hours) |
Total number of cells |
BrdU+ |
BrdU− |
||||
|---|---|---|---|---|---|---|---|
| Pax7+/MyoD− | Pax7+/MyoD+ | Pax7−/MyoD+ | Pax7+/MyoD− | Pax7+/MyoD+ | Pax7−/MyoD+ | ||
| Early | |||||||
| 2 | 575 | 6.09 ± 2.82 | 41.91 ± 3.91 | 0.35 ± 0.18 | 1.57 ± 0.38 | 45.22 ± 2.03 | 4.87 ± 1.31 |
| 6 | 569 | 8.08 ± 1.27 | 51.84 ± 1.45 | 0.00 | 9.14 ± 1.89 | 28.30 ± 0.24 | 2.64 ± 1.13 |
| Late | |||||||
| 2 | 2875 | 0.31 ± 0.31 | 14.58 ± 0.65 | 1.01 ± 0.51 | 2.82 ± 0.58 | 48.90 ± 1.15 | 32.37 ± 1.01 |
| 6 | 2592 | 0.96 ± 0.20 | 11.69 ± 1.23 | 8.49 ± 1.06 | 2.39 ± 0.55 | 31.44 ± 3.27 | 45.02 ± 3.28 |
| Pax7+/Myf5− | Pax7+/Myf5+ | Pax7−/Myf5+ | Pax7+/Myf5− | Pax7+/Myf5+ | Pax7−/Myf5+ | ||
|---|---|---|---|---|---|---|---|
| Early | |||||||
| 2 | 453 | 17.22 ± 2.12 | 38.41 ± 3.85 | 0.00 | 26.27 ± 4.51 | 17.22 ± 2.20 | 0.88 ± 0.12 |
| 6 | 572 | 44.23 ± 5.22 | 25.52 ± 2.31 | 0.00 | 20.63 ± 1.54 | 8.39 ± 0.90 | 1.22 ± 1.05 |
| Late | |||||||
| 2 | 1450 | 11.45 ± 2.01 | 8.90 ± 0.60 | 0.00 | 48.00 ± 2.74 | 29.79 ± 4.27 | 1.86 ± 0.36 |
| 6 | 1365 | 26.30 ± 3.38 | 8.57 ± 1.23 | 0.22 ± 0.20 | 40.95 ± 2.51 | 23.00 ± 2.58 | 0.95 ± 0.44 |
| Pax7+/Myog− | Pax7+/Myog+ | Pax7−/Myog+ | Pax7+/Myog− | Pax7+/Myog+ | Pax7−/Myog+ | ||
|---|---|---|---|---|---|---|---|
| Early | |||||||
| 2 | 479 | 44.05 ± 2.37 | 13.57 ± 0.39 | 0.00 | 26.10 ± 2.03 | 15.24 ± 0.40 | 1.04 ± 0.40 |
| 6 | 563 | 59.50 ± 2.50 | 7.64 ± 2.93 | 0.00 | 18.83 ± 0.44 | 11.01 ± 0.24 | 3.02 ± 0.82 |
| Late | |||||||
| 2 | 2728 | 12.13 ± 2.30 | 1.32 ± 0.27 | 0.62 ± 0.22 | 44.10 ± 1.47 | 10.37 ± 1.16 | 31.45 ± 1.58 |
| 6 | 2291 | 18.29 ± 3.91 | 1.79 ± 0.09 | 0.31 ± 0.03 | 33.96 ± 2.54 | 12.70 ± 1.20 | 32.96 ± 2.22 |
Cultures fixed at early (day 4) and late (day 10) stages were initiated at 5 × 104 and 1 × 103 cells per 35-mm plates, respectively. Cells were monitored by triple-immunofluorescence with antibodies against Pax7, BrdU and MRF (MyoD, Myf5 or myogenin). The total number of cells is based on the number of stained nuclei in each immunostaining scheme as indicated. Twenty arbitrary fields were monitored per culture using a 40× objective and studies were repeated three times with similar results.
In the late stage, there was a drastic decline in the overall percentage of BrdU+ cells with a concomitant increase in percentage of BrdU− cells for each MRF combination analyzed compared to early stage (Fig. 2, Table 1). Consistent with the early stage, the highest percentage of the BrdU+/MRF+ cell population out of total myogenic cells in the late stage was represented by Pax7+/MyoD+ cells, followed by Pax7+/Myf5+ cells, and the lowest percentage was the Pax7+/myogenin+ cells. The respective percentages of BrdU+/Pax7+/MRF+ cells for 2 and 6 hour BrdU pulse were: ~15 and 12 (MyoD+), ~9 and 9 (Myf5+), ~1 and 2 (myogenin+).
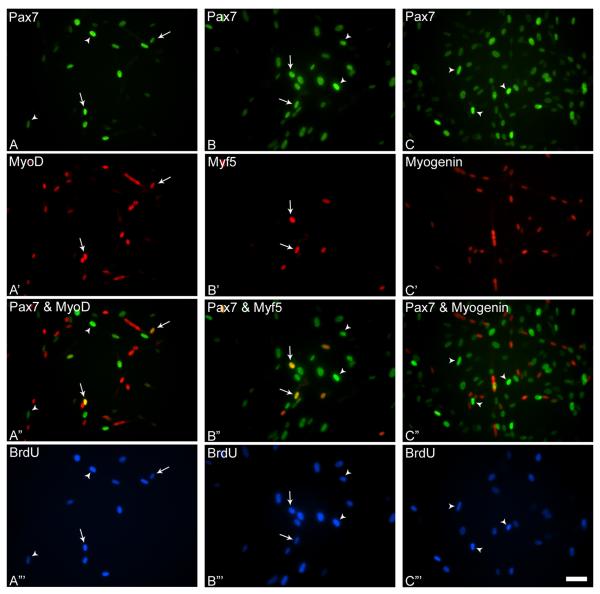
Figure 2.
Representative photomicrographs of day 10 cultures incubated with BrdU and triple-stained with antibodies against Pax7 and MyoD (A, A’), Pax7 and Myf5 (B, B’), Pax7 and myogenin (C, C’), and BrdU (A”’- C”’). Merged images of Pax7 and MRF immunostained cultures are depicted in A”- C”. Arrows indicate triple-stained nuclei in parallel images, and arrowheads indicate nuclei double-stained for Pax7 and BrdU. Scale bar, 15μM.
Essentially all Myf5+ cells (~98% of cells stained with Myf5 antibody) were within the Pax7+ pool at both early and late stages, while many of the MyoD+ and myogenin+ cells were within the BrdU−/Pax7− cell pool at the late stage (~30% or greater of the myogenic cells for each MRF) (Table 1). Such Pax7−/MyoD+ or Pax7−/myogenin+ cells represented mostly differentiated myoblasts or nuclei in myotubes (Halevy et al., 2004; Shefer et al., 2006). The only instance of BrdU incorporation into Pax7-/MRF+ cells was in the Pax7−/MyoD+ population at late stage that increased to ~8% with the 6h pulse (Table 1). For all other groups, Pax7−/MRF+ cells rarely labeled with BrdU at either stage regardless of pulse duration.
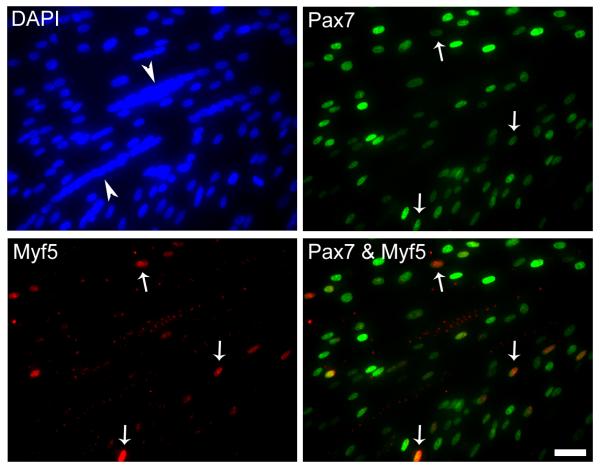
The reduced frequency of Myf5+ cells compared to MyoD+ cells within the Pax7+ population (in both early and late stage cultures) either reflected more restricted dynamics of Myf5 expression during the cell cycle, or represented a distinct Myf5+ subpopulation. To discern whether Myf5 was specifically expressed only within progeny of some progenitors, clonal cultures were established and grown until late stage for Pax7 and Myf5 immunostaining (Fig. 3). A similar approach was previously described for the detection of MyoD+ or myogenin+ cells in myogenic clones (Halevy et al., 2004). Myf5+ cells were detected in all 25 myogenic clones analyzed. Therefore, the dynamics of Myf5 detection is not reflective of restricted Myf5 expression within progeny of select satellite cell subpopulations, but indicates a temporal expression pattern within myoblasts at early and late stages in culture.
Figure 3.
Representative photomicrographs of a day 10 clone double-stained with antibodies against Pax7 and Myf5 and counter-stained with DAPI to monitor all nuclei. Merged panel is also depicted. Arrows mark double-stained cells in parallel and merged images. Arrowheads point to myotubes. Scale bar, 15μM.
Our initial analysis (Table 1) resulted in a nonparallel representation of the percentage of BrdU+/MRF+ cells within the Pax7+ population for Myf5 versus MyoD or myogenin immunostaining schemes because MyoD and myogenin were both detected within Pax7+ and Pax7− cells at late stage, while Myf5 was essentially detected only within Pax7+ cells. Therefore, the distribution of MRF+ and BrdU+ cells was evaluated only within the Pax7+ population to effectively compare the S phase detection pattern at both culture stages (Table 2).
Table 2.
Evaluated frequency (% ± SEM) of BrdU±/MRF± cells within Pax7+ cells in early and late stage cultures§
| BrdU pulse (hours) |
Early |
Late |
||
|---|---|---|---|---|
| BrdU+ |
BrdU− |
BrdU+ |
BrdU− |
|
| MyoD+ |
||||
| 2 | 44.22 ± 3.97 | 47.71 ± 1.62 | 21.89 ± 1.01 | 73.41 ± 1.79 |
| 6 | 53.25 ± 2.06 | 29.06 ± 0.58 | 25.15 ± 2.75 | 67.63 ± 1.53 |
| MyoD− |
||||
|---|---|---|---|---|
| 2 | 6.42 ± 3.02 | 1.65 ± 0.43 | 0.47 ± 0.47 | 4.23 ± 0.84 |
| 6 | 8.30 ± 1.23 | 9.39 ± 1.84 | 2.07 ± 0.29 | 5.15 ± 0.98 |
| Myf5+ |
||||
|---|---|---|---|---|
| 2 | 38.75 ± 3.84 | 17.37 ± 2.23 | 9.07 ± 0.63 | 30.36 ± 4.41 |
| 6 | 25.84 ± 2.63 | 8.50 ± 0.98 | 8.67 ± 1.19 | 23.28 ± 2.49 |
| Myf5− |
||||
|---|---|---|---|---|
| 2 | 17.37 ± 2.12 | 26.50 ± 4.58 | 11.67 ± 2.02 | 48.91 ± 2.61 |
| 6 | 44.78 ± 4.86 | 20.88 ± 1.76 | 26.61 ± 3.48 | 41.44 ± 2.66 |
| Myogenin+ |
||||
|---|---|---|---|---|
| 2 | 13.71 ± 0.34 | 15.40 ± 0.36 | 1.94 ± 0.37 | 15.27 ± 1.50 |
| 6 | 7.88 ± 3.01 | 11.36 ± 0.22 | 2.68 ± 0.10 | 19.03 ± 2.50 |
| Myogenin− |
||||
|---|---|---|---|---|
| 2 | 44.51 ± 2.43 | 26.37 ± 2.04 | 17.86 ± 3.10 | 64.92 ± 3.31 |
| 6 | 61.36 ± 2.50 | 19.41 ± 0.34 | 27.40 ± 5.27 | 50.88 ± 3.71 |
See footnote in Table 1.
Myogenic regulatory factor detection within Pax7+ myoblasts
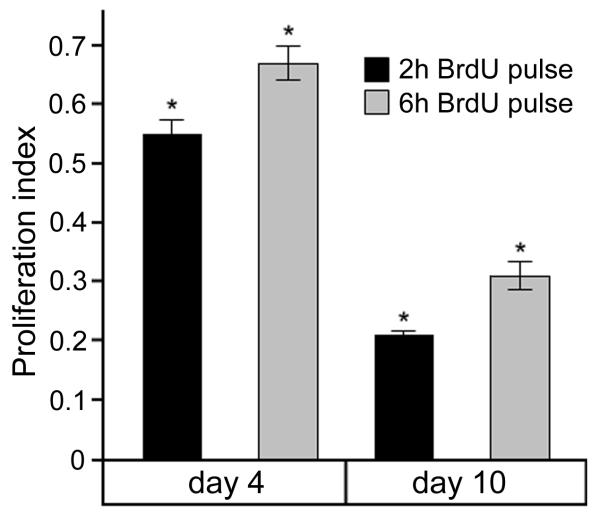
The frequency of BrdU+ cells within the Pax7+ population (proliferation index) at early and late culture stages was calculated by determining the mean number of nuclei that incorporated BrdU within Pax7+ cells among individual MRF immunostaining schemes (Fig. 4). The percent of Pax7+ nuclei that incorporated BrdU at early stage was double compared to late stage regardless of BrdU pulse duration, and there was a significant increase with the longer BrdU pulse at each culture stage when compared to the shorter pulse (~10%).
Figure 4.
Proliferation index for Pax7+ cells in day 4 and day 10 primary cultures. The frequency of BrdU+ cells within the Pax7+ population at early and late culture stages was calculated by determining the mean number of nuclei that incorporated BrdU within Pax7+ cells among individual MRF immunostaining schemes. Results are means (± SEM, n=3). Two-way ANOVA indicated a significant effect of BrdU incorporation with incubation time (2h and 6h), and culture stage (day 4 and day 10). Tukey post-hoc pairwise comparisons showed significant differences as indicated by asterisks above corresponding bars (P ≤0.025).
Assessment of MRF detection and BrdU incorporation within Pax7+ cells revealed very little, if any, changes in MRF distribution in early culture stage (~95% of all myogenic cells expressed Pax7). The distribution of Myf5+ cells also did not change at either stage (compare Tables 1-2). However, the distribution of MyoD+ and myogenin+ cells at late stage was altered; over 30% of the total myogenic cells were excluded from the evaluation as they were within the Pax7− population. The respective evaluated percentages at late stage for MyoD+ cells at 2 and 6 hour BrdU pulse were: ~22 and 25 (BrdU+/Pax7+/MyoD+) and ~73 and 68 (BrdU−/Pax7+/MyoD+). The frequency of the BrdU+/Pax7+/MyoD+ population was approximately half of the corresponding population at the early stage (Table 2). The respective evaluated percentages at late stage for myogenin+ cells at 2 and 6 hour BrdU pulse were: ~2 and 3 (BrdU+/Pax7+/myogenin+) and ~15 and 19 (BrdU−/Pax7+/myogenin+). These data indicate that the population of proliferating myogenin+ cells was far reduced compared to that of MyoD+ cells. In comparison to MyoD and myogenin populations, the respective percentages at late stage for Myf5+ cells at 2 and 6 hour BrdU pulse were: ~9 and 9 (BrdU+/Pax7+/Myf5+) and ~30 and 23 (BrdU−/Pax7+/Myf5+). Thus, the frequency of BrdU+/Myf5+ and BrdU−/Myf5+ populations is lower than that of the corresponding MyoD populations and higher than myogenin+ populations observed at both stages.
The data in Table 2 indicate that Pax7 and MyoD are essentially coexpressed throughout S phase at both early and late cultures stages (~92-95% of Pax7+ cells) with a small residual population (~5-7%) that expressed Pax7 alone. The reduced frequency of Myf5+ cells compared to MyoD+ cells at both early and late stages (regardless of BrdU pulse duration) indicates that Myf5 is present during a narrower period compared to MyoD. The collective data further suggest that Myf5 may be expressed during the beginning of S phase, but then declines as the cell cycle proceeds while MyoD presence persists. The population of BrdU+/Myf5+ cells was reduced in late stage compared to early stage cultures, and the longer BrdU pulse results in a decline in percent values of BrdU+/Myf5+ cells in early stage, but not in late stage (Table 2). The BrdU+/MyoD+ population was also similarly reduced by ~50% between early and late stages, and was altered very little with the longer BrdU pulse in late stage cultures (Table 2).
DISCUSSION
Our analysis of early and late stage cultures with BrdU incorporation revealed novel insights into MRF detection patterns as myoblasts proceeded through S phase. The early stage corresponds with a highly proliferative phase when differentiated myoblasts are rare (Fig. 5A), and the late stage represents a balance between active differentiation and proliferation (Fig. 5A, B). Hence, late stage cultures may closely resemble the phase of posthatch growth where proliferation and fusion occurs side by side to enlarge myofibers (Halevy et al., 2004; Allouh et al., 2008).
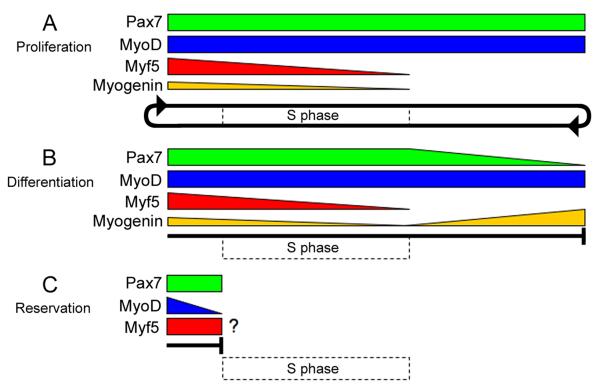
Figure 5.
A model depicting Pax7 and MRF detection profiles at S phase during various stages of posthatch myogenesis. Active proliferating cells (in early culture stage) maintain Pax7 and MyoD while Myf5 and myogenin levels decrease as they progress through S phase; sustained Pax7 and MyoD levels allow myoblasts to continue into a subsequent round of cell division (circular arrow) (A). The decision to differentiate involves decreased Pax7 and Myf5 levels, maintenance of MyoD, and induction of myogenin beyond S phase (or at least beyond our long BrdU pulse); cells will differentiate when myogenin levels sufficiently increase and Pax7 levels decrease as they exit the cell cycle outside of S phase as indicated by the block line (B). Pax7+/MyoD− reserve cells may emerge after proliferation when MyoD levels decrease within Pax7+/MyoD+ myoblasts to enter quiescence as shown by the block line; the question mark denotes potential Myf5 levels in myoblasts destined for reservation (C). Dotted-line rectangle depicts the interval of S phase that spans the BrdU pulses used at early and late culture stages.
The temporal presence of Myf5 during S phase in Pax7+ myoblasts at the early culture stage, and its absence in differentiated cells reinforces the notion that Myf5 has a unique role apart from MyoD that is linked to cell cycle kinetics (Fig. 5A-C). A previous study showed that Myf5 and MyoD levels oscillated at different phases of the cell cycle in mouse myoblasts where Myf5 was highest in quiescent, non-differentiated cells (i.e., G0) (Kitzmann et al., 1998; Kitzmann and Fernandez, 2001). Other studies demonstrated that phosphorylated Myf5 was degraded between mid S to mitosis while MyoD levels were maintained through mitosis and degraded at late G1 (Lindon et al., 1998; Tintignac et al., 2000; Batonnet-Pichon et al., 2006). Furthermore, by blocking Myf5 degradation, the passage of cells through cell division was perturbed (Lindon et al., 2000; Doucet et al., 2005). Thus, decreased levels of Myf5 during S phase may be regulated by proteolysis for potential entry into differentiation. Myf5 levels may also be highest before S phase to prevent continuation of myoblasts through the cell cycle and direct myoblasts toward reservation and quiescence (Fig. 5A-C). A minor Pax7+/MyoD− population at late culture stage may also represent the cells entering reservation where MyoD levels have decreased in late G1 (Fig. 5C) (Tintignac et al., 2000; Halevy et al., 2004; Zammit et al., 2004; Perez-Ruiz et al., 2008). However, it remains unclear if the Pax7+/MyoD− cells also express Myf5; Myf5 was detected in a proposed reserve pool in primary cultures, although Pax7 was not examined (Friday and Pavlath, 2001). A recent finding that Pax7 directly regulates Myf5 expression is also in accordance with our detection of Myf5 protein strictly within Pax7+ cells (McKinnell et al., 2008).
The evaluation of BrdU±/Pax7±/MRF± populations with regard to the length of BrdU incubation period also revealed a population of myoblasts actively entering differentiation at late culture stage. A relatively small change with pulse duration at late compared to early stage cultures in the BrdU−/Pax7+/MyoD+ population suggests that many of the Pax7+/MyoD+ cells at late stage are not entering S phase, but may be preparing to differentiate as verified by the increase in the BrdU−/Pax7−/MyoD+ population with longer pulse (compare Tables 1-2). Moreover, the increase in BrdU−/Pax7−/MyoD+ cells with longer pulse suggests that this population must also be myogenin− as the BrdU−/Pax7−/myogenin+ population was relatively unaltered with longer pulse (Table 1). Therefore, a transitional state (Pax7+/MyoD+ to Pax7−/MyoD+) may exist during differentiation in which myogenin upregulation in Pax7−/MyoD+ cells occurs beyond our 6h BrdU pulse. This demonstrates that MyoD may set the pace of differentiation by inducing myogenin expression as Pax7 levels decrease (Fig. 5B). The delayed appearance of myogenin+ cells in myogenic cultures from MyoD−/− mice promotes this concept (Yablonka-Reuveni et al., 1999).
Our model supports that maintenance of Pax7 is required for cell cycle continuity. Pax7 was detected in the cytoplasm of individual mouse myoblasts even during mitosis (Shinin et al., 2006). Pax7-deficient satellite cells also do not appear to appropriately progress in the cell cycle, and may alternatively initiate apoptosis (Relaix et al., 2006). The presence of a minor BrdU+/Pax7+/myogenin+ population during early stage that decreased with pulse time suggests that myogenin is incompatible with S phase, and the elimination of Pax7 outside of S phase is just as critical to complete differentiation as is the induction of myogenin (Table 1-2, Fig. 5B). Notably, sustained Pax7 expression with upregulation of myogenin in Rb-deficient myoblasts prevented their terminal differentiation (Huh et al., 2004). Thus, final cell cycle withdrawal may not be complete until the cell cycle inhibitor, p21, is present at a sufficient level (Andres and Walsh, 1996). In addition, the potential negative reciprocal inhibition between Pax7 and myogenin also suggests that their individual levels may influence fate decisions between renewal and differentiation (Olguin et al., 2007).
Overall, the unique patterns of Pax7 and MRF detection within myoblasts at S phase may reflect continuity of proliferation, terminal differentiation, or reservation during posthatch myogenesis. The early stage sustains proliferation by maintaining a continuous supply of active, Pax7+/MyoD+ progeny of satellite cells for enhancing muscle growth. Alternatively, the late stage may provide an environment where decisions are made as differentiation appeared in balance with proliferation. Discerning the specific function of Myf5 in the cell cycle during myogenesis may require genetic engineering in mice such as the expression of proteolysis-resistant Myf5 or MyoD in parallel null backgrounds. Further elucidation of the relationships between cell cycle kinetics and the MRFs that control myoblast proliferation, entry into differentiation, or reservation may provide valuable insights for cell-based therapies of muscle disorders and allow manipulation of growth for agricultural purposes.
EXPERIMENTAL PROCEDURES
Animal Procedure and Cell Cultures
Posthatch chickens (White Leghorn, 9-day old) were kindly provided by Drs. O. Bermingham-McDonogh and E. Rubel (University of Washington). Animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Washington. The pectoralis muscle was removed and used for preparation of primary myogenic cultures using Pronase digestion according to our published procedure (Halevy et al., 2004). Cells were cultured on gelatin coated 35-mm tissue culture plates using MEM-based medium containing 10% horse serum and 5% chicken embryo extract (Yablonka-Reuveni and Paterson, 2001; Shefer and Yablonka-Reuveni, 2005). Cultures were initiated at a density of 5×104 and 1×103 per plate for early (day 4) and late (day 10) stages, respectively; the lower cell density ensured that cultures were not overgrown at the later time points, to facilitate cell counts of immunostained cells. Parallel cultures were seeded with each density from 3 independent animals.
BrdU labeling
BrdU stock (10 mg/mL in PBS) was diluted into fresh medium at a final concentration of 5 μM and incubated with cultures for either 2 or 6 hours. Cultures were then rinsed with MEM and fixed in 4% paraformaldehyde/phosphate buffered saline/1% sucrose solution (Shefer and Yablonka-Reuveni, 2005). Fixed cultures were washed in Tris buffered saline (TBS, pH 7.4) and permeabilized for 10 min in 0.5% Triton-X-100 in TBS. Plates were then incubated with 4N HCl for 7 min and rinsed with TBS for 5 min each. Cultures were blocked overnight in 1% normal goat serum/TBS at 4°C prior to immunostaining.
Immunofluorescence and Antibodies
Cultures exposed to BrdU and fixed as described above were reacted with primary and secondary antibodies according to our routine immunofluorescence labeling protocol (Day et al., 2007). The following primary antibodies were used: rabbit polyclonal antibodies against chicken Myf5 (1:800), MyoD (1:400) and myogenin (1:1500), (developed and characterized as detailed in Yablonka-Reuveni and Paterson, 2001); mouse monoclonal antibody against chicken Pax7 (ascites fluid, 1:2000, Developmental Studies Hybridoma Bank, University of Iowa); rat monoclonal antibody against BrdU (Abcam, 1:2000). Secondary antibodies used were all AlexaFluor conjugated and produced in goat (Invitrogen, 1:1000 dilution): anti-mouse IgG1 – 488 (for detection of Pax7), anti-rat IgG – 350 (for detection of BrdU incorporation), anti-rabbit IgG – 568 (for detecting MRFs). For amplified detection of blue fluorescence signal (BrdU labeling), primary and secondary antibody staining was performed twice. Otherwise, all immunostaining procedures were performed as in previous publications (Halevy et al., 2004), except DAPI staining was omitted for use of the blue channel for the triple immunostaining technique. Clones were double immunostained for Pax7 and Myf5 with DAPI staining (BrdU labeling was omitted). Controls for immunolabeling specificity of the nuclei included immunostaining with omission of primary antibodies, staining with each of the primary antibodies alone followed by all secondary antibodies, staining with each of the primary antibodies alone followed by the reciprocal secondary antibodies, and immunostaining with the multiple primary antibodies followed by each of the secondary antibodies alone.
Microscopy and Imaging
An inverted fluorescent microscope (Nikon eclipse, TE2000-S) was used to acquire images with a Qimaging Retiga 1300i Fast 1394 monochrome CCD camera. The CCD camera drive and color acquisition were controlled by MetaVue Imaging System (Universal Imaging Corporation). Images were acquired using a 40× objective and cells were counted from these images as previously described (Halevy et al., 2004; Shefer et al., 2006). In brief, 20 arbitrary microscopic fields were counted from each triple immunostained plate and values were pooled from parallel duplicate plates per each time point in each experiment. Composites of photomicrographs were assembled using Adobe Photoshop software.
Data Analysis
Proliferation indices (percent BrdU+ cells within Pax7+ cells) were subjected to an arcsin of square root-transformation for percentages and ratios to meet the criteria of the ANOVA method before data were analyzed by two-way ANOVA. P values less than 0.05 were considered significant.
ACKNOWLEDGEMENTS
We thank Daniel Van de Mark for excellent technical assistance. We are additionally grateful to Dr. Gabi Shefer for her valuable input to this manuscript. This work was supported by grants from the USDA Cooperative State Research, Education and Extension Service (NRI, 2003-35206-12843, Z.Y.R), the National Institute on Aging (AG021566, Z.Y.R), and the Genetic Approaches to Aging Training Grant program (K.D.). Z.Y.R. acknowledges additional support during the course of this study from the National Institute on Aging (AG13797).
Grant sponsor: Cooperative State Research, Education and Extension Service-United States Department of Agriculture, National Research Initiative; Grant number: 2003-35206-12843 (Z.Y.R.); Grant sponsor: National Institutes of Health; Grant numbers; AG 21566; AG13798; (Z.Y.R.); Genetic Approaches to Aging Training Grant program (K.D.)
REFERENCES
- Allen RE, Merkel RA, Young RB. Cellular aspects of muscle growth: myogenic cell proliferation. J Anim Sci. 1979;49:115–127. doi: 10.2527/jas1979.491115x. [DOI] [PubMed] [Google Scholar]
- Allouh MZ, Yablonka-Reuveni Z, Rosser BW. Pax7 reveals a greater frequency and concentration of satellite cells at the ends of growing skeletal muscle fibers. J Histochem Cytochem. 2008;56:77–87. doi: 10.1369/jhc.7A7301.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batonnet-Pichon S, Tintignac LJ, Castro A, Sirri V, Leibovitch MP, Lorca T, Leibovitch SA. MyoD undergoes a distinct G2/M-specific regulation in muscle cells. Exp Cell Res. 2006;312:3999–4010. doi: 10.1016/j.yexcr.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G, Molinaro M. Cell heterogeneity in the myogenic lineage. Curr Top Dev Biol. 1987;23:185–208. doi: 10.1016/s0070-2153(08)60625-0. [DOI] [PubMed] [Google Scholar]
- Day K, Shefer G, Richardson JB, Enikolopov G, Yablonka-Reuveni Z. Nestin-GFP reporter expression defines the quiescent state of skeletal muscle satellite cells. Dev Biol. 2007;304:246–259. doi: 10.1016/j.ydbio.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C, Gutierrez GJ, Lindon C, Lorca T, Lledo G, Pinset C, Coux O. Multiple phosphorylation events control mitotic degradation of the muscle transcription factor Myf5. BMC Biochem. 2005;6:27. doi: 10.1186/1471-2091-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton VR, Roy RR. Regulation of skeletal muscle fiber size, shape and function. J Biomech. 1991;24(Suppl 1):123–133. doi: 10.1016/0021-9290(91)90383-x. [DOI] [PubMed] [Google Scholar]
- Friday BB, Pavlath GK. A calcineurin- and NFAT-dependent pathway regulates Myf5 gene expression in skeletal muscle reserve cells. J Cell Sci. 2001;114:303–310. doi: 10.1242/jcs.114.2.303. [DOI] [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Grounds MD. Muscle regeneration: molecular aspects and therapeutic implications. Curr Opin Neurol. 1999;12:535–543. doi: 10.1097/00019052-199910000-00007. [DOI] [PubMed] [Google Scholar]
- Grounds MD, Yablonka-Reuveni Z. Molecular and cell biology of skeletal muscle regeneration. Mol Cell Biol Hum Dis Ser. 1993;3:210–256. doi: 10.1007/978-94-011-1528-5_9. [DOI] [PubMed] [Google Scholar]
- Halevy O, Piestun Y, Allouh MZ, Rosser BW, Rinkevich Y, Reshef R, Rozenboim I, Wleklinski-Lee M, Yablonka-Reuveni Z. Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev Dyn. 2004;231:489–502. doi: 10.1002/dvdy.20151. [DOI] [PubMed] [Google Scholar]
- Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J Cell Biol. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell Mol Life Sci. 2001;58:571–579. doi: 10.1007/PL00000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon C, Albagli O, Domeyne P, Montarras D, Pinset C. Constitutive instability of muscle regulatory factor Myf5 is distinct from its mitosis-specific disappearance, which requires a D-box-like motif overlapping the basic domain. Mol Cell Biol. 2000;20:8923–8932. doi: 10.1128/mcb.20.23.8923-8932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon C, Montarras D, Pinset C. Cell cycle-regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J Cell Biol. 1998;140:111–118. doi: 10.1083/jcb.140.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170:421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- Mozdziak PE, Schultz E, Cassens RG. Myonuclear accretion is a major determinant of avian skeletal muscle growth. Am J Physiol. 1997;272:C565–571. doi: 10.1152/ajpcell.1997.272.2.C565. [DOI] [PubMed] [Google Scholar]
- Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177:769–779. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. Embo J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ruiz A, Ono Y, Gnocchi VF, Zammit PS. beta-Catenin promotes self-renewal of skeletal-muscle satellite cells. J Cell Sci. 2008;121:1373–1382. doi: 10.1242/jcs.024885. [DOI] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Schultz E, Chamberlain C, McCormick KM, Mozdziak PE. Satellite cells express distinct patterns of myogenic proteins in immature skeletal muscle. Dev Dyn. 2006;235:3230–3239. doi: 10.1002/dvdy.20976. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev Biol. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol Biol. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- Smith TH, Block NE, Rhodes SJ, Konieczny SF, Miller JB. A unique pattern of expression of the four muscle regulatory factor proteins distinguishes somitic from embryonic, fetal and newborn mouse myogenic cells. Development. 1993;117:1125–1133. doi: 10.1242/dev.117.3.1125. [DOI] [PubMed] [Google Scholar]
- Smith CK, 2nd, Janney MJ, Allen RE. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J Cell Physiol. 1994;159:379–385. doi: 10.1002/jcp.1041590222. [DOI] [PubMed] [Google Scholar]
- Stockdale FE. Myogenic cell lineages. Dev Biol. 1992;154:284–298. doi: 10.1016/0012-1606(92)90068-r. [DOI] [PubMed] [Google Scholar]
- Tintignac LA, Leibovitch MP, Kitzmann M, Fernandez A, Ducommun B, Meijer L, Leibovitch SA. Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp Cell Res. 2000;259:300–307. doi: 10.1006/excr.2000.4973. [DOI] [PubMed] [Google Scholar]
- Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z. Development and postnatal regulation of adult myoblasts. Microsc Res Tech. 1995;30:366–380. doi: 10.1002/jemt.1070300504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rivera AJ. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev Biol. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Day K, Vine A, Shefer G. Defining the transcriptional signature of skeletal muscle stem cells. J Anim Sci. 2008;86:E207–216. doi: 10.2527/jas.2007-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Paterson BM. MyoD and myogenin expression patterns in cultures of fetal and adult chicken myoblasts. J Histochem Cytochem. 2001;49:455–462. doi: 10.1177/002215540104900405. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006;54:1177–1191. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]