Abstract
In all eukaryotes, the ligation of newly synthesized DNA, also known as Okazaki fragments, is catalyzed by DNA ligase I1. An individual with a DNA ligase I deficiency exhibited growth retardation, sunlight sensitivity and severe immunosuppression2, likely due to accumulation of DNA damage. Surprisingly, not much is known about the DNA damage response (DDR) in DNA ligase I-deficient cells. Because DNA replication and DDR pathways are highly conserved in eukaryotes, we utilized Saccharomyces cerevisiae as a model system to address this question. We uncovered a novel pathway, which facilitates ubiquitination of lysine 107 of proliferating cell nuclear antigen (PCNA). Unlike ubiquitination at lysine 164 of PCNA in response to UV irradiation, which triggers translesion synthesis3, modification of lysine 107 is not dependent on the ubiquitin conjugating enzyme (E2) Rad64 nor the ubiquitin ligase (E3) Rad185, but requires the E2 variant Mms26 in conjunction with Ubc47 and the E3 Rad58,9. Surprisingly, DNA ligase I-deficient cdc9-1 cells that carry a PCNAK107R mutation are inviable, because they cannot activate a robust DDR. Furthermore, we show that ubiquitination of PCNA in response to DNA ligase I-deficiency is conserved in humans, yet the lysine that mediates this modification remains to be determined. We propose that PCNA ubiquitination provides a “DNA damage code” that allows cells to categorize different types of defects that arise during DNA replication.
Previous studies have suggested that DNA ligase I-deficient cdc9 mutants arrest in G2 phase after completing DNA synthesis10,11 because of the accumulation of nicked DNA12. These observations implied that S phase proceeds normally despite single-stranded breaks in nascent DNA, suggesting that cells may not be able to efficiently sense this type of DNA damage during S phase. To reinvestigate whether DNA ligase I is required for S phase progression, we have analyzed three different temperature sensitive alleles of CDC9 (Supplementary Information, Fig. S1a and b online, and Supplementary Information, Fig. S2a and b online). One of these alleles is a thermo labile degron mutant13 (cdc9-td in Fig. 1a). To ensure that ligase activity was sufficiently inactivated in these cells, we performed replication initiation point mapping and did not detect any measurable ligation of Okazaki fragments over the yeast origin ARS1 (Supplementary Information, Fig. S2c online) as previously demonstrated for the cdc9-1 allele14. Whereas DNA ligase I was not required for entry into S phase (Fig. 1b), we found it to be necessary to release from a hydroxyurea (HU) block (Fig. 1c). This was also true for two additional temperature sensitive alleles, cdc9-1 and cdc9-215 (Fig. 1d). This result was surprising as earlier reports suggested that DNA ligase I-deficient cells could complete DNA synthesis without joining Okazaki fragments to each other10,11. As expected, cell cycle delay in S phase was dependent on the DDR gene RAD910,11 because cdc9-1 rad9Δ mutants progressed farther than cdc9-1 cells (Supplementary Information, Fig. S3a online). Importantly, however, the mediator of the replication checkpoint (Mrc)1 appeared to contribute equally to Rad9 (Supplementary Information, Fig. 3a and b online). Mrc1 has been shown to have two roles, one in DNA replication and one in activating Rad53 after replication fork stalling, which results in exposure of single-stranded DNA16. Rad53 is a downstream target of the mitotic entry checkpoint gene MEC1, a homolog of the ATM/ATR checkpoint kinases in humans17. Complementation of cdc9-1 mrc1Δ double mutants with the S phase checkpoint-deficient mrc1AQ allele16 failed to induce cell cycle arrest (Supplementary Information, Fig. S3c online), suggesting that the S phase checkpoint and not the replication function of Mrc1 is important to delay S phase progression. Furthermore, the finding that both Mrc1 and Rad9 are activated in cdc9-1 cells at the non-permissive temperature implies that the DNA substrate recognized contains single stranded DNA at stalled replication forks as well as physical damage, which may have arisen from the lack of Okazaki fragment ligation12. Besides triggering a checkpoint response, certain types of DNA damage at replication forks have also been shown to cause ubiquitination of PCNA18. PCNA can be either mono-ubiquitinated or poly-ubiquitinated. Mono-ubiquitination of PCNA triggers the error-prone repair pathway through translesion polymerases, whereas PCNA poly-ubiquitination is needed for error-free repair3,19,20. Mono-ubiquitination depends on Rad6 and Rad18 and is a pre-requisite for poly-ubiquitination, which in turn is mediated by the ubiquitin conjugating complex Ubc13/Mms2 and Rad53. Interestingly, ubiquitin is linked through lysine 63 in these poly-ubiquitin chains3,21. To explore whether loss of DNA ligase I leads to PCNA ubiquitination, we examined the status of PCNA in whole cell extracts. We utilized an antibody specific for yeast PCNA22, which displays multiple non-specific bands in undiluted extracts (Supplementary Information, Fig. S4 online), but produces clean immunoblots with diluted extracts (Fig. 2a). Both cdc9-1 and cdc9-2 mutants exhibited a modified form of PCNA of approximately 39 kDa when shifted to the non-permissive temperature (Fig. 2a). Co-immunoprecipitation (Co-IP) experiments with strains that expressed Myc-tagged ubiquitin23,24 identified this 39 kDa band as ubiquitinated PCNA, which we did not observe when we mixed cell extracts only with beads (Fig. 2b). Curiously, we observed a non-specific band slightly above the 49 kDa marker, which was especially obvious in extracts from cdc9-1 cells. Unfortunately, our co-IP studies did not allow us to draw any conclusions about the nature of this band, although we cannot exclude that it represents poly-ubiquitinated PCNA sticking non-specifically to the beads. Therefore, we overexpressed different ubiquitin mutants, including a G75,76A double mutant, specifically designed to interfere with mono-ubiquitination. Another mutant carried substitutions in all seven lysines (Ub-KO) and was thus defective in forming poly-ubiquitin chains. Analysis of trichloroacetic acid-precipitated whole cell extracts confirmed that the 39 kDa form of PCNA represented mono-ubiquitinated protein (Fig. 2c). Furthermore, two forms of PCNA that represented poly-ubiquitinated protein (at ~52- and 76 kDa), because they disappeared in cells overexpressing the Ub-KO mutant, were visible (Fig. 2d). A distinct ladder of ubiquitinated PCNA with similar molecular weight distribution has also been reported by van der Kemp et al.25. Surprisingly, poly-ubiquitin chains on PCNA were not linked through K6321, as they are in response to other forms of replication stress3, but rather through K29 (Fig. 2d). Based on this result, we predicted that at least some of the genes that are known to play a role in K63 linked poly-ubiquitination (namely RAD6, RAD18, MMS2, UBC13, RAD5)3 are dispensable for the ubiquitination of PCNA in cdc9-1 mutants. Indeed, whereas MMS2 and RAD5 were required for mono- as well as poly-ubiquitination, UBC13 and RAD18 were not (Fig. 3a and Supplementary Information, Fig. S5a online). Moreover, deletion of RAD6 did not have any effect (Supplementary Information, Fig. S5b online), suggesting that PCNA ubiquitination in response to Okazaki fragment ligation defects differs from that triggered by other types of DNA damage. Our results also indicated that Mms2 likely cooperates with other ubiquitin conjugating enzymes besides Ubc1321. To determine Mms2's potential partner in this reaction, we took advantage of the fact that lysine 29 linkages are catalyzed by UbcH5A in mammalian cells26. UbcH5A's homolog in yeast is Ubc4, which is 93% identical to Ubc57. Unlike Ubc4, which is highly expressed in cycling cells, steady state levels of Ubc5 are low and are upregulated in stationary phase7. Ubc4 and 5 have previously been implicated in the degradation of unfolded proteins in yeast7, but not in PCNA ubiquitination. When we deleted UBC4 from cdc9-1 mutants and performed a temperature shift experiment, PCNA ubiquitination was drastically reduced, whereas deletion of UBC5 had no effect (Supplementary Information, Fig. S5c online). Because we were unable to generate an ubc4Δ ubc5Δ double mutant in the cdc9-1 background, we attached a nuclear export sequence onto UBC4 in a cdc9-1 ubc5Δ strain. Again, this resulted in a 95% reduction of PCNA mono-ubiquitination (Fig. 3b). These results suggested to us that the nuclear fraction of Ubc4 cooperates with Mms2 and Rad5 to ubiquitinate PCNA in DNA ligase I-deficient cells.
Figure 1.
DNA ligase I is required for S phase progression. (a) Asynchronous cultures of ABy010 (GAL-UBR1 CDC9) and ABy008 (GAL-UBR1 cdc9-td) were induced with galactose at 28°C for 30 min and subsequently shifted to 37°C. (b) Strains were arrested in G1 phase at 28°C and shifted to 37°C in 2% galactose and α-factor. After 90 min, cells were released from G1 phase either at 37°C in galactose or at 28°C in glucose. (c) Strains were arrested in G1 and released into S phase at 28°C in the presence of HU. Once arrested in S phase, cultures were shifted to 37°C in the presence of 2% galactose and HU. After 90 min, cells were transferred into nocodazole either at 37°C or 28°C. (d) Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1), and SSL613 (cdc9-2) were shifted to 35°C. In a-d, DNA content was monitored by flow cytometry and the vertical line indicates a 2C DNA content.
Figure 2.
S. cerevisiae PCNA is mono-ubiquitinated in cdc9 mutants. (a) Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1), and SSL613 (cdc9-2) were shifted to 35°C for 3 h. Total protein was TCA-precipitated, diluted and probed with a yeast specific PCNA antibody (S871). (b) Cells were shifted to 35°C for 3 h in the presence of copper sulfate to induce expression of Myc-tagged ubiquitin. Whole cell extracts were immunoprecipitated with an anti-Myc antibody and PCNA was detected with anti-PCNA antibody (S871). The right panel shows the beads control with or without the cell lysate. 5% of the input was loaded. (c) PCNA is mono-ubiquitinated in cdc9 mutants. Wild-type or mutant ubiquitin expression was induced in asynchronously growing cultures. Total protein was TCA-precipitated and diluted. Unmodified and mono-ubiquitinated PCNA was detected by PCNA antibody (S871). (d) PCNA poly-ubiquitination is linked through lysine 29 in cdc9 mutants. Undiluted TCA-precipitated protein samples from c were probed with anti-PCNA antibody (S871) to detect poly-ubiquitinated PCNA. In b, d, asterisks indicate non-specific bands. In d, filled circles (right side of the band) indicate poly-ubiquitinated forms of PCNA. In a, c, d, α-tubulin served as a loading control.
Figure 3.
PCNA mono-ubiquitination in cdc9 mutants is mediated by Mms2, Rad5 and Ubc4 but not Ubc13. (a) Cultures were grown asynchronously and shifted to 35°C for 3 h. (b) Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1) and ABy579 (cdc9-1 ubc5Δ UBC4-NES-3HA) were grown at 25°C and shifted to 35°C for 3 h. In a-b, unmodified and mono-ubiquitinated PCNA was detected using PCNA antibody (S871). α-tubulin served as a loading control.
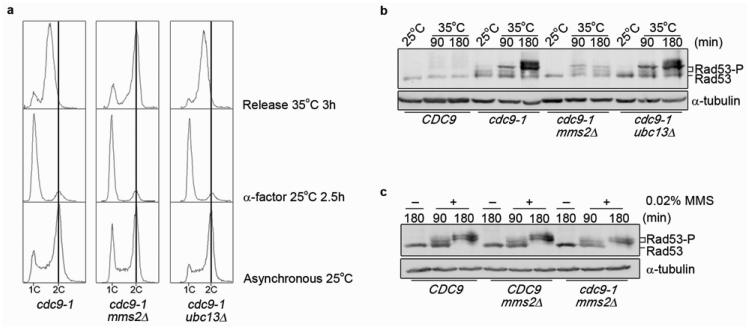
To further discern the function of PCNA ubiquitination in DNA ligase I-deficient yeast, we monitored cell cycle progression upon release from G1. Specifically, we compared cdc9-1 cells to cdc9-1 mms2Δ and cdc9-1 ubc13Δ double mutants. Consistent with our finding that UBC13 is not involved in the ubiquitination of PCNA (Fig. 3a), cdc9-1 ubc13Δ cells arrested in S phase similar to the cdc9-1 strain (Fig. 4a). In contrast, cdc9-1 mms2Δ mutants that fail to ubiquitinate PCNA at the non-permissive temperature (Fig. 3a), readily progressed through S phase (Fig. 4a). The observed lack of PCNA ubiquitination and S phase arrest went hand-in-hand with the inability to phosphorylate Rad53 (Fig. 4b). Importantly, this was not due to a general defect in checkpoint activation, as we detected Rad53 phosphorylation in all three strains after exposure to MMS (Fig. 4c). To address whether mono-ubiquitination is sufficient for Rad53 activation, we disrupted poly-ubiquitination by overexpressing a ubiquitin mutant in which lysine 29 was substituted with arginine. This did not significantly affect Rad53 activation (Supplementary Information, Fig. S6a and b online). These results argue that mono- rather than poly-ubiquitination of PCNA is a prerequisite for checkpoint activation of DNA ligase I-deficient yeast cells. Next, we set out to determine which lysine in PCNA was ubiquitinated. PCNA has a total of 18 lysines, nine of which are exposed and easily accessible27. We mutated these nine lysines at positions 107, 108, 117, 127, 164, 168, 183, 242 and 253. Wild-type or mutated forms of PCNA were introduced on a plasmid before the endogenous copy was deleted. When we monitored mono-ubiquitination of PCNA in cdc9-1 cells after they were shifted to the non-permissive temperature, the only mutant that lacked the characteristic 39 kDa band was pol30K107R (Fig. 5a). However, further examination revealed that this particular mutant had a second site suppressor mutation, which resulted in elevated DNA ligase I protein levels that were comparable to those in wild-type cells (Supplementary Information, Fig. S7a online). Moreover, the strain was no longer temperature sensitive (Supplementary Information, Fig. S7b online). We attempted several times to make the double mutant but these efforts remained unsuccessful, suggesting that the mutations were synthetically lethal. To prove this point, we performed a plasmid shuffle assay. We introduced pol30K107R into the chromosomal copy of PCNA in wild-type and cdc9-1 mutants, and expressed POL30 from a plasmid. Cells were forced to abandon the plasmid by re-streaking them onto 5-fluoroorotic acid-containing medium. Whereas CDC9 pol30K107R cells were viable and formed colonies, cdc9-1 pol30K107R mutants did not, indicating that K107 of PCNA becomes essential in this background (Supplementary Information, Fig. S8 online). This was further confirmed by tetrad analysis (data not shown), which seemingly yielded viable double mutants, but all of these strains had elevated DNA ligase I levels due to a second site suppressor mutation (data not shown). However, we were able to isolate a single double mutant in which Cdc9 levels were only slightly increased compared to cdc9-1 cells (Supplementary Information, Fig. S9a online). We designated this mutant cdc9-1* pol30K107R. In fact, the double mutant retained its temperature sensitivity (Supplementary Information, Fig. S9b online) and failed to ubiquitinate PCNA under non-permissive conditions (Fig. 5b). Importantly, PCNA ubiquitination in response to MMS was still functional (Fig. 5c). At this point, it is worthwhile to point out that we detected ubiquitinated and unmodified PCNA at an apparent ratio of approximately 1:1. However, it is not clear that the antibody binds these two forms of PCNA with equal affinity, and therefore it is difficult to make quantitative assessments. In addition, we take the fact that we could readily generate cdc9-1 mms2Δ and cdc9-1 rad5Δ, but not cdc9-1 pol30K107R double mutants as an indication that a redundant ubiquitination pathway might exist that targets K107 with low efficiency, thereby ensuring cell survival. To confirm that K107 is the target of PCNA ubiquitination in DNA ligase I-deficient cells, we introduced the pol30K107 mutation into cdc9-td cells. As expected, cdc9-td pol30K107R double mutants retained viability, because CDC9 is expressed from an inducible copper promoter13. Under non-permissive conditions, the ratio of mono-ubiquitinated to unmodified PCNA in cdc9-td pol30K107R cells resembled that of the DNA ligase I-proficient control strain, which carries the same mutation in POL30 (Fig. 5d). The same was true for poly-ubiquitinated PCNA (Fig. 5e). Furthermore, cdc9-td pol30K107R cells failed to exhibit robust Rad53 phosphorylation in the absence of DNA ligase I, whereas they displayed Rad53 activation in response to MMS (Fig. 5f). From these results we conclude that ubiquitination of PCNA at lysine 107 is a prerequisite for checkpoint activation of DNA ligase I-deficient yeast cells. This is in stark contrast to the previously described ubiquitination of PCNA at lysine 164 in response to UV irradiation or DNA alkylating agents3. Although exposure to UV light or DNA alkylating agents triggers both Rad53 phosphorylation and ubiquitination of PCNA at lysine 164, these processes are thought to occur independently and belong to separate genetic pathways28. We demonstrate that this cannot be generalized, but that PCNA ubiquitination in response to DNA ligase I deficiency must occur before Rad53 can be fully activated. It is worth noting that K107 is conserved in S. pombe and C. elegans, but higher eukaryotes have a conserved lysine residue at position 110, raising the possibility that this PCNA ubiquitination pathway is not only specific for S.cerevisiae (Supplementary Information, Fig. S10 online). To test whether human cells depleted for DNA ligase I exhibit PCNA modifications, we generated several U2OS cell lines expressing short hairpin RNAs (shRNAs) that interfere with DNA ligase I expression (Supplementary Information, Fig. S11 online). In the chromatin-bound protein fractions of these cells, we detected a slower migrating form of PCNA with the same mobility of mono-ubiquitinated PCNA induced by UV irradiation29 (Fig. 5g, upper panel and Supplementary Information, Fig. S11 online). In contrast, cells expressing control shRNA behaved like untreated U2OS cells (Fig. 5g, upper panel). Importantly, when we analyzed one representative DNA ligase I-specific shRNA clone side by side with a PCNA- or an ubiquitin-specific antibody, both antibodies recognized bands at the exact same position, consistent with the notion that PCNA ubiquitination in response to DNA ligase I-deficiency is conserved in humans, although the lysine residue of PCNA that is ubiquitinated in these cells is still under investigation (Fig. 5g, upper and middle panel).
Figure 4.
MMS2 but not UBC13 is required for S phase checkpoint activation in cdc9 mutants. (a) Cells were arrested in G1 phase at 25°C and released at 35°C for 3 h. DNA content was monitored by flow cytometry. The vertical line indicates a 2C DNA content. (b) Cells were grown asynchronously at 25°C and then shifted to 35°C for the indicated time period. Total protein was TCA-precipitated and Rad53 was detected by an anti-Rad53 antibody. (c) Asynchronous cultures were split and shifted to the non-permissive temperature of 35°C in the presence of methyl methane sulfonate (MMS) for 90 and 180 min or left untreated for 180 min. Rad53 was detected as described in b. In b-c, α-tubulin served as a loading control.
Figure 5.
PCNA mono-ubiquitination occurs at lysine 107 in DNA ligase I mutants and is required for Rad53 activation. (a) PCNA lysine mutants in DNA ligase I deficient cells (cdc9-1) were grown asynchronously and then shifted to 35°C for 3 h. (b) Cultures of SSL204 (CDC9), ABy685 (CDC9 pol30-K107R), SSL612 (cdc9-1), ABy782 (cdc9-1* pol30-K107R) were grown asynchronously and then shifted to 30°C for 3 h. (c) Asynchronous cultures were grown at 25°C and shifted to 30°C in the presence of MMS for the indicated time. Cultures not treated with MMS served as negative controls. (d) Cultures were induced with galactose at 28°C for 30 min and subsequently shifted to 37°C for 3 h. In a-d, TCA-precipitated protein samples were diluted and PCNA was detected with anti-PCNA antibody (S871). (e) Undiluted TCA-precipitated samples from d were run on SDS-PAGE and poly-ubiquitinated PCNA was detected with anti-PCNA antibody (S871). (f) Cells were grown asynchronously at 28°C and then shifted to 37°C in the presence or absence of MMS for 3 h. Rad53 and Cdc9-td-HA were detected with anti-Rad53 and anti-HA antibodies, respectively. In b, c, e, f, α-tubulin served as a loading control. In e-f, the asterisks indicate non-specific bands. (g) U2OS cells were either untreated or treated with 60 J/m2 UV and harvested 2 h later along with control shRNA (shCON) and DNA Ligase I shRNA (shLIG1 A) cell lines. Chromatin fractions were prepared, fractionated on SDS-PAGE and analyzed with the indicated antibodies. Samples on the PCNA and ubiquitin blots were analyzed side by side. Unmodified PCNA served as a loading control for the PCNA blot and histone H3 served as a loading control for the ubiquitin blot.
In the course of our studies, we also explored rad27Δ and dna2-1 mutants in S. cerevisiae, because both Rad27 and Dna2 have been implicated in lagging strand synthesis30. However, these mutants did not ubiquitinate PCNA (Supplementary Information, Fig. S12 online). This suggested that PCNA ubiquitination at K107 might be specific for DNA ligase I-deficiency. In summary, whereas ubiquitination at lysine 164 is responsible for activation of translesion synthesis, and is thus a result of DNA damage in the template strand, ubiquitination of lysine 107 in DNA ligase I-deficient cells might provide a signal for DNA damage residing in freshly synthesized, nascent DNA. Most importantly, our study demonstrates that PCNA provides a DNA damage-specific code via the ubiquitination of different lysines.
Methods
Strains and plasmids
All strains are isogenic derivatives of W303-1a or SSL204. A complete list of strains can be found in Supplementary Information, Table 1 online. The W303-1a strain carries the rad5-535 mutation (http://wiki.yeastgenome.org/index.php/CommunityW303.html), whereas SSL204 derivatives (CDC9, cdc9-1, cdc9-2) harbor wild type RAD5, which was confirmed by sequencing. SSL204 exhibits severe temperature sensitivity at 37°C and therefore the temperature shift experiments were performed at 35°C. To construct the cdc9-td strain, the first 510 base pairs of CDC9 were inserted into pPW66R (a gift from J. F. X. Diffley) using HindIII restriction sites13. The resulting pPW66R-CDC9 plasmid was linearized with BclI and transformed into YKL83, which contains UBR1 under the control of the GAL1 promoter at the endogenous UBR1 locus31. To delete various genes (MRC1, RAD9, UBC4, UBC5, RAD6, RAD18, MMS2, UBC13, RAD5) in SSL204 and SSL612, standard one-step PCR gene replacement was used32 and correct integration was confirmed by sequencing. Two independent colonies of each strain were isolated.
The UCB4-NES-3HA fragment containing a nuclear export sequence (NES)33 and three hemagglutinin (HA) tags was generated by PCR. This was inserted into the pRS404 plasmid (a gift from D. Koepp) using KpnI and SacI restriction sites. The resulting integration plasmid was linearized using BglII and transformed into two independent cdc9-1 ubc5Δstrains. Correct integration was confirmed by sequencing and immunoblotting with an anti-HA (16B12, Covance) antibody.
For the MRC1 complementation experiments, pAO138 and pMRC1 (gifts from S. J. Elledge) were transformed into ABy287 (CDC9 mrc1Δ) and ABy293 (cdc9-1 mrc1Δ). pAO138 contains wild-type MRC1 expressed from its endogenous promoter and pMRC1 expresses a mrc1 mutant that has 17 putative SQ or TQ phosphorylation sites switched to AQ (mrc1AQ mutant) and is also expressed from its endogenous promoter16. pRS316 served as the empty vector control.
mrc1Δ rad9Δ cells were derived from a strain containing a high copy number plasmid expressing RNR1 from its endogenous promoter (a gift from D. J. Clarke) to maintain cell viability.
PCNA lysine mutants were generated using plasmid YCplac22-POL30 (a gift from S. Jentsch)3. YCplac22-POL30 expresses full length PCNA from its endogenous promoter. Lysine to arginine substitutions in PCNA were introduced at positions 107, 108, 117, 168, 183, 253 by QuikChange mutagenesis (Stratagene). Four other lysine to arginine mutations (K127R, K164R, K127/164R, K242R) used in this study were a gift from the Jentsch laboratory3. As described previously32, the endogenous POL30 gene was subsequently deleted by replacement with either a URA3 or LEU2 marker in SSL204 and YKL83 strains, respectively.
To determine the cell viability of the double mutant cdc9-1 pol30-K107R, a plasmid expressing POL30 from its endogenous promoter (pRS316-POL30, a gift from D. M. Livingston) was first transformed into SSL204 (CDC9) and cdc9-1. PCNA lysine K107R mutant was generated using plasmid pCH1572 (a gift from L. Prakash). The resulting Leu2.PCNA-K107R PCR product was subsequently transformed into strains containing pRS316-POL30 plasmid. PCNA-K107R mutation at the endogenous locus was confirmed by sequencing. Strains were streaked out onto SC-Ura plate and 5'-FOA (2 mg/ml) plates. All plates were incubated at 25°C for 2-3 days.
For immunoprecipitation of ubiquitinated PCNA, YEp105 plasmid, expressing a synthetic Myc-tagged ubiquitin gene from a copper-inducible promoter (a gift from M. Hochstrasser)23 was transformed into SSL204 (CDC9), SSL612 (cdc9-1), SSL613 (cdc9-2). For the expression of ubiquitin mutants carrying lysine to arginine substitutions at positions 6, 11, 27, 29, 33, 48, 63, YEp105 was altered by QuikChange mutagenesis. The C-terminal glycine to arginine mutations in ubiquitin at positions 75 and 76 were constructed in YEp105 by QuikChange mutagenesis.
Yeast culture conditions
Temperature shift experiments were carried out in YPD, unless stated otherwise. For a typical temperature shift experiment, cells were grown to mid-log phase (OD600 = 0.6) at 25°C and then shifted to the restrictive temperature of 35°C for 3 hours or as indicated.
The degron strain, cdc9-td, was grown overnight at 28°C in YP plus 2% raffinose and supplemented with 10 μM CuSO4 to induce CDC9 gene expression. Copper sulfate was omitted when cells were shifted to 37°C. To increase the efficiency of Cdc9-td degradation, Ubr1 was overexpressed from a galactose-inducible promoter in the presence of 2% galactose.
For all experiments in which PCNA or ubiquitin were expressed from a plasmid, two independent colonies of each strain were grown in synthetic complete medium lacking tryptophan. In ubiquitin overexpressing strains, ubiquitin was induced over the time period of three generations by addition of 0.1 mM CuSO4 at a low cell density (OD600 = 0.1) as described earlier23.
Co-Immunoprecipitation
Whole cell extracts from exponentially growing cells were prepared using glass beads as described earlier34. A cocktail of freshly prepared protease inhibitors (pepstatin, leupeptin, benzamidine and phenylmethyl sulfonyl fluoride) and N-ethylmaleimide were used to preserve ubiquitinated proteins by inhibiting deubiquitinating enzymes24. For immunoprecipitation, 4 μg of anti-Myc antibody (9E11, Thermo scientific) was added to the extract for 2 hours at 4°C. PCNA was detected by Western blotting with an anti-PCNA antibody (clone S871, a gift from Z. Zhang)22.
Protein Preparation and Western Blot Analysis
Total protein extracts were prepared from cycling yeast cultures using TCA precipitation35 (see Supplementary Information, Fig. S1b online) and proteins were detected by Western blot analysis. Cdc9-td-HA was detected with an anti-HA antibody (16B12, Covance), histone H3 was detected with an anti-histone H3 antibody (Abcam), endogenous Cdc9 was detected with an anti-Cdc9 antibody (gift from A. E. Tomkinson) and endogenous Rad53 was detected with an anti-Rad53 antibody (gift from J. F. X. Diffley). α-tubulin served as a loading control. In some experiments shown in the Supplementary Information, Ponceau S staining of the nitrocellulose membrane prior to blotting served as a loading control.
Detection of PCNA ubiquitination
Unmodified and mono-ubiquitinated PCNA were analyzed by Western blotting with an anti-yeast PCNA antibody (clone S871, a gift from Z. Zhang)22. To detect mono-ubiquitination with the polyclonal S871 antibody, we diluted WCEs 10-fold. Poly-ubiquitinated PCNA was detected with the same antibody in undiluted, TCA-precipitated whole cell extracts. Please note that this antibody was not able to detect the 76 kDa poly-ubiquitinated form of PCNA in immunoprecipitated samples (Fig. 2b).
Cell Synchrony, FACS Analysis, and Microscopy
To arrest cells in G1, alpha-factor was added to a final concentration of either 50 ng/ml (bar1Δ cells) or 15 μg/ml (BAR1 cells). Cells were blocked in S phase with the addition of 200 mM hydroxyurea (HU). For G2/M arrest, nocodazole was added to a final concentration of 10 μg/ml. Cell cycle progression was monitored using flow cytometry as described earlier36. DNA was stained with propidium iodide in case of YKL83 strains, whereas Sytox Green was used for all other strains. All FACS samples were analyzed using a Becton Dickinson FACSCalibur.
MMS sensitivity
Cells were grown to mid-log phase (OD600 = 0.5-0.6) at 25°C for cdc9 strains and at 28°C for YKL83 derived strains. Cells were then incubated with 0.02% MMS3 at 30°C for cdc9-1 strains and at 37°C for YKL83 strains for the indicated time period. For YKL83 strains, 2% galactose was added for 30 minutes prior to shifting the cells to 37°C. After each time point, sodium thiosulfate (10%) was added to the treated cells to inactivate the MMS. Yeast cells were pelleted, washed with distilled water and total protein was TCA-precipitated.
Generation of stable shRNA expressing cell lines
U2OS cells were maintained in RPMI 1640 supplemented with 10% heat inactivated FBS and 1% penicillin/streptomycin at 37°C in 5% CO2 (v/v). To generate stable DNA ligase I knockdown cell lines, DNA ligase I specific lentiviral shRNA plasmids and control shRNA were purchased from Open Biosystems. The sequences are shLIG#1 TGCTGTTGACAGTGAGCGCGCTTTCACCTGCGAATACAAATAGTGAA GCCACAGATGTATTTGTATTCGCAGGTGAAAGCTTGCCTACTGCCTCGGA shLIG#2 TGCTGTTGACAGTGAGCGACCTGTTTGTACCGGAAGCAAATAGTGAA GCCACAGATGTATTTGCTTCCGGTACAAACAGGCTGCCTACTGCCTCGGA. We also used a non-silencing control shRNA TGCTGTTGACAGTGAGCGATCTCGCTTG GGCGAGAGTAAGTAGTGAAGCCACAGATGTACTTACTCTCGCCCAAGCGAGA GTGCCTACTGCCTCGGA. These vectors have been described previously37. Stable cell lines were constructed as described38. Briefly, U2OS cells were transfected with 10 μg of shRNA plasmid (control or ligase I) with Fugene (Roche) per manufacturer's instructions. 48 h later cells were trypsinized and plated in media containing 2 μg /ml puromycin at different dilutions for colony formation. 12 days later drug-resistant colonies were picked and expanded and screened by Western blot for DNA ligase I expression.
Chromatin fractionation
To obtain the chromatin fraction, one 10 cm plate for each cell line was harvested and chromatin fractions were prepared exactly as described39. Briefly, cells were harvested, extracted to release soluble proteins and nuclear (insoluble) proteins pelleted. The nuclear pellet was then sonicated to release chromatin bound proteins. Proteins were fractionated on SDS polyacrylamide gels and analyzed by Western blots. Anti-DNA ligase I- (Santa Cruz), anti-histone H3- (Abcam), anti-PCNA- (Labvision), anti-ubiquitin- (Millipore), and anti-tubulin antibodies (Sigma) were used in this study.
Mammalian Cell Lysate and Western blotting
Cells were lysed with NETN (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 % Nonidet P-40 plus protease inhibitors) on ice and then rocked for 10 min at 4°C. Crude cell lysates were then centrifuged at 14,000 rpm for 10 min and cleared lysates were collected. Samples were boiled in 2X Laemmli buffer and fractionated on SDS-PAGE. Membranes were blocked in 5% milk-TBST and then probed with antibodies as indicated.
Supplementary Material
Acknowledgements
We thank Drs. D. J. Clarke, J. F. X. Diffley, D. Durocher, S. J. Elledge, M. Hochstrasser, S. Jentsch, D. Koepp and D. M. Livingston for strains and plasmids. We also thank Drs. J. F. X. Diffley, A. E. Tomkinson, and Z. Zhang for antibodies against Rad53, Cdc9 and PCNA. We acknowledge the assistance of the Flow Cytometry Core Facility at the University of Minnesota. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). J. C. Haworth was supported by NIH training grant CA009138. This work was supported by a Grant-in-Aid from the University of Minnesota and NIH grant (GM074917) to AKB. AKB is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Ellenberger T, Tomkinson AE. Eukaryotic DNA Ligases: Structural and Functional Insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster AD, Barnes DE, Arlett CF, Lehmann AR, Lindahl T. Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet. 1992;339:1508–1509. doi: 10.1016/0140-6736(92)91266-b. [DOI] [PubMed] [Google Scholar]
- 3.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 4.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 5.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 6.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–97. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiestl RH, Reynolds P, Prakash S, Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol. Cell. Biol. 1989;9:1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston LH, Nasmyth KA. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 13.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 14.Bielinsky AK, Gerbi SA. Chromosomal ARS1 has a single leading strand start site. Mol. Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 15.Ireland MJ, Reinke SS, Livingston DM. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics. 2000;155:1657–1665. doi: 10.1093/genetics/155.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez Y, et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 18.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 20.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 23.Ellison MJ, Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 24.Das-Bradoo S, Ricke RM, Bielinsky AK. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 2006;26:4806–4817. doi: 10.1128/MCB.02062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Kemp PA, Padula MD, Burguiere-Slezak G, Ulrich HD, Boiteux S. PCNA monoubiquitylation and DNA polymerase-eta ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2549–2559. doi: 10.1093/nar/gkp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Pickart CM. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 2005;24:4324–4333. doi: 10.1038/sj.emboj.7600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Frampton J, et al. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol. Biol. Cell. 2006;17:2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 30.Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 2004;279:15014–24. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
References for Methods
- 31.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz MC, et al. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 33.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 34.Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Ricke RM, Bielinsky AK. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J. Biol. Chem. 2006;281:18414–18425. doi: 10.1074/jbc.M513551200. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- 37.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genetics. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 38.Hannon GJ, Conklin DS. RNA interference by short hairpin RNAs expressed in vertebrate cells. Methods Mol Biol. 2004;257:255–266. doi: 10.1385/1-59259-750-5:255. [DOI] [PubMed] [Google Scholar]
- 39.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication fork. PNAS. 2008;26:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.