Abstract
A recently identified chemokine, fractalkine, is a member of the chemokine gene family, which consists principally of secreted, proinflammatory molecules. Fractalkine is distinguished structurally by the presence of a CX3C motif as well as transmembrane spanning and mucin-like domains and shows atypical constitutive expression in a number of nonhematopoietic tissues, including brain. We undertook an extensive characterization of this chemokine and its receptor CX3CR1 in the brain to gain insights into use of chemokine-dependent systems in the central nervous system. Expression of fractalkine in rat brain was found to be widespread and localized principally to neurons. Recombinant rat CX3CR1, as expressed in Chinese hamster ovary cells, specifically bound fractalkine and signaled in the presence of either membrane-anchored or soluble forms of fractalkine protein. Fractalkine stimulated chemotaxis and elevated intracellular calcium levels of microglia; these responses were blocked by anti-CX3CR1 antibodies. After facial motor nerve axotomy, dramatic changes in the levels of CX3CR1 and fractalkine in the facial nucleus were evident. These included increases in the number and perineuronal location of CX3CR1-expressing microglia, decreased levels of motor neuron-expressed fractalkine mRNA, and an alteration in the forms of fractalkine protein expressed. These data describe mechanisms of cellular communication between neurons and microglia, involving fractalkine and CX3CR1, which occur in both normal and pathological states of the central nervous system.
Chemokines mediate the directed migration of a variety of leukocyte subsets and consist of at least four subfamilies based principally on the relative position of highly conserved cysteine residues in their amino acid sequences (1, 2). Most chemokine peptides are characterized as secreted proteins of ≈7–10 kDa. The recent discovery of a chemokine termed fractalkine has revealed additional distinctive structural features in this gene family. These features include a CX3C motif and a mucin-like stalk that tethers the chemokine domain to transmembrane (TM) spanning and short intracellular domains (3, 4). Evidence from transfected cell systems indicates that fractalkine can exist as membrane-anchored, pro-adhesive, and secreted, chemotactic forms. Furthermore, unlike most chemokine peptides, fractalkine expression is demonstrable in nonhematopoietic tissues including brain, kidney, lung, and heart. In particular, the relatively high levels of fractalkine in the brain raises questions related to the function of chemokines in the central nervous system (CNS).
G-protein coupled receptors for chemokine peptides have been characterized extensively in transfected cells and peripheral leukocytes (2). However, very little is known regarding chemokine receptor expression and function in the CNS. Some chemokine receptors, including CCR5, CCR3 (5–7), CXCR4 (7–10), CXCR1, and DARC (11) have been demonstrated to be expressed in either normal brain tissue or cells derived from the brain. The chemokine receptor-like gene RBS11 (12) and its human ortholog V28 (13, 14) are known to be expressed prominently in the CNS. Recently, V28 was identified as a receptor for human fractalkine based on binding and signaling characteristics in transfected cells (15). Following established rules for nomenclature of chemokine receptors, these investigators identified V28 as CX3CR1.
The relative levels of fractalkine and CX3CR1 mRNA in tissue extracts from the CNS prompted us to characterize the location and function of fractalkine and CX3CR1 in the CNS. Herein, we report on a detailed characterization of fractalkine and CX3CR1 in the rat CNS in which we use approaches from both in vitro and in vivo experimental paradigms. We show that fractalkine is found principally in neurons and functional CX3CR1 is expressed by microglia. Furthermore, we present evidence demonstrating that levels of fractalkine and CX3CR1 in the facial motor nucleus are altered in a dynamic manner after peripheral nerve injury. These data shed light on fundamental interactions between neurons and microglia in both the normal and diseased CNS.
MATERIALS AND METHODS
Molecular Cloning of Rat Fractalkine cDNA.
A rat brain cDNA library (Strategene) was screened by hybridization using a rat fractalkine cDNA probe. The hybridization probe was generated by PCR using primers derived from a mouse cDNA (GenBank accession no. R75309). The rat fractalkine cDNA was sequenced by standard methods, and the DNA and conceptualized protein sequence were made available to GenBank (accession no. AF030358). The conceptualized amino acid sequence derived from the cDNA predicts a 392-aa protein with a molecular mass of 42,161 Da. The amino acid sequence identities of rat fractalkine with human and murine forms are 64% and 81%, respectively. Analogous to these human and murine proteins, rat fractalkine contains a signal peptide, a chemokine module, a mucin-like stalk, TM spanning region, and a short intracellular C terminus.
An EcoRI/SalI DNA fragment containing sequences encoding full length rat fractalkine protein was subcloned to the mammalian cell expression vector pCDM8 (Invitrogen). DNA containing human fractalkine protein coding sequence was generated by PCR and was subcloned into pCDM8. To generate peptidase-resistant, membrane-anchored fractalkine, human and rat fractalkine mutant forms were prepared by PCR in which the putative dipeptidase site Arg-Arg, adjacent to the TM spanning domain, was mutated into Ser-Ala.
RNase Protection (RPA) and in situ Hybridization (ISH) Analysis.
RPA was performed (16) by using total RNA (5 μg) and [32P]-UTP-labeled riboprobes generated from in vitro transcription reactions using plasmid construct templates containing either a 0.4-kbp NcoI fragment of rat fractalkine cDNA or a 0.4-kbp SmaI/KpnI fragment of RBS11 cDNA.
ISH was carried as described (17). After ISH and before emulsion autoradiography, some sections were stained with isolectin from Griffonia simplicifolia (GSA I-B4) according to published procedures (18). Emulsion dipped slides were developed (D19, Kodak), fixed (Kodak), and counter-stained with hematoxylin, hematoxylin/eosin, or Nissl (Cresyl violet).
Functional Characterization of Rat CX3CR1 in Transfected Cells.
A cDNA containing the ORF of RBS11 was cloned to the EcoRV site of pcLDN-10b. CHO cells (grown in Ham’s F-12, 10% heat inactivated fetal bovine serum, penicillin, and streptomycin) were transfected with 10 μg of plasmid DNA by using LipofectAMINE reagent (Life Technologies, Grand Island, NY). G418-resistant clonal cells expressing RBS11 cRNA were characterized in binding and functional assays. HEK293T cells (grown in DMEM, 10% fetal bovine serum, pen, and strep) were transfected by using LipofectAMINE and 10 μg DNA/100 mm plate and were used 3 days post-transfection.
Synthetic chemokines were generated by native chemical ligation of peptides synthesized by solid-phase methods (Applied Biosystems 430A Peptide Synthesizer), were purified by reverse-phase HPLC, and were characterized by electrospray mass spectrometry (19). Purified synthetic chemokines were reconstituted in PBS and were frozen at −70°C.
Whole-cell binding analysis was performed on three million PBS-rinsed cells per 35-mm well. Cells were incubated (1 hr at 22°C) in Hanks’ balanced salt solution, 0.1% BSA, 125I-labeled human fractalkine/chemokine domain (100 Ci/mmol), and various concentrations of unlabeled chemokine ligands. At the end of the incubation, cells were rinsed three times with ice-cold Hanks’ balanced salt solution and 0.1% BSA. Radioactivity retained in the wells was removed with 0.2 M NaOH and was quantitated by gamma spectroscopy.
Intracellular calcium levels in fura-2 acetoxymethyl-loaded CHO-RBS11 cells were assayed (20) by using a spectrofluorimeter (Photon Technology International, Princeton). Cells were illuminated alternately with UV light (1/sec) of 340- and 380-nm wavelength and was monitored for fluorescence at 510 nm. Because of the uncertainties involved in the calculation of [Ca2+(i)], the signals are reported as changes in F340/F380, which gives a relative measure of [Ca2+(i)].
Functional Characterization of Rat Microglia.
Mixed glial cells were isolated from newborn rat cortex by mechanical dissociation (sequential passage through 18-gauge, 21-gauge, and 25-gauge needles) and were plated in T75 flasks (Costar) containing DMEM and 10% fetal calf serum. Confluent cells were maintained without medium change for 5–7 days to favor microglial cell proliferation. Mixed glial cells then were rotary shaken for 3–4 hr at 275 rpm. The supernatant, containing an enriched population of microglia, was passed through a 70-μm sieve, and the pelleted cells subsequently were replated at a density of 100,000 cells/cm2 in DMEM and 10% fetal calf serum. Two hours later, cells were shaken manually and the medium was replaced with DMEM and 10% fetal calf serum containing 200 units/ml granulocyte/macrophage colony-stimulating factor or macrophage colony-stimulating factor. Cells were cultured for 2 or more days before assay.
Chemotaxis assay was performed as described (21). Intracellular calcium levels were measured according to modifications of published methods (22). Fluorescence measurements were performed on indo-1 acetoxymethyl-loaded microglia with a spectrofluorimeter (Photon Technology International, Princeton) by using an excitation wavelength of 350 nm (3-nm bandwidth) with dual simultaneous monitoring of emission at 405 and 490 nm (5-nm bandwidth).
Facial Motor Nerve Transections.
Facial motor nerve transections on methoxyflurane-anesthetized male Sprague–Dawley rats (175–200 g) were carried out as described (23). 1, 4, 7, 14, and 21 days after axotomy, rats (n = 3 per time point) were overdosed (75 mg/kg sodium pentobarbital) and perfused transcardially with 1× PBS (pH 7.0) followed by 4% paraformaldehyde in 0.1 M PO4 buffer (pH 7.2). Brains were saturated in 15% sucrose in 0.1 M PO4 buffer (pH 7.2), were cryosectioned coronally (12 μm), and were thaw-mounted onto poly-l-Lys-treated slides, and ISH analysis was performed. Quantitation of areal grain densities of bilateral facial motor nuclei was performed on emulsion-dipped slides under dark field conditions by using a video imaging system (Imaging Technology, Bedford, MA) and image analysis program (Imaging Research, St. Catherine’s, ON, Canada) interfaced via a video camera (Sony XC77) connected to a Zeiss microscope (Axioscop 20). Statistical comparisons (one-way ANOVA) were made between ipsilateral and contralateral values within each post-lesion time point.
Total protein extracts were prepared by homogenization of microdissected rat facial motor nuclei into buffer (20 mM Hepes:Na (pH 7.4)/2 mM EGTA/2 mM dithioerythritol/1 mM phenylmethylsulfonyl fluoride/20 μg/ml aprotonin). Protein concentrations were determined (24) with BSA as the standard.
Preparation of Recombinant Proteins, Antibodies, and Western Blot Analysis.
DNA sequences encoding recombinant rat soluble fractalkine (amino acids 1 to 85) were generated by PCR. Primers were synthesized, with high frequency Escherichia coli codons substituted for the natural rat sequences, and were used to generate a synthetic gene by PCR. The modified DNA sequence of rat fractalkine was subcloned to pET20 (Novagen), was expressed in E. coli, and was refolded to generate biologically active peptide (25).
A recombinant bacterial fusion protein containing the N terminus of RBS11 (PTSFPELDLENFEYDDSAEAC) linked to mouse dihydrofolate reductase was expressed in E. coli, was affinity purified by Ni–nitrilotriacetic acid resin, and was used to immunize rabbits. Fluorescence-activated cell sorter (FACS) analysis was carried out on detached wild-type and CHO-RBS11 cells incubated sequentially with rabbit anti-RBS11 antibody, biotin-conjugated rat anti-rabbit IgG (Zymed), and phycoerythrin-conjugated streptavidin (PharMingen). Stained cells were analyzed (10,000 events) on a FACScan station (Becton Dickinson).
The purified rat recombinant fractalkine expressed in E. coli was affinity purified by a Ni–nitrilotriacetic acid affinity column and was used to immunize rabbits (25). Antiserum was used at a dilution of 1:3,000 in a Western blot analysis on SDS/PAGE-fractionated protein samples (25). The protein blot was incubated with anti-rat fractalkine antibody, antibody preadsorbed with the chemokine domain of recombinant rat fractalkine, or preimmune rabbit sera.
RESULTS
Localization of Fractalkine in Rat Brain.
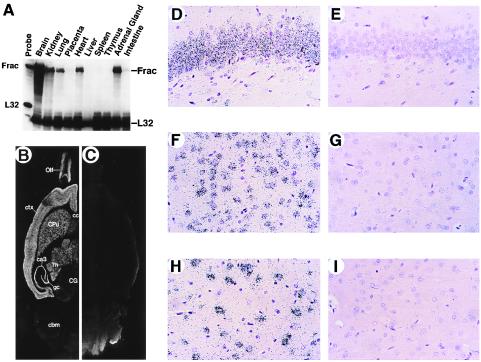
A cDNA encoding a protein of high similarity to human and murine fractalkines was isolated from a rat brain cDNA library. Fractalkine gene expression in normal adult rat tissues then was determined by using RPA. Of the tissues examined, rat brain was found to be the most abundant source of fractalkine mRNA (Fig. 1A), with significant levels present in kidney, lung, heart, and adrenal gland. The cellular source of fractalkine mRNA in the brain was assessed by ISH analysis. Fractalkine mRNA was detected readily in several discrete regions of adult rat brain, including cortex, hippocampus, caudate putamen, thalamus, and olfactory bulb (Fig. 1 B and C) and was significantly lower or absent in cerebellum, brainstem, and white matter regions, including corpus collosum and fimbria/fornix. This pattern of expression in the brain indicated that discrete subsets of neurons express the fractalkine gene. A higher resolution analysis revealed neurons as the principal cell type expressing fractalkine mRNA in the brain (Fig. 1 D–I).
Figure 1.
Localization of fractalkine mRNA in rat tissues and to neurons in the rat brain. (A) RPA of fractalkine mRNA in various tissues of adult rat. (B–I) Localization of fractalkine mRNA in the rat brain by using ISH analysis. Hybridization analysis using “anti-sense” (B, D, F, and H) and “sense” (C, E, G, and I) [35S]-UTP-labeled riboprobes were generated from rat fractalkine cDNA. Hybridized horizontal sections (B and C) were exposed to film for 18 hours (Olf, olfactory bulb; ctx, cortex; cc, corpus collosum; CPu, caudate putamen; Th, thalamus; ca3, ca3 region of the hippocampus; gc, granule cell layer of the hippocampus; CG, central gray; cbm, cerebellum) Hybridization signals (silver grains) reside principally over neurons in the granule cell layer of hippocampus (D), cortex (F), and thalamus (H).
Identification of Rat CX3CR1.
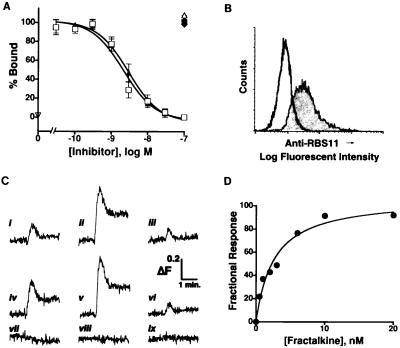
The relative abundance of fractalkine mRNA in normal rat brain suggests that its receptor is present constitutively in the CNS. The similarity of amino acid sequence of the rat chemokine receptor-like gene RBS11 to human CX3CR1 (formally known as V28 and CMKBRL1) suggests that RBS11 is a rat fractalkine receptor. To test this, we assessed the ability of RBS11-transfected cells to bind the chemokine domain of fractalkine. Radioligand binding analysis (Fig. 2A) of the CHO-RBS11 cells revealed specific fractalkine binding to the transfected cells; wild-type CHO cells did not display any significant binding. Inhibition binding constants (IC50s) of 2 and 3 nM for the chemokine domains of rat and human fractalkine, respectively, were determined. Members of the CC [macrophage inflammatory protein (MIP)-1α; regulated on activation, normal T-cell expressed and secreted (RANTES); and monocyte chemoattractant protein 1] and CXC (MIP-2, and stromal cell-derived factor 1) chemokine families did not compete for the binding of labeled fractalkine to the CHO-RBS11 cells, indicating a relative binding selectivity of RBS11 for chemokines containing the CX3C chemokine motif. A polyclonal antibody developed against a bacterial fusion protein containing the amino acid sequence of the N terminus of RBS11 stained the surface of the CHO-RBS11 cells but not the wild-type CHO cells (Fig. 2B).
Figure 2.
Identification of RBS11 as rat CX3CR1: binding of fractalkine and elevation of intracellular calcium by fractalkine in CHO-RBS11 cells. (A) Whole cell radioligand binding analysis of CHO-CX3CR1-expressing cells. Competition binding of 1 nM labeled peptide with the chemokine domain of rat fractalkine (□), the chemokine domain of human fractalkine (▴), rat monocyte chemoattractant protein 1 (■), rat MIP-1α (♦), rat regulated on activation, normal T-cell expressed and secreted (RANTES) (•), rat stromal cell-derived fator 1 (▾), and murine MIP-2 (▵). Wild-type CHO cells did not display any significant specific binding of 125I-fractalkine (▿). Data were calculated as the fraction of specific binding in the presence of the specified concentration of unlabeled chemokine peptides (Counts per minute bound per 3 million cells: total, 7945 ± 428; nonspecific: 2078 ± 231, n = 7). The results (from triplicate determinations) are presented as the mean ± SEM of three experiments (rat fractalkine) or four experiments (human fractalkine) or the mean of two experiments (all other chemokines). The calculated IC50s for inhibition of labeled peptide binding by the chemokine domains of either rat or human fractalkine are 2.2 nM (95% confidence interval: 0.9–5.3 nM) and 2.9 nM (95% confidence interval: 1.9–4.5 nM), respectively. (B) Rat CX3CR1 protein expression in CHO-CX3CR1-expressing (filled trace) and wild-type CHO (open trace) cells as determined by FACS analysis using anti-RBS11 polyclonal antibody. Neither preimmune rabbit sera nor immune sera preadsorbed with immunogen showed any increased reactivity to the CHO-CX3CR1-expressing cells (data not shown). (C) Representative traces of CHO-CX3CR1 expressing cells in response to application of either 293T cells transiently expressing rat (i, ii) or human fractalkine (iv, v) or the chemokine domain of rat (iii) or human (vi) fractalkine. Wild-type fractalkines (i, iv) or mutant fractalkines (ii, v) in which Arg-Arg residues adjacent to the TM domain were changed to Ser-Ala were expressed transiently in 293T cells. Lower traces depict application of mock transfected 293T cells (vii) to CHO-CX3CR1-expressing cells or addition of the chemokine domain of rat fractalkine (viii) or the wild-type membrane-tethered form of rat fractalkine (ix) to wild-type CHO cells. (D) Concentration-dependent effect of human fractalkine (chemokine domain) on intracellular calcium levels in CHO-CX3CR1-expressing cells. Data are plotted as the fraction of the maximal response seen in the presence of a high dose of fractalkine. Values are the mean of at least two determinations at each concentration. The calculated EC50 from these experiments is 3 nM.
The functional properties of RBS11 in the CHO cells were examined. Intracellular calcium levels in the CHO-RBS11 cells were elevated in response to application of either soluble or membrane-tethered forms of fractalkine (Fig. 2C). Two forms of either rat or human full-length fractalkines were expressed in 293T cells. One set of forms consisted of the wild-type fractalkines whereas the second set was expressed as mutant forms in which the putative dipeptidase cleavage residues (Arg-Arg) adjacent to the TM spanning domain were changed to Ser-Ala. The level of stimulation of intracellular calcium mobility by membrane-anchored forms of fractalkine was consistently higher than responses seen in the presence of just the chemokine domain. Furthermore, 293T cells expressing mutant forms of fractalkine displayed a greater calcium mobilizing activity than did cells expressing the wild-type forms of fractalkine, probably because of stabilization of the membrane-anchored form by the elimination of the proteolytic cleavage site. Despite the lower efficacy of the chemokine domain for stimulation of intracellular calcium release, titration by this form was achievable (Fig. 2D). An EC50 of 3 nM was determined, which is in good agreement with the binding data. The fractalkine specific binding and functional properties of the RBS11-expressing cells allow the identification of RBS11 as rat CX3CR1.
Functional Expression of CX3CR1 on Microglia.
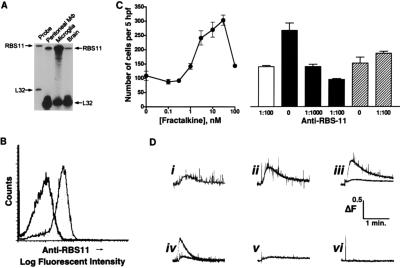
The presence of fractalkine in neurons of the CNS suggests that glia could be a target of this chemokine protein. RPA was used to determine the presence of CX3CR1 mRNA in primary cultures of rat microglia. CX3CR1 mRNA was relatively abundant in these cells (Fig. 3A) and was found to be at a level of ≥10-fold higher than either whole brain or peritoneal macrophages on the basis of relative L32 levels. Thus, the signal observed in whole brain RNA is likely to be accounted for by expression in microglia. A FACS analysis of the cultured microglia, by using anti-RBS11 antisera, revealed cell surface expression of rat CX3CR1 protein (Fig. 3B).
Figure 3.
Expression of CX3CR1 in primary cultures of rat microglia; fractalkine dependent responses mediated by CX3CR1. (A) RPA of CX3CR1 in peritoneal macrophage, cultured microglia, and brain by using RBS11 and L32 riboprobes. (B) Cell surface expression of CX3CR1 in cultured microglia assessed by FACS analysis by using anti-CX3CR1 antibody and reaction of preimmune (open trace) and immune sera (filled trace) to cultured microglia. Immune sera preadsorbed with immunogen did not react with the microglia (data not shown). (C) Chemotaxis of microglia in response to recombinant chemokine domain of fractalkine (left). Each point represents the mean ± SEM cell number in five high-power (×1,000) fields (hpf) from three experiments performed in quadruplicate. Inhibition of fractalkine-mediated chemotaxis by anti-RBS11 antibody is shown on the right. Unshaded histogram represents unstimulated migration in the presence of anti-RBS11 antibody (1:100 dilution). Black histograms represent dilution-related inhibition of fractalkine-induced microglial cell migration by using anti-RBS11 antibody. Hatched histograms represent MIP-1α-mediated microglial cell migration in the absence (0) or presence (1:100) of anti-RBS11 antibody. Each histogram represents the mean ± SEM cell number per five hpfs from three experiments performed in quadruplicate. (D) Effect of fractalkine forms on intracellular calcium levels in primary cultures of rat microglia. i–iii Show dose-dependent increases in intracellular calcium mediated by rat fractalkine expressed as a membrane-bound form in 293T cells. The lower trace of iii shows the effect of anti-RBS11 antibody (1:100). iv shows 293T-membrane-bound human fractalkine ± anti-RBS11 antibody (1:100). v shows wild-type 293T, and vi shows soluble rat fractalkine (50 nM). Representative traces from n = 3 experiments are depicted.
To determine whether the receptor protein was functional, microglia were examined for responses to application of soluble and membrane-anchored forms of fractalkine. Cultured rat microglia showed increased chemotactic activity in the presence of the chemokine domain of fractalkine (Fig. 3C). Microglia chemotaxis, induced by fractalkine but not MIP-1α, was blocked by pretreatment of the microglial cultures with anti-RBS11 antibodies, indicating that fractalkine-dependent chemotaxis is mediated by rat CX3CR1. The effect of fractalkine on stimulating an increase in the level of intracellular calcium also was determined. A marked increase in the intracellular calcium level was seen after application of 293T cells expressing membrane-tethered forms of either rat or human fractalkine (Fig. 3D). Analogous to the chemotaxis response, the elevation of intracellular calcium by fractalkine was inhibited by anti-RBS11 antibodies. Intracellular calcium levels within highly purified microglia were not affected by the fractalkine chemokine domain alone, indicating a potential requirement for other domains of the molecule in stimulating this signal transduction pathway.
In Vivo Localization of CX3CR1; Regulation of Receptor and Fractalkine After Motor Neuron Axotomy.
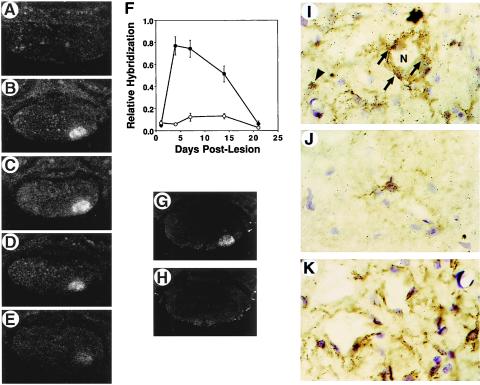
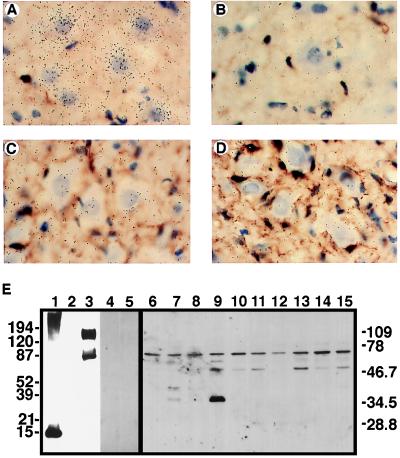
The widespread distribution of rat CX3CR1 in normal rat brain and its presence on cultured microglia indicate that the CX3CR1 gene also might be expressed on these cells in vivo. An animal model of peripheral nerve injury involving a microglial response was used to assess the in vivo localization and regulation of CX3CR1. After axotomy of the facial motor nerve, microglia within the facial motor nucleus undergo dramatic cellular and morphological changes that include a marked proliferative response (26, 27). ISH analysis was used to assess the expression of CX3CR1 in the facial motor nucleus over a 21-day period during which the facial motor neurons undergo regeneration. At time points of 4, 7, and 14 days post-axotomy, the levels of CX3CR1 hybridizing mRNA were elevated significantly in the injured facial motor nucleus (Fig. 4 A–F), as compared with the contralateral nucleus. Highest levels of CX3CR1 mRNA were found on days 4 and 7 post-axotomy. At the 21-day time point, CX3CR1 mRNA had returned to a level comparable to that detected in the contralateral nucleus. A higher resolution analysis revealed microglia as the source of CX3CR1 mRNA (Fig. 4 I and J) as concordant expression with a selective microglial cell reactive stain (lectin GSA-I-B4) was evident. CX3CR1 expressing microglia in the neuropil were present in both contralateral and ipsilateral facial motor nuclei. However, in the ipsilateral facial motor nucleus, CX3CR1-expressing cells were increased markedly and occupied perineuronal positions at 4 days after axotomy (Fig. 4I).
Figure 4.
ISH analysis of CX3CR1 expression in the rat facial nucleus after motor neuron axotomy. ISH was performed on rat brainstem sections from animals killed at 1 (A), 4 (B), 7 (C), 14 (D), and 21 (E) days after nerve transection by using [35S]UTP-labeled riboprobe. F depicts a quantitative analysis of the hybridization signal on the operated (•) and contralateral (○) sides. Results are presented as the mean ± SEM of data derived from at least three sections from three different animals. The hybridization signal was specific for CX3CR1 because no increase in signal was observed by using a sense riboprobe (H) when compared with a corresponding anti-sense riboprobe hybridized-section (G). Hybridization signals (silver grains) from anti-sense [33P]UTP-labeled riboprobe hybridized-sections are found principally over lectin (GSA-I-B4)-staining microglia in the neuropil of the unoperated facial motor nucleus (J) or lesioned facial motor nucleus (arrowhead in I) whereas several CX3CR1 mRNA-containing microglial cells are found perineuronally (arrows in I) surrounding axotomized motor neurons (N, motor neuron cell body). (K) Sense [33P]UTP-labeled riboprobe-hybridized section showing only a few scattered silver grains.
The expression of fractalkine mRNA and protein in the facial nucleus after axotomy also was examined. ISH analysis demonstrated that the motor neurons were the primary source of fractalkine mRNA (Fig. 5 A–D). A decrease in the hybridization signal in motor neurons 4 days after axotomy was also evident. Polyclonal antibodies recognizing the chemokine domain of rat fractalkine were developed and used in Western blot analysis to probe for the presence of fractalkine protein in extracts of the facial motor nucleus over the time course of motor neuron regeneration (Fig. 5E). The anti-fractalkine sera are highly specific for this protein, as antibody reactivity was blocked completely by preadsorption with recombinant fractalkine protein, and there was no reactivity with preimmune sera. Immunoreactive protein was detected in both the control and lesioned facial motor nuclei. A protein band with a molecular mass of 65 kDa was present in all samples whereas smaller bands of 50 and 36 kDa were detected in extracts of the lesioned facial motor nuclei. The 50-kDa band was present on day 4 and persisted for at least 21 days after axotomy. The 36-kDa form first appeared on day 1 and was most abundant in the 4-day extract. Forms of molecular mass 40 and 56 kDa were also present in the day 1 (axotomized) and day 4 (axotomized) samples, respectively.
Figure 5.
Fractalkine expression in the rat facial nucleus after motor neuron axotomy. ISH was performed on rat brainstem sections from animals killed 4 days after nerve transection. Shown is hybridization analysis of the contralateral (A and B) and ipsilateral (C and D) facial motor nuclei by using anti-sense (A and C) and sense (B and D) [33P]UTP-labeled riboprobes generated from rat fractalkine cDNA. Hybridization signals are found principally over the motor neuron cell bodies. (E) Western blot analysis of recombinant (bacterial- and mammalian cell-expressed) fractalkines, as well as protein extracts from the facial motor nucleus from the control and lesioned sides after peripheral nerve transection. Fractalkine forms were subject to SDS/PAGE and include bacterial-expressed fusion protein (10 ng) containing the chemokine module of rat fractalkine (lane 1), and cell lysates prepared from COS cells transfected with either antisense (lane 2) or sense (lanes 3–5) constructs containing membrane-anchored forms of rat fractalkine. In addition, protein extracts (10 μg/lane) from the facial motor nucleus from the control (lanes 6, 8, 10, 12, and 14) and lesioned (lanes 7, 9, 11, 13, and 15) sides 1 (lanes 6 and 7), 4 (lanes 8 and 9), 7 (lanes 10 and 11), 14 (lanes 12 and 13), and 21 (lanes 14 and 15) days after axotomy were subject to SDS/PAGE. Immunoblot analysis was performed by using anti-rat fractalkine rabbit sera (lanes 1–3, 6–15), preimmune sera (lane 4), or immune sera preadsorbed with recombinant rat fractalkine, chemokine domain (lane 5).
DISCUSSION
Recent observations indicate that chemokines mediate additional actions other than chemotaxis of peripheral leukocytes (2). The presence of chemokines and chemokine receptors in cells of the CNS raises major questions concerning the role of these genes in neurophysiology. The recent discovery of a gene encoding the chemokine fractalkine and the demonstration that it is expressed abundantly in the brains of humans and rodents prompted our investigation to localize the expression of this gene to specific cells of the CNS. In addition, we sought to determine the nature of the receptor in the CNS responsible for mediating the actions of fractalkine. Our results demonstrate that neurons are the primary source of expression of the fractalkine gene and that microglia are the principal fractalkine-responding cells in the CNS. Furthermore, we present evidence that indicates that fractalkine and its receptor CX3CR1 are involved intimately in the facial motor neuron response to peripheral nerve injury. Our data have implications in understanding fundamental interactions between neurons and microglia in both normal and pathological states of the CNS.
ISH analysis revealed neurons as the principal source of fractalkine mRNA in the brain. Neuronal structures in a variety of discrete regions of the rat brain were found to contain intense hybridization signals. This is a clear demonstration of the in vivo expression of a chemokine gene in neurons of the CNS and is in stark contrast to described data in which immunoreactive fractalkine protein was found in both normal and activated microglia (4). In probing cultured microglia, we were unable to detect either fractalkine mRNA (by using either RPA or a more sensitive reverse transcription–PCR-based assay) or fractalkine protein (by Western blot or FACS analysis; data not shown). The technique of ISH analysis more definitively demonstrates the source of expression of a gene, in this case fractalkine, versus an immunohistochemical analysis that can only detect where the protein is and not necessarily where it is synthesized. Furthermore, our demonstration that microglia express fractalkine receptor indicates that, in the CNS, a paracrine interaction between neurons and microglia exists that involves fractalkine and CX3CR1. The coexpression of fractalkine and CX3CR1, implicated from the Pan et al. study (4), would imply an autocrine interaction.
The relative abundance of fractalkine in the CNS indicates that its receptor is likely to be expressed in nervous tissue as well. Although some known chemokine receptors have been shown to be expressed in the CNS, the relative abundance of the rat chemokine receptor-like gene RBS11 in the CNS, coupled with additional fractalkine-concordant expression in other peripheral tissues including lung, kidney, and gut, prompted us to consider the RBS11 gene as a candidate receptor for fractalkine. Recently the human ortholog of RBS11, V28, was identified as a receptor for human fractalkine based on binding and stimulation of intracellular calcium mobilization by a soluble form of fractalkine (15). Our specific binding and signaling properties of receptor transfected cells confirms RBS11 as rat CX3CR1. However, unlike the Imai et al. study (15), our antibody-blocking experiments provide direct evidence that the fractalkine receptor on native cells (microglia in our instance) is CX3CR1. Because chemokines are known to be redundant in their actions and their receptors promiscuous with respect to chemokine binding, the data from Imai et al. (15) do not rule out the possibility that additional receptors mediate fractalkine-dependent responses on freshly isolated leukocytes.
We found prominent microglia expression of CX3CR1, both in terms of mRNA and protein. This microglial cell surface-expressed receptor was functional, mediating fractalkine-dependent chemotaxis and intracellular calcium mobilization. The presence of CX3CR1 on cultured microglia suggests that these cells express this gene in vivo. The localization of CX3CR1 to lectin GSA-I-B4 staining cells in the facial nucleus is clear demonstration that resident microglial cells express this receptor. Thus, experiments using cultured microglia are likely to provide useful correlates to the study of CX3CR1-dependent phenomena associated with microglia in vivo. We also provide additional compelling evidence indicating that the form of fractalkine, membrane vs. soluble, determines the response of the fractalkine-responsive cell. Membrane-anchored forms of fractalkine induced a greater calcium mobilizing activity in receptor transfected cells, as compared with soluble forms. Thus, the role of the membrane-anchored form is not exclusively as a mediator of receptor-expressing cell adhesion, as shown (15). Our data indicate that intracellular calcium levels are elevated, in CX3CR1-expressing cells, during the adhesion process. Analogous to the receptor-transfected CHO cell model, differential stimulation of microglia intracellular calcium mobilization by fractalkine depended on the form present. Membrane-anchored forms of fractalkine stimulated robust changes in intracellular calcium levels whereas a soluble form of fractalkine had no effect. These data have implications for neuronal regulation of microglia activity in vivo and suggest that the neurons can differentially regulate intracellular levels of calcium within microglia, depending on the form of fractalkine they provide to the microenvironment. The physiological significance of these relative intracellular calcium levels in microglia remains to be determined.
The regulation of CX3CR1 and fractalkine in the facial motor nucleus after peripheral nerve injury is intriguing. The time course of transient CX3CR1 mRNA expression parallels the transient increase in microglial cell numbers after axotomy. It also is known that, at this time point, the majority of axotomized motor neurons are ensheathed tightly by the perineuronal microglial cells (26, 27). An additional distinguishing feature of this experimental paradigm, which contrasts to other CNS inflammatory models, including experimental allergic encephalomyelitis, is the notable absence of an inflammatory cell infiltrate. Therefore, cellular and molecular changes taking place within the facial nucleus are responses of resident cells, and the interpretation of the cellular localization of expression of the relevant genes is not confounded by the presence of an inflammatory cell infiltrate. Our data indicate that one response of the motor neurons to peripheral nerve injury is a reduction in fractalkine mRNA and an increase in the levels of lower molecular mass forms of fractalkine protein. The antibody we used in this study does not allow us to determine the specific forms of fractalkine—i.e., membrane-anchored vs. secreted—expressed in the facial motor nucleus. However, it is reactive against the chemokine domain of rat fractalkine, and thus, all bands should at least contain this region of the protein. The larger (65 kDa) form is presumably the membrane-anchored form whereas the smaller injury-regulated protein bands are consistent with them being secreted forms. These latter forms may be mediating neuronally directed chemotaxis of microglia in the lesioned facial motor nucleus. Once adjacent to the injured neuron, the microglia then would encounter the membrane-anchored form of fractalkine, thus taking up a perineuronal position. The apparent increase in lower molecular mass protein forms as well as no change in the 65-kDa form was evident despite the decrease in fractalkine mRNA observed in the ISH analysis. In the absence of strict quantitative measurements, it is difficult to assess whether total fractalkine protein levels have changed. However, it is clear that the lower molecular weight forms are increased after motor neuron axotomy. The precise mechanism underlying this increase is worthy of investigation and may reflect an altered level(s) or activity of the protease(s) that is(are) responsible for generating the shed forms. The apparent maintenance in the level of the 65-kDa form may reflect the necessity of the motor neurons to provide a constant level of fractalkine protein on the surface of the cell body. In addition, the higher molecular mass form may represent a nonregulatable pool of fractalkine and may not be the source of the smaller forms.
The dynamic regulation of motor neuron fractalkine expression, coupled with increased numbers of CX3CR1-expressing microglia in the injured facial motor nucleus, indicates that fractalkine- and CX3CR1-dependent signaling mechanisms are part of the intrinsic cellular response that occurs during motor neuron regeneration. In addition, the constitutive expression of these genes in many diverse functional areas of the brain indicates that they are involved in fundamental processes of communication between neurons and microglia. Phenotypic analysis of fractalkine and/or CX3CR1 knockout mice may reveal the function of this ligand receptor pair in neurophysiology. An examination of these molecules in other neuropathological situations, including cerebral ischemia or stroke, and neurodegenerative diseases may yield further insights into the molecular basis of diseases of the CNS. Moreover, the development of therapeutic agents targeting fractalkine and CX3CR1 may alter pathological outcomes associated with these disease processes.
Acknowledgments
We thank L. Belardinelli for CHO cells, D. Camerini for 293T cells, M. A. Siani for preparing synthetic human fractalkine, and C. H. Gelband for valuable comments. This work was supported by National Institutes of Health Grants NS34901 (J.K.H.) and DK49832 (L.F.) and an equipment grant from the Howard Hughes Medical Institute Research Resources Program to the University of Florida (J.K.H.). All animal procedures conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
ABBREVIATIONS
- TM
transmembrane
- CNS
central nervous system
- RPA
RNase protection
- ISH
in situ hybridization
- FACS
fluorescence-activated cell sorter
- CHO
Chinese hamster ovary
- MIP
macrophage inflammatory protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF030358).
References
- 1.Baggiolini M, Dewald B, Moser B. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 2.Baggiolini M, Dewald B, Moser B. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 3.Bazan J F, Bacon K B, Hardiman G, Wang W, Soo K, Rossi D, Greaves D R, Zlotnik A, Schall T J. Nature (London) 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 4.Pan Y, Lloyd C, Zhou H, Dolich S, Deeds J, Gonzalo J A, Vath J, Gosselin M, Ma J, Dussault B, et al. Nature (London) 1997;387:611–617. doi: 10.1038/42491. [DOI] [PubMed] [Google Scholar]
- 5.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 6.Rottman J B, Ganley K P, Williams K, Wu L, Mackay C R, Ringler D J. Am J Pathol. 1997;151:1341–1351. [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y, Salafranca M N, Adhikari S, Xia Y, Feng L, Sonntag M K, deFiebre C M, Pennell N A, Streit W J, Harrison J K. J Neuroimmunol. 1998;86:1–12. doi: 10.1016/s0165-5728(98)00005-8. [DOI] [PubMed] [Google Scholar]
- 8.Wong M-L, Xin W W, Duman R S. Mol Psychiatry. 1996;1:133–140. [PubMed] [Google Scholar]
- 9.Tanabe S, Heesen M, Yoshizawa I, Berman M A, Luo Y, Bluel C C, Springer T A, Okuda K, Gerard N, Dorf M E. J Immunol. 1997;159:905–911. [PubMed] [Google Scholar]
- 10.Lavi E, Strizki J M, Ulrich A M, Zhang W, Fu L, Wang Q, O’Connor M, Hoxie J A, Gonzalez-Scarano F. Am J Pathol. 1997;151:1035–1042. [PMC free article] [PubMed] [Google Scholar]
- 11.Horuk R, Martin A W, Wang Z-X, Schweitzer L, Gerassimides A, Guo H, Lu Z-H, Hesselgesser J, Perez H D, Kim J, et al. J Immunol. 1997;158:2882–2890. [PubMed] [Google Scholar]
- 12.Harrison J K, Barber C M, Lynch K R. Neurosci Lett. 1994;169:85–89. doi: 10.1016/0304-3940(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 13.Raport C J, Schweickart V L, Eddy R L, Jr, Shows T B, Gray P W. Gene. 1995;163:295–299. doi: 10.1016/0378-1119(95)00336-5. [DOI] [PubMed] [Google Scholar]
- 14.Combadiere C, Ahuja S K, Murphy P M. DNA Cell Biol. 1995;14:673–680. doi: 10.1089/dna.1995.14.673. [DOI] [PubMed] [Google Scholar]
- 15.Imai T, Hieshima K, Haskell C, Baba M, Nagira M, Kakizaki M, Takagi S, Nomiyama H, Schall T J, Yoshie O. Cell. 1997;91:521–530. doi: 10.1016/s0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 16.Feng L, Xia Y, Wilson C B. J Biol Chem. 1994;269:2342–2348. [PubMed] [Google Scholar]
- 17.McNamara R K, Routtenberg A. Mol Brain Res. 1995;33:22–28. doi: 10.1016/0169-328x(95)00083-5. [DOI] [PubMed] [Google Scholar]
- 18.Streit W J. J Histochem Cytochem. 1990;38:1683–1686. doi: 10.1177/38.11.2212623. [DOI] [PubMed] [Google Scholar]
- 19.Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 20.Dunstan C-A N, Salafranca M N, Adhikari S, Xia Y, Feng L, Harrison J K. J Biol Chem. 1996;271:32770–32776. doi: 10.1074/jbc.271.51.32770. [DOI] [PubMed] [Google Scholar]
- 21.Bacon K B, Camp R D, Cunningham F M, Woolard P M. Br J Pharmacol. 1988;95:966–974. doi: 10.1111/j.1476-5381.1988.tb11727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacon K B, Premack B A, Gardner P, Schall T J. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 23.Streit W J, Graeber M B, Kreutzberg G W. Glia. 1988;1:301–307. doi: 10.1002/glia.440010502. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Feng L, Xia Y, Yoshimura T, Wilson C B. J Clin Invest. 1995;95:1009–1017. doi: 10.1172/JCI117745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreutzberg G W. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 27.Streit W J. Neurotoxicology. 1996;17:671–678. [PubMed] [Google Scholar]