Abstract
Glutathione-S-transferases (GSTs) are ubiquitous detoxification enzymes that catalyse the conjugation of electrophilic substrates to glutathione. Here, we present the crystal structures of Gtt2, a GST of Saccharomyces cerevisiae, in apo and two ligand-bound forms, at 2.23 Å, 2.20 Å and 2.10 Å, respectively. Although Gtt2 has the overall structure of a GST, the absence of the classic catalytic essential residues—tyrosine, serine and cysteine—distinguishes it from all other cytosolic GSTs of known structure. Site-directed mutagenesis in combination with activity assays showed that instead of the classic catalytic residues, a water molecule stabilized by Ser129 and His123 acts as the deprotonator of the glutathione sulphur atom. Furthermore, only glycine and alanine are allowed at the amino-terminus of helix-α1 because of stereo-hindrance. Taken together, these results show that yeast Gtt2 is a novel atypical type of cytosolic GST.
Keywords: Saccharomyces cerevisiae, glutathione-S-transferase, crystal structure, catalytic type, site-directed mutagenesis
Introduction
Many xenobiotics have cytotoxic and/or genotoxic properties and can damage cells in various ways. To eliminate toxic xenobiotics, organisms have evolved a series of sophisticated detoxification strategies. In general, the detoxification process can be described as a three-phase reaction. In phase one, xenobiotics are activated by the introduction of reactive functional groups. In phase two they are neutralized by conjugation to chemical constituents through the reactive groups. In phase three, the conjugated xenobiotics are pumped out of cells to be metabolized through downstream pathways and eventually eliminated (Sheehan et al, 2001).
The glutathione-S-transferases (GSTs; Enzyme Commission number 2.5.1.18) have a crucial role in phase two of enzymatic detoxification (Hayes et al, 2005). These enzymes catalyse the reaction of xenobiotics with the thiolate group of glutathione (GSH), thereby neutralizing their electrophilic sites and raising the water solubility of the products (Habig & Jakoby, 1981). Eukaryotes usually contain several GST paralogues with different catalytic activities and a wide range of cellular functions. In mammals, cytosolic GSTs are dimeric proteins that are grouped into eight main strain classes—Alpha, Kappa, Pi, Mu, Theta, Zeta, Sigma and Omega—primarily by sequence alignment. In addition, studies of non-mammalian species have shown several new classes such as the Beta class in bacteria, the Phi and Tau classes in plants, and the Delta class in insects (Armstrong, 1997; Sheehan et al, 2001; Allocati et al, 2009). Fungi also have several GSTs, but most cannot be grouped into the pre-existing classes (McGoldrick et al, 2005). In addition to the sequence-based classification, the nomenclature of cytosolic GSTs can refer to the essential catalytic residue. All cytosolic GSTs of known structure are classified into one of three catalytic types, tyrosine, serine or cysteine, depending on the residue that drives the conjugation of GSH to a xenobiotic through lowering the pKa of GSH, and stabilizing the thiolate anion through a hydrogen bond (Armstrong, 1997; Sheehan et al, 2001; Allocati et al, 2009). Mutations of the essential residue usually result in a substantial, if not complete, inactivation of the GST (Stenberg et al, 1991; Board et al, 1995).
In the yeast Saccharomyces cerevisiae, seven proteins have GST activity (Gtt1 and Gtt2; Gto1, Gto2 and Gto3, and Grx1 and Grx2; Collinson & Grant, 2003). However, the structures of Grx1 and Grx2 show that they are not bona fide GSTs, but only correspond to the GST amino-terminal domain (Yu et al, 2008). Of the five other homologous proteins, Gto1–Gto3 are classified into the Omega GST class (Garcera et al, 2006), whereas Gtt1 and Gtt2 are not categorized into any existing classes. Although the physiological and biochemical functions of these five yeast GSTs have been studied extensively, no three-dimensional structures have been reported. We began a systematic characterization of the yeast GSTs by determining the crystal structures of Gtt2 in apo form at 2.23 Å, and in two ligand-bound forms at 2.20 Å and 2.10 Å. Gtt2 is distinct from the three classic catalytic types, because a water molecule fixed by two residues in the carboxy-terminal domain is responsible for activity. Furthermore, only glycine (Gly) and alanine (Ala) are allowed at the N-terminus of helix-α1 because of stereo-hindrance. These results enabled us to define a new catalytic cytosolic GST type, which we call the atypical type.
Results And Discussion
Overall structure
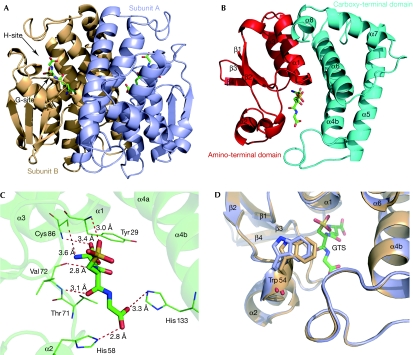
Gtt2 shares a sequence identity of less than 28% with GSTs of known structure, but clearly adopts the canonical fold of cytosolic GSTs. The overall structure of the Gtt2 dimer with two molecules of the GSH analogue glutathione sulphonate (GTS) is illustrated in Fig 1A. The protein is characterized by two spatially distinct domains that are connected by an eight-residue linker. In addition, it contains an 18-residue signal peptide at the N-terminal that targets the mitochondrial matrix. This region is too flexible to be traced in the electron density map.
Figure 1.
Overall structure of Gtt2. (A) Schematic representation of the Gtt2 homodimer. GTS molecules are shown as stick models and coloured according to their atom type (C, green; S, yellow; O, red). The G-site and H-site of subunit B are indicated by arrows. (B) The GTS-bound Gtt2 monomer, with the two domains in cyan and red. (C) Hydrogen bonds between GTS and Gtt2. (D) Superposition of apo-bound (light blue) and GTS-bound Gtt2 (light orange) showing the different position of the loop after helix-α2 upon GTS binding. Cys, cysteine; G-site, GSH-binding site; GTS, glutathione sulphonate; H-site, hydrophobic substrate binding site; His, histine; Thr, threonine; Trp, tryptophan; Tyr, tyrosine; Val, valine.
Despite the low level of sequence identity to other GSTs, the tertiary structure of the N-terminal domain resembles the typical βαβαββα motif of a thioredoxin fold (Fig 1B, red). The all-α C-terminal domain of helices α4–α8 is wound into a right-handed super-helical structure (Fig 1B, cyan). Inter-subunit contacts are mainly mediated by hydrophobic residues in β4-strands, as are contacts between helix-α3 of one subunit and the antiparallel helix-α4 of the other.
The GSH-binding site (G-site) of each subunit binds to a molecule of GTS, which is well fitted in the electron-density map. The interactions between Gtt2 and GTS are much the same as for previous structures (Fig 1C). Gtt2 also undergoes induced-fit upon GTS binding, as reported for some other cytosolic GSTs (Neuefeind et al, 1997). For example, the loop after helix-α2 was observed in a closer position to GTS, as shown by Trp 54 in Fig 1D. The H-site, which is the binding site of the hydrophobic substrate, is located adjacent to the G-site and could be described as a shallow hydrophobic tunnel, to the bottom of which the sulphonate moiety of GTS exactly protrudes from the G-site (Fig 1A).
Gtt2: a new catalytic type of cytosolic GST
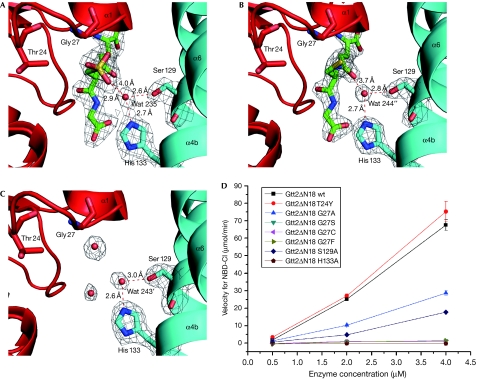
Previous biochemical experiments showed that Gtt2 conjugates GSH with the GST substrate 1-chloro-2,4-dinitrobenzene. We found that the pKa of GSH on binding to Gtt2 was 6.08±0.15, which showed that Gtt2 could deprotonate GSH at physiological pH, similarly to the classic GSTs (supplementary Fig S1A,B online). According to these data, one of the three classic catalytic residues should be close to GSH to stabilize the thiolate anion. Surprisingly, we found that not only were tyrosine, serine and cysteine absent near to the GTS sulphur atom, but that other polar groups were also lacking, except for the water molecule Wat 235. These results indicated that Wat 235 might be a crucial element in the GST activity of Gtt2. In the GTS-bound structure, Wat 235 was found to be fixed by Ser 129Oγ, His 133Nɛ and the oxygen atom of the sulphonate group, with hydrogen bonds of 2.6 Å, 2.7 Å and 2.9 Å, respectively (Fig 2A). The sulphur atom of GTS is 4.0 Å away from Wat235, whereas the corresponding distance from the sulphur atom of GSH to Wat244″ is 3.7 Å in the GSH-bound form (Fig 2B). In the apo form, a corresponding water molecule (Wat 243′) was also observed at 3.0 Å to Ser 129Oγ and 2.6 Å to His 133Nɛ (Fig 2C). Residues Ser 129 and His 133 could function as a ‘clamp' to grip Wat235 in position to form a hydrogen bond with the sulphonate group. We hypothesized that any mutations that destroyed the ‘clamp' would inactivate the enzyme. Subsequent site-directed mutagenesis in combination with activity assays supported this hypothesis (Fig 2D). Compared with the wild type, the activity of the S129A mutant is clearly impaired, and the H133A mutation completely abolishes activity. This implies various roles for Ser129 and His133, and supports the mechanism we propose below.
Figure 2.
Residues crucial for Gtt2 activity. (A) The sulphur atom of GTS is located over the amino-terminus of helix-α1, at Gly 27. Ser 129 and His 133 form a ‘clamp' to grip water molecule Wat 235, which forms a hydrogen bond with the sulphur atom. The electron density maps of GTS, Wat 235, Ser 129 and His 133 (2Fo−Fc map) were contoured at 1.5 σ. (B) The active site of GSH in the GSH-bound form is located close to that of GTS in the GTS-bound form. (C) The active site of apo-form Gtt2. A water molecule (Wat 243′) locates at about the same position as Wat 235 in the GTS-bound structure, whereas several water molecules take the place of GTS. (D) The relative activity of the wild-type and seven Gtt2 mutants. GSH, glutathione; GTS, glutathione sulphonate.
After the catalytic essential residues of Gtt2 were assigned, we investigated whether the activity of Gtt2 could be increased by the introduction of these residues of classic cytosolic GSTs. Three mutants were designed on the basis of structural superposition—mutants T24Y, G27S and G27C—and all were located in conserved positions for previously defined essential GST residues. As shown in Fig 2D, the T24Y mutant has activity similar to wild type, whereas the G27S and G27C mutants completely lose activity towards 4-chloro-7-nitro-2,1,3-benzoxadiazole (NBD-Cl).
Mutation of Gly27 at the N-terminus of helix-α1 gave a surprising result. In an α-helix, the alignment of the peptide dipoles parallel to the helix axis creates a helix dipole of considerable strength (Hol, 1985; Porter et al, 2006). Several GSTs were reported to have helix dipoles that stabilize the thiolate anion (Chen et al, 1988). In our structure, the cysteinyl sulphur atom of GTS is located almost directly over the N-terminus of helix-α1. After deprotonation, the thiolate anion of GSH might be stabilized by the positive charge around the N-terminus of helix-α1, possibly accompanied by a hydrogen bond with the main-chain nitrogen atom of Tyr29. Thus, we reasoned that pushing the thiolate group of GSH away from the N-terminus of helix-α1 would destabilize the thiolate anion and result in lower reaction efficiency. To test this proposal, mutants G27A and G27F were constructed. The G27A mutation apparently weakens activity, and the G27F mutant is completely inactive (Fig 2D). The decreased activity correlates to the increase in side-chain size from glycine to alanine/serine/cysteine/phenylalanine, clearly indicating that spatial accessibility to the N-terminus of helix-α1 is another crucial determinant of the GST activity. All protein samples were quality controlled by circular dichroism, the results of which showed that the mutations did not introduce significant changes to the protein structures (supplementary Fig S2 online). On the basis of these characteristics, we defined Gtt2 as the first member of a new catalytic type of cytosolic GST family, named the atypical type, in contrast to the classic tyrosine, serine and cysteine types.
On the basis of the results from crystal structures, site-directed mutagenesis and enzymatic assays, we proposed the following catalytic mechanism for Gtt2. First, the side chains of Ser129 and His133 grasp Wat235 through two hydrogen bonds. Second, the relatively low pKa of His133 enables the ionization of the Nɛ atom at physiological pH. Third, after ionization, the His133Nɛ atom attracts one proton from Wat235, which makes the oxygen atom of Wat235 more electronegative. Fourth, on GSH binding, the electronegative oxygen atom of Wat235 attracts a proton from the thiolate group and forms a hydrogen bond. Meanwhile, Gly27 guarantees that the interaction between the negative thiolate group and the positive helix-α1 dipole is not disturbed by stereo-hindrance. At this point, the thiolate group of GSH is ionized and is ready to react with a xenobiotic. A previous report also proposed a water-assisted mechanism of GSH activation for an Alpha-class GST (Dourado et al, 2008). However, some characteristics of our proposed mechanism are distinct, such as the new active site, the redundancy of conformational change for GSH and the dual roles of the water molecule.
During revision of the paper, we determined the crystal structure of Gtt2 in complex with its natural substrate GSH at 2.10 Å resolution. Superposition of GSH-bound and GTS-bound forms gave a root-mean-square deviation of only 0.2 Å. GSH and GTS were well superimposed, with only a 0.3 Å variation between the two sulphur atoms. This indicated that the differences between the two structures were slight and the conclusions drawn from the GTS-bound form could be applied to the GSH-bound form.
Alignment analysis of atypical cytosolic GST
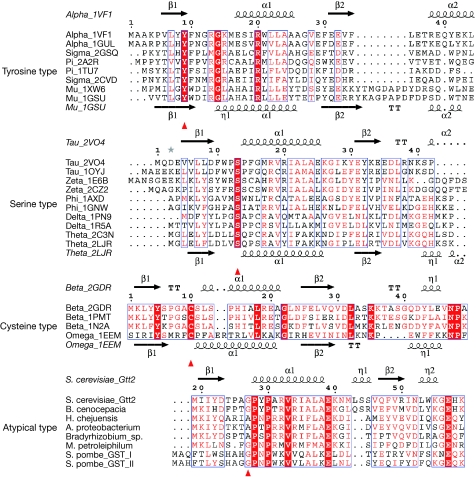
To find the probable origin and distribution of the atypical-type cytosolic GSTs, the sequence of Gtt2 was used in a BLAST search against the non-redundant protein sequences database (www.ncbi.nlm.nih.gov/BLAST). Almost all the top hits with high identity are from bacteria, except for two GSTs from the yeast Schizosaccharomyces pombe. These sequences have low identity to the classic GST classes. For example, the identity between Burkholderia cenocepacia GST (NCBI accession number YP_002095143) and the most similar classic GST (NCBI accession number ABA42223, Phi class) is 28%, and identity between S. pombe GSTI (NCBI accession number Q9Y7Q2) and the closest classic GST (NCBI accession number XP_002004610, Delta class) is 29%. Both identity values are lower than the threshold (40%) for classic sequence-based classification (Allocati et al, 2009). We also investigated a total of 254 entries in the Protein Data Bank (www.rcsb.org) that resulted from a search using ‘glutathione transferase' as the keyword. After removing 168 entries with greater than 90% identity, 27 without GST activity and four from the topologically distinct Kappa class and microsomal GSTs, 55 entries remained. Among these, we chose 22 representatives of various classes and distinct species for further analyses. To show more clearly the essential residue for various catalytic types, we compared the full-length sequences—only the first 50 residues of the alignment are shown in Fig 3A (Corpet, 1988; Gouet et al, 2003). In agreement with previous reports, residues tyrosine, serine and cysteine are highly conserved in their respective types. In the atypical-type GST family, the conserved residue was either glycine or alanine, both of which allowed the conjugation of NBD-Cl to GSH in our activity assays (Fig 2D). However, phenylalanine was also found at this site in some putative GSTs that had lost their GST activity, as shown by our enzymatic assays. Alternatively, distinct activities might have evolved for these enzymes, as in the cases of Ure2 and eEF1Bγ in S. cerevisiae, SspA from Yersina pestis and CLIC1 from Homo sapiens. The residues corresponding to Ser129 and His133 of Gtt2 show little conservation in the atypical-type GSTs among species. Several residues in the C-domain have side chains that protrude towards the thiolate group of GSH, bringing some polar side chains close enough to participate in first-sphere interactions (Xiao et al, 1996). Even if these polar side chains were not long enough to interact directly with the thiolate group, a nucleophilic water molecule might substitute, as seen in our structures. However, further enzymatic assays of atypical-type GSTs are needed to demonstrate the universal feature of this novel mechanism.
Figure 3.
Conservation of the amino-terminal regions of various types of GST. Sequences of tyrosine, serine and cysteine types are from Protein Data Bank (www.rcsb.org; PDB codes 1TU7, 2A2R, 1XW6, 1GSU, 2CVD, 2GSQ, 1VF1, 1GUL, 2VO4, 1OYJ, 1AXD, 1GNW, 1PN9, 1R5A, 2C3N, 2LJR, 1E6B, 2CZ2, 2GDR, 1N2A, 1PMT and 1EEM). Sequences of the atypical type are downloaded from the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov). Codes in parentheses are NCBI accession numbers: Saccharomyces cerevisiae Gtt2, Schizosaccharomyces pombe GST I (Q9Y7Q2), S. pombe GST II (O59827), Burkholderia cenocepacia PC184 GST (YP_002095143), Hahella chejuensis KCTC 2396 GST (YP_437484), Methylibium petroleiphilum PM1 GST (YP_001020104), Alpha proteobacterium HTCC2255 GST (ZP_01450455) and Bradyrhizobium sp. ORS278 GST (YP_001202246). GTS, glutathione sulphonate.
In addition to primary sequence alignment, comparative structure analyses of Gtt2 was performed using the DALI Server (www2.ebi.ac.uk/dali; Holm & Sander, 1993). Most of the output entries are from the Delta, Beta, Theta and Zeta classes, which are all cysteine-type or serine-type GSTs, and form an early evolutionary branch of GST family before the tyrosine-type GSTs emerged (Frova, 2006). Although further definite classification is not available, the high structural identity of these proteins implies Gtt2 might have a similar evolutionary path and could have descended from a common archaic ancestor of the Delta, Beta, Theta and Zeta classes. So far, Gtt2 represents the only three-dimensional structure of fungal GST. More information on fungal GST is needed to provide adequate information for determining their origins.
Methods
Crystallization and structure determination. Wild-type Gtt2 and seven mutants were overexpressed in Escherichia coli and purified with Ni–NTA affinity chromatography followed by gel filtration. Crystals of apo-form and ligand-bound Gtt2 were grown by hanging-drop vapour diffusion at 16°C. After flash-freezing at 100 K in liquid nitrogen, data were collected from the crystals using a Rigaku MM007 X-ray generator (1.5418 Å) with a MarResearch 345 image-plate detector. All structures were determined by molecular replacement by using the GST structure (1JLV) from Anopheles dirus (Oakley et al, 2001) with Phaser (McCoy, 2007) in CCP4 (CCP4, 1994). Refinement was performed using REFMAC (Murshudov et al, 1997) and Coot (Emsley & Cowtan, 2004). The overall assessment of model quality was performed using MolProbity (Lovell et al, 2003). Atomic coordinates and structure factors were deposited in the Protein Data Bank (www.rcsb.org) under the accession codes 3ERF, 3ERG and 3IBH. The crystallographic parameters of the three structures are listed in Table 1. All structure figures were prepared with PyMOL (DeLano, 2002). Further details can be found in supplementary information online.
Table 1.
Crystal parameters, data collection and structure refinement
| Apo-form | GTS-bound form | GSH-bound form | |
|---|---|---|---|
| Data collection | |||
| Space group | P3121 | I222 | P3121 |
| Unit cell (Å, °) | 88.3, 88.3, 69.15 | 79.28, 94.09, 125.11 | 88.77, 88.77, 69.1 |
| 90, 90, 120 | 90, 90, 90 | 90, 90, 120 | |
| Molecules per asymmetric unit | 1 | 2 | 1 |
| Resolution range (Å) | 40–2.23 (2.33–2.23)* | 40–2.20 (2.32–2.2) | 40–2.1 (2.18–2.1) |
| Unique reflections | 15,617 (2,003) | 24,059 (3,447) | 18,606 (1,778) |
| Completeness (%) | 98.2 (87.7) | 99.8 (99.7) | 99.3 (96) |
| 〈I/σ(I)〉 | 27 (9.1) | 16.4 (5.9) | 15.5 (5.9) |
| Rmerge ‡ (%) | 4.2 (19.2) | 4.9 (19.4) | 7.7 (24.5) |
| Average redundancy | 6.9 | 4.1 | 4.8 |
| Structure refinement | |||
| Resolution range (Å) | 40–2.23 (2.28–2.23) | 40–2.20 (2.26–2.2) | 40–2.1 (2.15–2.1) |
| R-factor§/R-free∥ (%) | 21.2/24.8 | 21.8/25.4 | 20.8/24.7 |
| Number of protein atoms | 1,653 | 3,306 | 1,653 |
| Number of water atoms | 119 | 224 | 116 |
| RMSD¶ bond lengths (Å) | 0.013 | 0.014 | 0.013 |
| RMSD bond angles (°) | 1.3 | 1.3 | 1.4 |
| Mean B factors (Å2) | 38 | 27.5 | 30.5 |
| Ramachandran plot# | |||
| Most favoured (%) | 98.1 | 99 | 99 |
| Additional allowed (%) | 1.4 | 0.5 | 0.5 |
| Outliers (%) | 0.5 | 0.5 | 0.5 |
| Protein Data Bank entry | 3ERF | 3ERG | 3IBH |
| *The values in parentheses refer to statistics in the highest bin. | |||
| ‡Rmerge=∑hkl∑i∣Ii(hkl)−〈I(hkl)〉∣/∑hkl∑iIi(hkl), where Ii(hkl) is the intensity of an observation and 〈I(hkl)〉 is the mean value for its unique reflection; summations are over all reflections. | |||
| §R-factor=∑h(Fo(h)–Fc(h))/∑hFo(h), where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively. | |||
| ∥R-free was calculated with 5% of the data excluded from the refinement. | |||
| ¶RMSD from ideal values. | |||
| #Categories were defined by MolProbity. | |||
| GSH, glutathione; GTS, glutathione sulphonate; RMSD, root-mean-square deviation. | |||
Enzymatic assays. Wild-type Gtt2ΔN18 and mutants were used for qualitative activity assays with NBD-Cl (4-chloro-7-nitro-2,1,3-benzoxadiazole) as co-substrate. Absorbance of the product, GS-NBD, was measured at 419 nm (ɛ419=14.5 mM−1 cm−1) with a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA, USA) equipped with a cuvette holder fixed at 25°C. Experiments were performed in 200 μl (final volume) of 0.1 M sodium acetate buffer, pH 5.0, containing 2 mM GSH, 0.5 mM NBD-Cl and 0.5–4 μM protein (Ricci et al, 1994). Reactions were monitored at 14 s intervals for a total of 140 s.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Jiang Yu at University of Science and Technology of China for data collection. We are grateful to all the developers of the CCP4 Suit, ESPript, MolProbity and PyMOL. This work was supported by the National Natural Science Foundation of China (Program 30870490) and the Ministry of Science and Technology of China (Projects 2006CB910202 and 2006CB806501).
Footnotes
The authors declare that they have no conflict of interest.
References
- Allocati N, Federici L, Masulli M, Di Ilio C (2009) Glutathione transferases in bacteria. FEBS J 276: 58–75 [DOI] [PubMed] [Google Scholar]
- Armstrong RN (1997) Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol 10: 2–18 [DOI] [PubMed] [Google Scholar]
- Board PG, Coggan M, Wilce MC, Parker MW (1995) Evidence for an essential serine residue in the active site of the Theta class glutathione transferases. Biochem J 311: 247–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chen WJ, Graminski GF, Armstrong RN (1988) Dissection of the catalytic mechanism of isozyme 4-4 of glutathione S-transferase with alternative substrates. Biochemistry 27: 647–654 [DOI] [PubMed] [Google Scholar]
- Collinson EJ, Grant CM (2003) Role of yeast glutaredoxins as glutathione S-transferases. J Biol Chem 278: 22492–22497 [DOI] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Dourado DF, Fernandes PA, Mannervik B, Ramos MJ (2008) Glutathione transferase: new model for glutathione activation. Chemistry 14: 9591–9598 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Frova C (2006) Glutathione transferases in the genomics era: new insights and perspectives. Biomol Eng 23: 149–169 [DOI] [PubMed] [Google Scholar]
- Garcera A, Barreto L, Piedrafita L, Tamarit J, Herrero E (2006) Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem J 398: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31: 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig WH, Jakoby WB (1981) Assays for differentiation of glutathione S-transferases. Methods Enzymol 77: 398–405 [DOI] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR (2005) Glutathione transferases. Annu Rev Pharmacol Toxicol 45: 51–88 [DOI] [PubMed] [Google Scholar]
- Hol WG (1985) Effects of the alpha-helix dipole upon the functioning and structure of proteins and peptides. Adv Biophys 19: 133–165 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50: 437–450 [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick S, O'Sullivan SM, Sheehan D (2005) Glutathione transferase-like proteins encoded in genomes of yeasts and fungi: insights into evolution of a multifunctional protein superfamily. FEMS Microbiol Lett 242: 1–12 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neuefeind T, Huber R, Dasenbrock H, Prade L, Bieseler B (1997) Crystal structure of herbicide-detoxifying maize glutathione S-transferase-I in complex with lactoylglutathione: evidence for an induced-fit mechanism. J Mol Biol 274: 446–453 [DOI] [PubMed] [Google Scholar]
- Oakley AJ, Harnnoi T, Udomsinprasert R, Jirajaroenrat K, Ketterman AJ, Wilce MC (2001) The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B. Protein Sci 10: 2176–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter MA, Hall JR, Locke JC, Jensen JH, Molina PA (2006) Hydrogen bonding is the prime determinant of carboxyl pKa values at the N-termini of alpha-helices. Proteins 63: 621–635 [DOI] [PubMed] [Google Scholar]
- Ricci G, Caccuri AM, Lo Bello M, Pastore A, Piemonte F, Federici G (1994) Colorimetric and fluorometric assays of glutathione transferase based on 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole. Anal Biochem 218: 463–465 [DOI] [PubMed] [Google Scholar]
- Sheehan D, Meade G, Foley VM, Dowd CA (2001) Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J 360: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg G, Board PG, Mannervik B (1991) Mutation of an evolutionarily conserved tyrosine residue in the active site of a human class alpha glutathione transferase. FEBS Lett 293: 153–155 [DOI] [PubMed] [Google Scholar]
- Xiao G, Liu S, Ji X, Johnson WW, Chen J, Parsons JF, Stevens WJ, Gilliland GL, Armstrong RN (1996) First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase. Biochemistry 35: 4753–4765 [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang NN, Yin PD, Cui PX, Zhou CZ (2008) Glutathionylation-triggered conformational changes of glutaredoxin Grx1 from the yeast Saccharomyces cerevisiae. Proteins 72: 1077–1083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information