Abstract
Tumour cannibalism is a characteristic of malignancy and metastatic behaviour. This atypical phagocytic activity is a crucial survival option for tumours in conditions of low nutrient supply, and has some similarities to the phagocytic activity of unicellular microorganisms. In fact, Dictyostelium discoideum has been used widely as a model to study phagocytosis. Recently, phg1A has been described as a protein that is primarily involved in the phagocytic process of this microorganism. The closest human homologue to phg1A is transmembrane 9 superfamily protein member 4 (TM9SF4). Here, we report that TM9SF4 is highly expressed in human malignant melanoma cells deriving from metastatic lesions, whereas it is undetectable in healthy human tissues and cells. TM9SF4 is predominantly expressed in acidic vesicles of melanoma cells, in which it co-localizes with the early endosome antigens Rab5 and early endosome antigen 1. TM9SF4 silencing induced marked inhibition of cannibal activity, which is consistent with a derangement of intracellular pH gradients, with alkalinization of acidic vesicles and acidification of the cell cytosol. We propose TM9SF4 as a new marker of malignancy, representing a potential new target for anti-tumour strategies with a specific role in tumour cannibalism and in the establishment of a metastatic phenotype.
Keywords: cancer, cannibalism, melanoma, oncogene, TM9SF4
Introduction
Phagocytic cells with cannibalistic behaviour were identified in malignant tumours more than a century ago (Steinhaus, 1891; Stroebe, 1892). Cannibal tumour cells, however, might be described as tumour cells containing engulfed material of different origin in large vacuoles that often push the nucleus to the periphery, therefore giving these cells a crescent-shaped form and prompting names such as ‘bird-eye cells' or ‘signet-ring cells' (Fais, 2007). However, the significance and mechanisms underlying tumour cannibalism are mostly unknown. Detection of cannibal cells has been related to poor prognosis in human tumours of various histology, including breast carcinoma (Marin-Padilla, 1977), haematological malignancies (Kadin et al, 1981), bladder cancer (Kojima et al, 1998), medulloblastoma (Youness et al, 1980), gastric adenocarcinomas (Caruso et al, 2002), melanoma and skin carcinomas (Monteagudo et al, 1997; Breier et al, 1999). We have recently seen that the cannibalism of apoptotic or live lymphocytes (Lugini et al, 2003, 2006) is exclusive to metastatic melanomas, which are able to feed on the ingested material (Fais, 2007). Interestingly, the tumour cannibalism shows many similarities to the phagocytic activity of unicellular microorganisms, such as amoebas. The cellular slime mould Dictyostelium discoideum has been previously used as a model organism to study phagocytosis (Duhon & Cardelli, 2002; Maniak, 2003). D. discoideum dysphagia mutants revealed that the phg1 gene has a crucial role in the phagocytic process (Cornillon et al, 2000; Benghezal et al, 2003). The closest human homologue to phg1 is transmembrane 9 superfamily protein member 4 (TM9SF4). Phg1A/TM9SF4 functions have been conserved throughout evolution because defective phagocytosis was also highlighted in circulating plasmatocytes of phg1A/TM9SF4-null mutant Drosophila (Bergeret et al, 2008).
TM9SF4 belongs to the transmembrane 9 superfamily, a highly conserved family of proteins that are characterized by the presence of a large variable hydrophilic amino-terminal domain and 9–10 putative transmembrane domains. According to secondary structure provisional models, the additional transmembrane domain is located at the N terminus before the hydrophilic domain and is predicted to be a signal peptide, probably with a cleavage site (Chluba-de Tapia et al, 1997; Schimmöller et al, 1998). TM9SF4 function and localization in human cells has not yet been described. In this study, we show that: TM9SF4 is highly expressed in the early endosomal compartment of melanoma cells, whereas it is undetectable in healthy skin tissues and peripheral blood lymphocytes; this protein has a crucial role in the phagocytic/cannibal behaviour of metastatic melanoma cells; and it is involved in the regulation of intracellular pH.
Results And Discussion
Detection and characterization of TM9SF4
Hydropathy analysis of TM9SF4 using the TopPred prediction server (Kyte & Doolittle, 1982) revealed a mostly hydrophilic, N-terminal portion that extends up to amino acid 262, whereas the remaining portion of the protein is extremely hydrophobic and contains nine potential transmembrane domains.
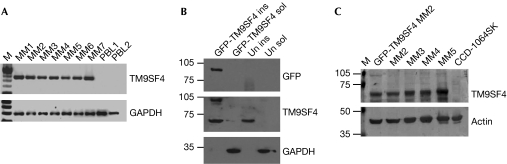
TM9SF4 expression was analysed by reverse transcriptase PCR (RT–PCR) performed on a panel of human melanoma cell lines (MM1–MM7). MM1–MM7 cells are derived from metastatic lesions and have been previously characterized for their ability to cannibalize other cells (Lugini et al, 2006), as compared with human peripheral blood lymphocytes (PBL1 and PBL2). The results showed that TM9SF4 amplicons were detectable in human melanoma cells but undetectable in PBL (Fig 1A). This set of data allowed us to presume that TM9SF4 expression could be related to the metastatic phenotype of melanoma cells. To support this finding, we used RT–PCR to analyse the expression of TM9SF4 transcripts in metastatic melanoma cells (MM1 and MM2), as compared with two primary melanoma cell lines (PM1 and PM2), previously described as non-phagocytic (Lugini et al, 2003, 2006), and compared with normal melanocytes. The results showed that TM9SF4 mRNA, expressed in both metastatic melanoma cell lines, was undetectable in primary melanoma cells and in normal melanocytes (supplementary Fig S1 online), therefore supporting the hypothesis that TM9SF4 might represent a new marker of malignancy in human melanomas.
Figure 1.
Detection of TM9SF4. (A) RT–PCR analysis of TM9SF4 and GAPDH in seven metastatic melanoma cell lines (MM1–MM7) and peripheral blood lymphocytes from two donors (PBL1 and PBL2). (B) Western blot analysis of GFP-TM9SF4 and GAPDH in Triton-insoluble (lane 1) and Triton-soluble (lane 2) fractions of GFP-TM9SF4-transfected MM1 cells, and in Triton-insoluble (lane 3) and Triton-soluble (lane 4) fractions of untransfected MM1 cells. (C) TM9SF4 western blot analysis of GFP-TM9SF4-transfected MM2 cells, four metastatic melanoma cell lines (MM2–MM5), and CCD-1064SK human skin fibroblasts. The loading amount was controlled by immunodetection of actin. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein; ins, insoluble; M, molecular size marker; MM, metastatic melanoma; RT–PCR, reverse transcriptase PCR; sol, soluble; TM9SF4, transmembrane 9 superfamily protein member 4; Un, untransfected.
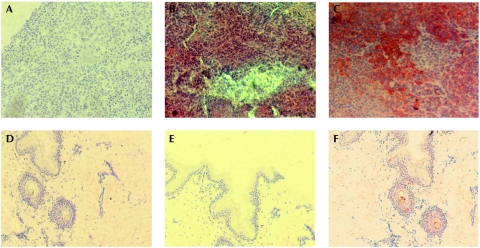
To characterize the protein, we cloned complementary DNA derived from MM1 cells in bacterial expression vectors to obtain TM9SF4 amino acids 1–265 fused to a 6His N-terminal tag (6H-Nt-TM9SF4). Western blot analysis of recombinant protein resulted in a translation product of about 30 kDa absent in control bacterial whole lysates. Consequently, 6H-Nt-TM9SF4 was used to immunize mice and generate TM9SF4 antibodies. The reactivity of antibodies was evaluated by western blot analysis of the purified 6H-Nt-TM9SF4 fusion protein immunoblotted with both 6His and TM9SF4 antibodies (supplementary Fig S2 online). TM9SF4 antibodies were subsequently used for western blot analysis of Triton-insoluble and Triton-soluble fractions of both MM1 cells transfected with a green fluorescent protein (GFP)-tagged full-length TM9SF4 (GFP-TM9SF4) and the non-transfected control. The use of a GFP antibody revealed a single specific translation product in the 100 kDa range, whereas TM9SF4 antibodies recognized both the GFP-tagged and endogenous TM9SF4 corresponding to an approximately 70 kDa protein detectable in both cell lines (Fig 1B). These results were supported by western blot analysis of four melanoma cell lines (MM2–MM5), as compared with GFP-TM9SF4-transfected MM2 cells and healthy skin fibroblasts (CCD-1064SK) that were used as positive and negative controls, respectively. TM9SF4 was detectable in melanoma cells but not detectable in skin cells (Fig 1C). These results were supported by immunocytochemical analysis that detected TM9SF4 in melanoma cells (supplementary Fig S3A online) but not in PBL or macrophages (supplementary Fig S3B,C online). Non-reactive pre-immune mouse serum was included as a negative control of the antibody (supplementary Fig S2D–F online). Furthermore, immunohistochemical analysis of malignant melanoma tissues showed a clear positive staining for TM9SF4 (Fig 2B) and for the positive control marker GP100 (Fig 2C; Cormier et al, 1998). Healthy skin did not show any detectable TM9SF4 staining (Fig 2E), whereas it stained positively for Ezrin, which is known to be expressed in normal skin (Fig 2F; Ilmonen et al, 2005). Pre-immune mouse serum staining was negative in both tissues (Fig 2A,D). The results were confirmed by five consecutive experiments that did not show TM9SF4 staining in normal skin. Furthermore, immunocytochemical experiments revealed an absence of TM9SF4 staining in cultures of normal human melanocytes (data not shown), thus confirming the data from RT–PCR analysis (supplementary Fig S1 online) and data obtained using normal human leucocytes (supplementary Fig S3 online).
Figure 2.
Immunohistochemical analysis of TM9SF4. Immunohistochemical analysis of malignant melanoma tissues stained with (A) pre-immune mouse serum, (B) mouse anti-TM9SF4 serum and (C) anti-GP100. Immunohistochemical analysis of healthy skin stained with (D) pre-immune mouse serum, (E) mouse anti-TM9SF4 serum and (F) Ezrin antibody. Nuclei were stained with Mayer's haematoxylin. Original magnification, ×10. Results are representative of five experiments. TM9SF4, transmembrane 9 superfamily protein member 4.
Subcellular localization of TM9SF4-expressing cells
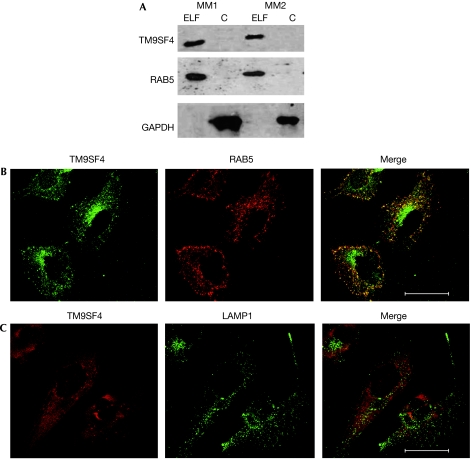
Preliminary indications about subcellular distribution of TM9SF4 were suggested by the western blot result shown in Fig 1B, in which TM9SF4 was detectable in Triton-insoluble fractions of whole-cell extracts: the glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-negative fraction corresponded to the portion of extracts enriched in cytoskeleton proteins and cytoskeleton-associated membrane proteins. For a more detailed analysis of TM9SF4 subcellular localization, MM2 cells were subjected to subcellular fractionation on sucrose density gradients and the distribution of TM9SF4 was analysed by western blotting (Fig 3A). The results revealed that TM9SF4 was mainly recovered in the endo-lysosomal fraction of melanoma cells, whereas it was undetectable in the remaining fraction. By using an adapted protocol of the Qproteome kit (Qiagen), which allowed us to separate cytosol, organelles and plasma membrane fractions, we showed that TM9SF4 localized in the organellar fraction of melanoma cells (supplementary Fig S4 online). To support this finding, the co-localization of TM9SF4 with the following subcellular markers was assessed by confocal laser scanning microscopy analysis: Rab5 and early endosome antigen 1 (EEA1; early endosomal markers that are also involved in phagocytic processes; Alvarez-Dominguez & Stahl, 1999; Nakaya et al, 2006), lysomal-associated membrane protein 1 (Lamp1; late endosomes and lysosomes) and mitochondrial marker MitoTracker. The results showed that TM9SF4 co-localized with Rab5 (Fig 3B) and EEA1 (data not shown), whereas it did not co-localize with LAMP1 (Fig 3C) or MitoTracker (supplementary Fig S5 online).
Figure 3.
Subcellular localization of TM9SF4. (A) Western blot analysis of TM9SF4, GAPDH and RAB5 for detection in sucrose gradient-separated subcellular fractions. ELF and C fractions of MM1 cells (lanes 1 and 2) and MM2 cells (lanes 3 and 4). (B) CLSM analysis of TM9SF4 (green) and RAB5 (red). (C) CLSM analysis of TM9SF4 (red) and LAMP1 (green). Yellow/orange areas in merged picture indicate co-localization. Scale bars, 47.62 μm. C, cytosolic; ELF, endo-lysosomal fraction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; CLSM, confocal laser scanning microscopy; MM, metastatic melanoma; TM9SF4, transmembrane 9 superfamily protein member 4.
TM9SF4 is a tumour cannibalism-associated protein
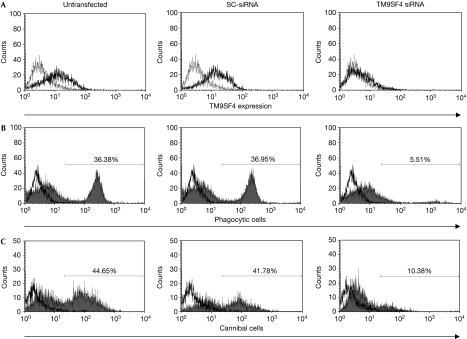
By using an analogy with the D. discoideum Phg1A, we hypothesized a possible role for TM9SF4 in the phagocytic/cannibal activity of human metastatic melanoma cells. To test this hypothesis, we transfected MM2 and MM3 cells with either small interfering RNA (siRNA) for TM9SF4 (TM9SF4 siRNA) or a negative control scrambled sequence (SC-siRNA). The effective knockdown of TM9SF4 was confirmed by a marked reduction of TM9SF4 expression when analysed by fluorescence-activated cell sorting (FACS) 48 h after transfection (Fig 4A). To assess the phagocytic activity of cells knocked down for TM9SF4, fluorescein isothiocyanate (FITC)-stained Saccaromyces cerevisiae cells or dihydrorhodamine 123-stained living lymphocytes were added 48 h after siRNA transfection. The levels of phagocytic activity (against yeasts) and cannibal activity (against lymphocytes) were assessed by FACS, as previously described (Lugini et al, 2003, 2006). The results showed that knocking down TM9SF4 virtually abolished the phagocytic and cannibal activities of melanoma cells (Fig 4 and Table 1). For this reason, we decided to propose a new name for TM9SF4, which is tumour cannibalism associated protein 1 (TUCAP1).
Figure 4.
TM9SF4 silencing and phagocytosis. (A) FACS analysis of TM9SF4 expression in untransfected, SC-siRNA-transfected or TM9SF4 siRNA-transfected MM2 cells. (B) FACS analysis of phagocytic activity in untransfected, SC-siRNA-transfected or TM9SF4 siRNA-transfected MM2 cells incubated with FITC-stained yeast cells. (C) FACS analysis of cannibal activity in untransfected, SC-siRNA-transfected or TM9SF4 siRNA-transfected MM2 cells incubated with DHR123-stained lymphocytes. Only melanoma cell fluorescence emission was evaluated. Numbers reported are the percentage of cells containing FITC-stained yeast cells or DHR123-stained lymphocytes. DHR123, dihydrorhodamine 123; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; MM, metastatic melanoma; SC, scrambled; siRNA, short interfering RNA; TM9SF4, transmembrane 9 superfamily protein member 4.
Table 1.
The role of TM9SF4 in phagocytosis/cannibalism
| Yeasts | Live lymphocytes | |||
|---|---|---|---|---|
| SC-RNAi | TM9SF4 RNAi | SC-RNAi | TM9SF4 RNAi | |
| MM2 | 37.5±0.8 | 3.8±2.0 | 39.8±13.5 | 11.8±7.5 |
| MM3 | 34.3±3.2 | 7.2±5.6 | 44.0±7.2 | 12.5±1.6 |
| Phagocytic/cannibal activity of scrambled siRNA transfected (SC-siRNA) and TM9SF4 silenced (TM9SF4 siRNA) MM2 and MM3 cells against FITC-stained yeast cells and DHR123-stained live lymphocytes. The phagocytic activity was expressed as a percentage of phagocytic cells. Numbers are mean±s.d. of four experiments. | ||||
| DHR123, dihydrorhodamine 123; FITC, fluorescein isothiocyanate; MM, metastatic melanoma; RNAi, RNA interference; SC, scrambled; siRNA, short interfering RNA; TM9SF4, transmembrane 9 superfamily protein member 4. | ||||
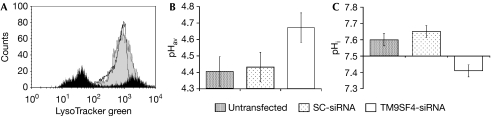
TM9SF4 and endosomal pH
The presence of an increased number of acidic vesicles has been reported to be associated with the malignant behaviour of tumour cells (Montcourrier et al, 1994; Luciani et al, 2004). Ion channels are main regulators of endosomal acidification in phagocytic cells (Moreland et al, 2006; Carrithers, 2007) and the predicted structure of TM9SF4 makes it possible to hypothesize a role for this molecule as an ion channel or an ion channel regulatory protein that is involved in pH regulation of intracellular vesicles. To test this hypothesis, untransfected, SC-siRNA-transfected and TM9SF4-silenced cells were stained with the acidotropic probe LysoTracker (Invitrogen) green and analysed by FACS. TM9SF4 silencing significantly decreased the internal accumulation of LysoTracker, compared with SC-siRNA-transfected and untransfected control melanoma cells (Fig 5A). To determine the pH of acidic vesicles (pHav) in response to TM9SF4 silencing, we used a FITC-dextran staining and FACS, as described by Nilsson et al (2003) and detailed in the Methods section. The results showed that TM9SF4 silencing induced an increase of the intravesicular pH of melanoma cells (from 4.44±0.051 to 4.67±0.047), whereas the pH of control SC-siRNA transfected cells did not vary significantly (Fig 5B). To further support this finding, we analysed the role of TM9SF4 in cytosolic pH regulation by using the fluorescent probe 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein (BCECF-AM; Nilsson et al, 2003). The results showed that the cytosolic pH (pHi) in TM9SF4 silenced cells significantly decreased from 7.51±0.82 to 7.23±0.94 (n=6), whereas the pHi of SC-siRNA-transfected cells did not vary significantly (Fig 5C).
Figure 5.
TM9SF4 silencing and intracellular pH. (A) FACS analysis of LysoTracker DND-26 staining of untransfected (black line), SC-siRNA-transfected (grey area) and TM9SF4 siRNA-transfected (black area) MM2 cells. This graph is representative of four experiments. (B) pHav measurement in untransfected and SC-siRNA-transfected cells compared with TM9SF4-silenced MM2 cells loaded with the pH-sensitive fluorescent dextrans. (C) BCECF-AM-based pHi measurement in untransfected and SC-siRNA-transfected cells compared with TM9SF4-silenced MM2 cells. BCECF-AM, 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein; FACS, fluorescence-activated cell sorting; MM2, metastatic melanoma 2; pHav, pH value for acidic vesicles; pHi, cytosolic pH; SC, scrambled; TM9SF4, transmembrane 9 superfamily protein member 4.
Mechanisms controlling the pH of intracellular organelles have a crucial role in regulating endocytic and phagocytic processes (Liu et al, 2002; Moreland et al, 2006). Here, we suggest that human TM9SF4, in comparison with other multispanning proteins (for example, Na+/H+ exchanger NHE1, calcium and sodium channels, and the anion channel C3), might have a role in regulating the pH of tumour cell intracellular vesicles. TM9SF4 is highly expressed and fully active in human metastatic cancer cells, suggesting that this protein exerts its specific role in contributing to set up a metastatic phenotype of tumour cells. In fact, it is feasible that the ability to cannibalize other cells, and possibly the ability to ‘nibble' the extracellular matrix, are exclusive properties of highly invasive and metastatic tumour cells, allowing them to remain alive in an often hostile microenvironment with low nutrient and blood supplies. Here, we show that knocking down TM9SF4 leads to a clear inhibition of the phagocytic and cannibal activity of metastatic melanoma cells which is consistent with a decreased acidification of internal vesicles, such as early endosomes, suggesting that it contributes to acidification of these vesicles. TM9SF4 knockdown significantly changes the pH gradients between the extracellular microenvironment and intracellular compartments, suggesting its role in regulating proton fluxes in metastatic melanoma cells. In fact, TM9SF4 is virtually undetectable in normal cells, tissues of different origins (leucocytes, melanocytes and skin) and more differentiated tumour cells, such as those deriving from primary lesions. This suggests an exclusive role of TM9SF4 in metastatic or particularly aggressive cancers. Interestingly, this protein is co-expressed in unicellular microorganisms, with a comparable role in regulating phagocytic activity. It is therefore conceivable that amoebas and extremely malignant tumour cells share some functions and proteins helping survival in hostile conditions (Fais, 2007).
Methods
RT–PCR, TM9SF4 cloning, protein expression and antibody generation. Primers used for TM9SF4 RT–PCR amplification were: 5′-TGTGTGAAACAAGCGCCTTC-3′ and 5′-ATGAGGTGGACGTAGTAGT-3′; and for detecting GAPDH: 5′-CCATGGAGAAGGCTGGGG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′. Primers used to direct TM9SF4 His-tagged N-terminal domain synthesis were: 5′-GAATTCATGTGTGAAACAAGCGCCTT-3′ and 5′-GTCGACAGAAAACCAGTGGATCTG-3′. The PCR product was cloned into a pTopo vector (Invitrogen, Milan, Italy), excised with EcoRI and SalI, and ligated into pTrcHis2 (Invitrogen, Milan, Italy). Recombinant protein was purified using Ni2+–NTA agarose resin (Qiagen, Milan, Italy) and was then used to immunize mice. Primers used to direct GFP-tagged full-length TM9SF4 were: 5′-GAATTCATGTGTGAAACAAGCG-3′ and 5′-GTCGATGTCTATCTTCACAGCATA-3′. The PCR product was cloned into a pTopo vector, excised with EcoRI and SalI, and ligated into a pEGFPN1 vector (Clontech, Saint-Germain-en-Laye, France). Rabbit TM9SF4 polyclonal antibodies were developed by immunizing rabbits with the peptide NH2-adekssctlpegtns-COOH corresponding to TM9SF4 amino acids 221–235.
Western blot analysis of sucrose gradient subcellular fractionation. MM2 cells were homogenized in 250 mM sucrose, 5 mM EGTA and 20 mM HEPES-KOH (pH 7.2) in a Dounce homogenizer and centrifuged for 15 min at 800g. Supernatants were subjected to differential ultracentrifugation (50,000g for 7 min) to obtain a pellet enriched in endolysosomes (P50; Gardella et al, 2002). This pellet was re-suspended in Akt buffer (150 mM NaCl, 20 mM TRIS (pH 7.4), 1% NP-40, 10% glycerol and protease inhibitors) and incubated for 15 min on ice. Cytosol was purified from P50 supernatants by 100,000g ultracentrifugation. Aliquots of subcellular fractions and cell lysates were resolved on 12% sodium dodecyl sulphate polyacrylamide gel electrophoresis. Filters were stained with mouse TM9SF4 antibody, rabbit anti-RAB5 (Santa Cruz Biotechnology, Heidelberg, Germany) and mouse anti-GAPDH (Santa Cruz Biotechnology).
TM9SF4 silencing and transfection. The siRNA duplexes targeting human TM9SF4 were: 5′-GUAUGAUCCUCAUCGUCAUTT-3′ and 5′-AUGACGAUGAGGAUCAUACAG-3′; 5′-AGCGGAUCACAGAAGACUATT-3′ and 5′-UAGUCUUCUGUGAUCCGCUCG-3′; 5′-CGGGUACCAUCGGCUUCUATT-3′ and 5′-UAGAAGCCGAUGGUACCCGTT-3′; and 5′-GGAGCCUUCUGUACGGCAATT-3′ and 5′-UUGCCGUACAGAAGGCUCCTT-3′. siRNAs were obtained from Qiagen and used according to the manufacturer's instructions. AllStars Negative control siRNA (Qiagen) was used as SC-siRNA.
For transfection, MM2 and MM3 melanoma cells were seeded in six-well plates and incubated in normal growth conditions. After 24 h, cells were transfected with 5 nM TM9SF4 siRNA (a pool of the four oligos) or SC-siRNA mixed to HiPerFect Transfection Reagent (Qiagen), according to the manufacturer's instructions. At 48 h after transfection, cells were permeabilized and the level of TM9SF4 expression was evaluated by FACS using rabbit anti-TM9SF4 and Alexa Fluor 488-conjugated anti-rabbit IgG.
Phagocytic/cannibal activity of tumour cells. Melanoma cells were transfected as above. At 48 h after transfection, SC-siRNA-transfected or TM9SF4 siRNA-transfected MM2 and MM3 melanoma cells were incubated at 37°C under 5% CO2 with live lymphocytes stained with 10 μM dihydrorhodamine 123 (Molecular Probes, Milan, Italy; ratio, 1:10) or with FITC-stained yeast cells (ratio, 1:60). After 6 h, medium was removed and melanoma cells were washed, collected and analysed. At least 10,000 events were acquired on a FACSCalibur and analysed using the CellQuestPro software (Becton-Dickinson, Buccinasco, Italy). Melanoma cells showing green fluorescence were considered as phagocytic/cannibal.
Flow cytometric analysis of endo-lysosomal pH. The pHav value was measured by flow cytometry as described previously (Nilsson et al, 2003). Untransfected, SC-siRNA-transfected and TM9SF4 siRNA-transfected cells were collected and pellets were exposed to 0.1 mg/ml FITC–dextran 40,000 (Sigma, St Louis, MO, USA) for 45 min at 37°C under 5% CO2. Cells and pellets were washed once with HBSS and incubated in RPMI medium for an additional 30 min. After washing, pellets were placed on ice with cold Hanks' balanced salt solution (HBSS). To estimate endolysosomal pH values, a standard curve was created for each cell condition, incubating an aliquot of cells with an appropriate volume of modified Britton-Robinson buffer (pH 4.0–7.0) plus 2.5 M sodium azide, 50 mM 2-deoxy-D-glucose and, for each pH, Nigericin was added at a final concentration of 10 μM. A total of 10,000 events for each sample were acquired on a FACSCalibur equipped with a 488 nm argon laser and analysed using CellQuestPro. The pHav was calculated using FL1/FL2 fluorescence mean values, the standard curve and linear equations.
Measurement of intracellular pH. The pHi value was calculated by flow cytometry according to the protocol of Nilsson et al (2003), using the pH probe BCECF-AM. After overnight culture of untransfected, SC-siRNA-transfected and TM9SF4 siRNA- transfected cells, the medium was replaced with a fresh medium containing 20 μM BCECF-AM and cells were incubated for 30 min at 37°C under 5% CO2. Cells were then washed in HBSS and placed on ice. An aliquot of BCECF-AM-loaded cells was incubated with different potassium phosphate buffers (pH 5.5–7.5) in the presence of Nigericin to obtain a calibration curve. A total of 10,000 events for each sample were acquired on a FACSCalibur and analysed using CellQuestPro. The pHi value was calculated using FL1/FL2 fluorescence mean values and standard curve and linear equations.
Other standard procedures are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary information
Acknowledgments
We thank Giulietta Venturi for helpful discussion. TM9SF4 (transmembrane 9 superfamily protein member 4)/TUCAP1 (tumour cannibalism associated protein 1) antibodies and related products are patented (provisional patent US 61/062,453).
Footnotes
The authors declare that they have no conflict of interest.
References
- Alvarez-Dominguez CM, Stahl PD (1999) Increased expression of rab5a correlates directly with accelerated maturation of Listeria monocytogenes phagosomes. J Biol Chem 271: 11459–11462 [DOI] [PubMed] [Google Scholar]
- Benghezal M, Cornillon S, Gebbie L, Alibaud L, Bruckert F, Letourneur F, Cosson P (2003) Synergistic control of cellular adhesion by transmembrane 9 proteins. Mol Biol Cell 14: 2890–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeret E, Perrin J, Williams M, Grunwald D, Engel E, Thevenon D, Taillebourg E, Bruckert F, Cosson P, Fauvarque MO (2008) TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci 121: 3325–3334 [DOI] [PubMed] [Google Scholar]
- Breier F, Feldmann R, Fellenz C, Neuhold N, Gschnait F (1999) Primary invasive signet-ring cell melanoma. J Cutan Pathol 10: 533–536 [DOI] [PubMed] [Google Scholar]
- Carrithers MD (2007) Expression of the voltage-gated sodium channel NaV1.5 in the macrophage late endosome regulates endosomal acidification. J Immunol 178: 7822–7832 [DOI] [PubMed] [Google Scholar]
- Caruso RA, Muda AO, Bersiga A, Rigoli L, Inferrera C (2002) Morphological evidence of neutrophil-tumor cell phagocytosis (cannibalism) in human gastric adenocarcinomas. Ultrastruct Pathol 26: 315–321 [DOI] [PubMed] [Google Scholar]
- Chluba-de Tapia J, de-Tapia M, Jäggin V, Eberle AN (1997) Cloning of a human multispanning membrane protein cDNA: evidence for a new protein family. Gene 197: 195–204 [DOI] [PubMed] [Google Scholar]
- Cormier JN, Hijazi YM, Abati A, Fetsch P, Bettinotti M, Steinberg SM, Rosenberg SA, Marincola FM (1998) Heterogeneous expression of melanoma-associated antigens and HLA-A2 in metastatic melanoma in vivo. Int J Cancer 75: 517–524 [DOI] [PubMed] [Google Scholar]
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Bruckert F, Cosson P (2000) phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J Biol Chem 275: 34287–34292 [DOI] [PubMed] [Google Scholar]
- Duhon D, Cardelli J (2002) The regulation of phagosome maturation in Dictyostelium. J Muscle Res Cell Motil 23: 803–808 [DOI] [PubMed] [Google Scholar]
- Fais S (2007) Cannibalism: a way to feed on metastatic tumors. Cancer Lett 258: 155–164 [DOI] [PubMed] [Google Scholar]
- Gardella S, Andrei C, Ferrera D, Lotti LV, Torrisi MR, Bianchi ME, Rubartelli A (2002) The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep 10: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmonen S, Vaheri A, Asko-Seljavaara S, Carpen O (2005) Ezrin in primary cutaneous melanoma. Mod Pathol 18: 503–510 [DOI] [PubMed] [Google Scholar]
- Kadin ME, Kamoun K, Lamberg J (1981) Erythrophagocytic T gamma lymphoma: a clinicopathologist entity resembling malignant histiocytosis. N Engl J Med 304: 648–653 [DOI] [PubMed] [Google Scholar]
- Kojima S, Sekine H, Fukui I, Ohshima H (1998) Clinical significance of ‘cannibalism' in urinary cytology of bladder cancer. Acta Cytol 42: 1365–1369 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Liu T, Mirschberger C, Chooback L, Arana Q, Dal Sacco Z, MacWilliams H, Clarke M (2002) Altered expression of the 100 kDa subunit of the Dictyostelium vacuolar proton pump impairs enzyme assembly, endocytic function and cytosolic pH regulation. J Cell Sci 115: 1907–1918 [DOI] [PubMed] [Google Scholar]
- Luciani F et al. (2004) Effect of proton pump inhibitor pretreatment on resistance of solid tumors to cytotoxic drugs. J Natl Cancer Inst 96: 1702–1713 [DOI] [PubMed] [Google Scholar]
- Lugini L et al. (2003) Potent phagocytic activity discriminates metastatic and primary human malignant melanomas: a key role of ezrin. Lab Invest 83: 1555–1567 [DOI] [PubMed] [Google Scholar]
- Lugini L et al. (2006) Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res 66: 3629–3638 [DOI] [PubMed] [Google Scholar]
- Maniak M (2003) Fusion and fission events in the endocytic pathway of Dictyostelium. Traffic 4: 1–5 [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M (1977) Erythrophagocytosis by epithelial cells of a breast carcinoma. Cancer 39: 1085–1089 [DOI] [PubMed] [Google Scholar]
- Montcourrier P, Mangeat PH, Valembois C, Salazar G, Sahuquet A, Duperray C, Rochefort H (1994) Characterization of very acidic phagosomes in breast cancer cells and their association with invasion. J Cell Sci 107: 2381–2391 [DOI] [PubMed] [Google Scholar]
- Monteagudo C, Jorda E, Carda C, Illueca C, Peydro A, Llombart-Bosch A (1997) Erythrophagocytic tumour cells in melanoma and squamous cell carcinoma of the skin. Histopathology 31: 367–373 [DOI] [PubMed] [Google Scholar]
- Moreland JG, Davis PA, Bailey G, Nauseef WM, Lamb FS (2006) Anion channels, including clc-3, are required for normal neutrophil oxidative function, phagocytosis, and transendothelial migration. J Biol Chem 281: 12277–12288 [DOI] [PubMed] [Google Scholar]
- Nakaya M, Tanaka M, Okabe Y, Hanayama R, Nagata S (2006) Opposite effects of rho family GTPases on engulfment of apoptotic cells by macrophages. J Biol Chem 281: 8836–8842 [DOI] [PubMed] [Google Scholar]
- Nilsson C, Kagedal K, Johansson U, Ollinger K (2003) Analysis of cytosolic and lysosomal pH in apoptotic cells by flow cytometry. Meth Cell Sci 25: 185–194 [DOI] [PubMed] [Google Scholar]
- Schimmöller F, Díaz E, Mühlbauer B, Pfeffer SR (1998) Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene 216: 311–318 [DOI] [PubMed] [Google Scholar]
- Steinhaus J (1891) Ueber carcinoma-einschlusse. Virchows Arch 126: 533–535 [Google Scholar]
- Stroebe S (1892) Verschiedener cellularer vorgange und erscheinungen in geschwulsten. Beitr Pathol 11: 1 [Google Scholar]
- Youness E, Barlogie B, Ahearn M, Trujillo JM (1980) Tumor cell phagocytosis. Its occurrence in a patient with medulloblastoma. Arch Pathol Lab Med 104: 651–653 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information