Abstract
Mistranslation from confusion of serine for alanine by alanyl-tRNA synthetases (AlaRSs) has profound functional consequences1-3. Throughout evolution, two editing-checkpoints prevent disease-causing mistranslation from confusing glycine or serine for alanine at the active site of AlaRS. In both bacteria and mice, Ser poses a bigger challenge than Gly1,2. One checkpoint is the AlaRS editing center, while the other is from widely distributed AlaXps—free-standing, genome-encoded editing proteins that clear Ser-tRNAAla. The paradox of misincorporating both a smaller (glycine) and a larger amino acid (serine) suggests a deep conflict for nature-designed AlaRS. To understand the chemical basis for this conflict, kinetic and mutational analysis, together with nine crystal structures, provided snapshots of adenylate formation for each amino acid. An inherent dilemma is posed by constraints of a structural design that pins down the α–amino group of the bound amino acid using an acidic residue. This design, of more than 3 billion years, creates a serendipitous interaction with the serine OH that is difficult to avoid. Apparently not able to find better architecture for recognition of alanine, the serine misactivation problem was solved through free-standing AlaXps, which appeared contemporaneously with early AlaRSs. The results reveal unconventional problems and solutions arising from the historical design of the protein synthesis machinery.
The twenty (one for each amino acid) aminoacyl-tRNA synthetases play a fundamental role in establishing faithful translation, through close control over the two-step aminoacylation reaction that establishes the genetic-code-linkage between an amino acid and its cognate nucleotide triplet of the tRNA anticodon4,5. Even a mild deviation from the rigorous accuracy of the amino acid-nucleotide triplet relationship, caused by misaminoacylation, is toxic to bacteria and gives rise to serious pathology in the mouse1,2. By sequentially working through two ‘sieves’—first at the site for amino acid activation and then at the center for editing6-8 —overall amino acid specificity is the product of the discrimination achieved at each step9. For example, IleRS misactivates Val to produce Val-tRNAIle. When the valyl moiety is translocated to the center for editing, it snugly fits into a pocket that excludes the sterically larger Ile side chain10. Thus, Val-tRNAIle, but not Ile-tRNAIle, is cleared11. Conversely, ValRS cannot activate Ile, but misactivates the isosteric Thr and uses a separate editing site to clear Thr-tRNAVal,12. Paradoxically, this ‘steric-exclusion’ mechanism does not operate for AlaRS, which activates the smaller (than Ala) glycine, but also activates the sterically larger serine. This misactivation is seen in AlaRSs from bacteria to human2,13.
Two mechanisms correct misaminoacylation of Gly or Ser for Ala. One is associated with the editing center of AlaRS1. The second comes from separate, genome-encoded proteins (AlaXps) that are homologs of the editing domain of AlaRSs3,14. Thus, if misacylated Ser-tRNAAla is produced, it is cleared by the editing center of AlaRS or, failing that, can be cleared by an AlaXp15--either before translocation to or re-sampled from the ribosome16. AlaRS is the only tRNA synthetase that has a widely distributed (through all three kingdoms of life) homolog of its editing domain that is separately encoded by the genome14,17. This observation suggests that overcoming the confusion of Gly or Ser for Ala was a major challenge for nature. Toxicity from an editing defect in AlaRSs in both the mouse and bacteria appears to be more problematic with Ser than with Gly1,2. Consistent with these observations, while AlaRSs edit both mischarged Gly-tRNAAla and Ser-tRNAAla, 1, all investigated AlaXps can hydrolyze Ser-tRNAAla, but only some can hydrolyze Gly-tRNAAla,18 (Supplementary Fig. 2). Plausibly, the additional AlaXp-based editing mechanism developed to address the special challenge of serine for mistranslation.
Because serine is not simply bound, but activated by AlaRS to form seryl adenylate, we set out to obtain co-crystals of the three aminoacyl adenylates with the enzyme (See comment on previous work19 (Supplementary material)). The monomeric N-terminal aminoacylation domain (1-441) of E. coli AlaRS is responsible for synthesis of alanyl-tRNAAla. It is clearly separated from the C-terminal half that is homologous to the freestanding editing protein AlaXp-II. Pursuant to obtaining an effective interface that could enhance the assembly of a stable crystal lattice, a surface leucine-zipper mutation--H104L/Q108L/E112L--was introduced into AlaRS441. Nine structures were solved using this construct. The kinetic parameters for amino acid activation (adenylate synthesis) of the engineered and native protein were closely similar (see Supplementary Fig. 3 and 4 online). The structures reported here are generally referred to as AlaRS441-LZ and G237A AlaRS441-LZ, where G237A is a substitution that affects the shape of the amino acid binding pocket (see below). The AlaRS441-LZ structures with five different ligands were co-crystallized in two different crystal forms. Subsequently, G237A AlaRS441-LZ alone, and bound with 3 ligands, was also crystallized (Supplementary Table 1). All complex structures were solved at ~ 2 Å resolution (1.9-2.2 Å) with an estimated atomic coordinate error of ~ 0.2 Å, with one exception (the complex of G237A AlaRS441-LZ with the seryl-adenylate analog (2.8 Å)).
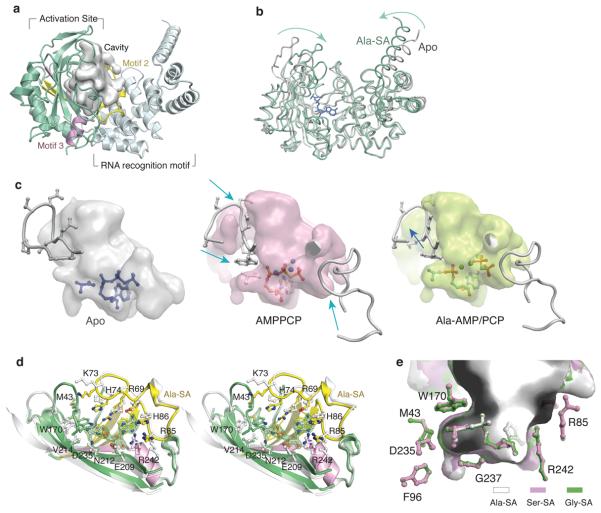
E. coli AlaRS441-LZ adopts a cradle-like shape consisting of the N-terminal (residues 1-249) seven-stranded β-sheet structure (with flanking α-helices) that is characteristic of class II tRNA synthetases, followed by the C-terminal α-helix bundle that forms a tRNA recognition motif (Fig. 1a). The three signature motifs (motif 1, 2, and 3) of class II enzymes are located at residues 5-21, 60-97 and 233-248, respectively. The structure of the E. coli apo-enzyme resembles that of A. aeolicus AlaRS (rmsd= 2.55 Å over 407 Cα positions). Three of the 5 complexes with wild-type AlaRS441-LZ included those with the ATP analog AMPPCP (adenyl 5′-(β,γ-methylene) triphosphonate) alone, with the alanyl-AMP adenylate and the PPi analog PCP ((β,γ-methylene)pyrophosphate) all bound together, and with the Ala-SA (5′-O-(N-(l -alanyl)-sulfamoyl-adenosine)) adenylate analog. The structures of AlaRS441-LZ in the complexes are closely similar to each other, with rmsd=0.35 Å between the two complexes with the adenylates, and a rms deviation of 0.67 Å between the adenylate complexes and the ATP analogue complex. On the other hand, much larger deviations (rmsd= 2.6- 2.8 Å, 418 Cα) were observed when comparing the complexes with the apo-enzyme. In particular, the enzyme undergoes an apparent ‘open to close’ transition upon ligand binding. This contraction involved not only the activation site but also the C-terminal tRNA recognition motif (Fig. 1b).
Figure 1. Plasticity of active site in AlaRS.
a, Overall structure of the N-terminal catalytic fragment AlaRS441-LZ of E. coli AlaRS, shown with the water-accessible cavity (grey).
b, Global movement of catalytic domain upon ligand binding.
c, Dynamic adjustments of active site pocket of AlaRS. Left, active site cavity of apo AlaRS441-LZ with l-alanine and ATP placed by modeling (blue). Model of apo AlaRS441-LZ is derived from structure of apo G237A AlaRS441-LZ by mutating Ala237 back to Gly. Middle, active site cavity of AMPPCP complex with bound AMPPCP and l-alanine placed by modeling (gray). Right, active site cavity of Ala-AMP/PCP complex with bound Ala-AMP and βγ-PCP. The movements of two active site loops are indicated by arrows.
d, Stereo view of ‘open to close’ active site conformational change upon Ala-SA binding. The difference map of the omitted Ala-SA ligand is contoured at 9.0 σ (green).
e, Amino acid pocket with three different ligands bound in EcAlaRS441-LZ. Three co-crystal structures of EcAlaRS441-LZ with Ala-SA, Ser-SA, and Gly-SA were superimposed. The ligand, active site residues and cavity of each complex structure are colored as indicated. The small cavity containing 3 water molecules is seen below the amino acid moiety of all three bound ligands.
The active site cavity is mainly formed by motif 2, motif 3, and two loop regions connecting β4-β5 and β5-β6, respectively (Fig. 1a, Supplementary Fig. 3). The active site cavities of the various enzyme structures were calculated for different states by the program Hollow19. The solvent accessible surface around the active site was computed with a probe radius of 1.4 Å (approximating a water molecule). This shell defined the ligand-binding space formed by the enzyme. From this calculation, the cavity of the apo-enzyme is seen clearly as wide open (Fig. 1c), with a space at the amino acid binding site much larger than that is required to fit the cognate amino acid (alanine). Binding of AMPPCP induced dramatic contraction of the active center that shrunk the amino acid and the adenylate binding sites (Fig. 1c). With this contraction, the space available for amino acid binding became even smaller than that needed to fit alanine. In particular, when bound to AMPPCP, the space for the amino group of alanine was partially occupied by the indole side chain of Trp170 (Fig. 1c). When alanine bound to the amino acid site together with ATP to form the Ala-AMP adenylate, Trp170 swung slightly back (Fig. 1c and 1d). Thus, the enzyme did not pre-form a rigid amino acid binding pocket, but rather was adjustable to the binding ligands. Replacing Ala-AMP with the Ala-SA analog resulted in a similar structure (rmsd=0.15 Å, for residues within 5 Å of the ligands).
Structures with Ser-SA and Gly-SA had similar resolutions (~1.9 Å) and the same space group (P41212) as the complex with Ala-SA. The structures of the 3 adenylate analog complexes closely resemble each other (max rmsd = 0.18 Å). The calculated active site cavities showed no apparent differences among these three structures, and all cavity-forming residues aligned to the same positions (Fig. 1e). After superimposing the three proteins, the adenylate parts of all three ligands--adenosine, ribose and sulfur--also aligned closely. Thus, all three amino acids bound to almost identical positions in the active site (rmsd=0.14 Å). Only Cβ of serine rotated 0.53 Å, when compared to Cβ of alanine. This rotation allowed the extra hydroxyl group of serine to fit snugly to the lower side of the amino acid pocket. A small cavity right under the amino acid site is partially filled by 3 stable bound water molecules (found in all 3 complex structures, Fig. 1e). No apparent change of pocket size upon binding of serine (no expansion) or glycine (no shrinkage) occurred.
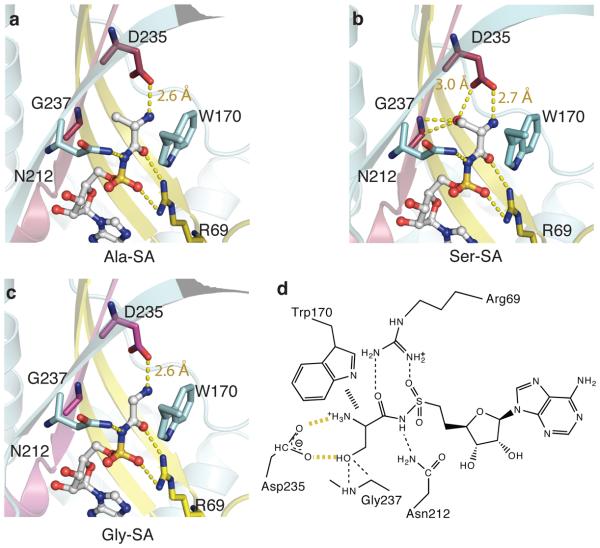
Similar as alanine, the α-amino group of serine forms a hydrogen bond with the carboxyl group of Asp235 (Fig. 2a, 2b and 2d). Also, the carboxyl group of serine forms a hydrogen bond with the class II invariant Arg69 on motif 2. The same Arg69 forms a salt-bridge with the α-sulfate of the adenylate analog, and this interaction is critical for catalyzing the activation reaction of class II enzymes20,21. A conserved Asn212 is hydrogen-bonded with the bridging nitrogen between the seryl moiety and AMP (substituted by O in the natural adenylate). When an amino acid was absent, the Asn212 side chain swung to bind to the α-phosphate of AMPPCP. These interactions are also observed in the structures of AlaRS441-LZ with Ala-SA and Gly-SA, with almost all coordinates of the AlaRS441-LZ residues within the binding pocket being the same, regardless of the bound amino acid (Fig. 2a and 2c). This common main chain conformation may explain why cognate and noncognate amino acids have similar kcat values and are mainly differentiated by their respective Km values22.
Figure 2. Alanine active site with intrinsic design defect that mis-binds serine.
a-c, Interactions with l-alanine, l-serine and glycine as shown in their ligand-bound structures. Two extra interactions are formed between AlaRS and the γ-OH of serine: Asp235(COO−) -- Ser(OH) and Gly237(N) -- Ser (OH). A contact between Gly237(Cα) -- Ser(OH) is also shown (3.3 Å). Asp235 also forms a common hydrogen bond with α-amino group of all three amino acids.
d, Diagram of the interactions between the amino acid moiety of Ser-SA and the synthetase. The aromatic ring of Trp170 stacks with the backbone of amino acid substrates.
In contrast to serine, α-aminobutyrate is not activated by AlaRS, possibly because the larger size of CH3 (compared with OH) blocks entry of α-aminobutyrate into the active site13. However, our structures show that serine forms interactions (with the enzyme) that are not possible with α-aminobutyrate. Two extra H-bonds pin down the -OH. One is from the G237 backbone NH, while the other is from the carboxyl of Asp235 (Fig. 2b). As mentioned before, the carboxyl of Asp235 (conserved in all AlaRSs) also holds the α–NH3+group of serine, alanine, and glycine. (This finding, in the present work, sheds light on the lethality of a previously described D235A substitution in E. coli AlaRS, which kinetic analysis suggested was due in large part to D235's role in transferring the aminoacyl group from the adenylate to tRNA23, probably by stabilizing the transition state24. ) So, the necessity of holding the α-amino group (of the cognate substrate alanine) through an acidic group creates the dilemma of not being able to avoid serendipitous binding to the serine OH. The interactions flowing from Asp235 are major determinants for serine binding to AlaRS (see below).
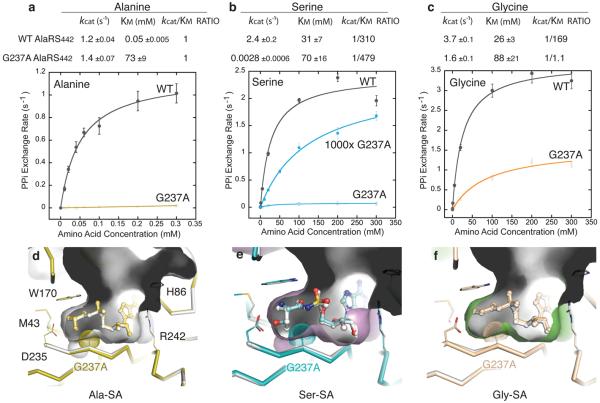
A close inspection of the structure showed that the serine OH is only 3.3 Å away from the Cα of G237 in motif 3, β7 (Fig. 2b). Binding of serine does not induce apparent movement of β7, which is the central β-strand in the class II-defining β-sheet cradle and is, therefore, the least flexible compared to other parts of the cradle. This lack of flexibility may explain why the Km for serine is 600-fold higher than that for alanine (Fig. 3a). Because of the close approach of the Ser OH to the Cα of G237, the pocket would likely be shrunk by substituting a slightly bulkier residue for G237. Pursuant to this idea, G237A AlaRS441 (without leucine zipper) was made.
Figure 3. Mutant enzyme with a smaller pocket retains serine binding affinity.
a-c, Kinetic parameters for amino acid activation of G237A and WT AlaRS441. kcat/KM ratios of noncognate serine and glycine were calculated against cognate alanine for each protein, separately. Error bars are s.d., n ≥ 3.
d-f, G237A AlaRS441-LZ structures in complex with three different ligands are compared with the corresponding WT AlaRS441-LZ complex structures (white). For clarity, only the active site cavities of WT AlaRS441-LZ structures are shown here.
In the standard amino acid dependent ATP-PPi exchange assay (Fig. 3a-c), which follows the rate of synthesis of the adenylate. G237A AlaRS441 had a sharply elevated Km for alanine (1,250-fold increase) (Fig. 3a). This sharp drop in affinity for alanine is expected, because the distance between the Cα of G237 and Cβ of alanine is 3.8 Å, and the extra β-CH3 present in G237A AlaRS441 would crowd the bound alanine. Next, we found that G237A AlaRS441 showed only a small change in the Km for glycine. This result is consistent with the introduced extra β-CH3 of G237A AlaRS441 affecting only the β-position of a bound amino acid substrate (Fig. 3c). Interestingly, although serine is the bulkiest among the three amino acids, G237A AlaRS441 bound to serine with a similar affinity as did wild-type AlaRS441 (Fig. 3b). Thus, within limits, the pocket size does not play a key role for serine binding.
To achieve further clarity on the reason for serine being insensitive to the shrinkage of the pocket size, we crystallized G237A AlaRS441-LZ bound to each analog--Ala-SA, Gly-SA, and Ser-SA (Fig. 3d-f). As expected, the G237A mutation introduced bulk into the alanine-binding pocket, with little other change in the pocket or surrounding atoms. The steric crowding of alanine with the introduced Cβ in G237A AlaRS441-LZ is apparent in the small separation of Cβ of alanine from Cβ of A237 (3.6 Å, Fig. 3d). This crowding easily explains the sharply elevated Km for alanine of G237A AlaRS441-LZ.
As expected, bound Gly-SA was little affected by the extra bulk from the β-methyl introduced at position 237 in G237A AlaRS441-LZ (Fig. 3f). For that reason, the Km for glycine is virtually unchanged. However, in the case of serine, crowding by the introduced Cβ of G237A AlaRS441-LZ is clear And yet, while crowding at the Cβ positions of A237 and serine occurs (which breaks the H-bond between backbone NH of G237A and the serine OH (extends separation to 4.0 Å)), the carboxyl of Asp235 remains H-bonded to the serine OH and the α-NH3+(Fig. 3e). Thus, the bifurcated interactions between Asp235 and serine appear to be a major cause for the inability of the enzyme to evolve to a higher discrimination against serine.
Inspection of the kcat and kcat/Km values for the three amino acids, with native AlaRS441 and G237A AlaRS441, further showed the difficulty of resolving the dilemma posed by the structural design of the enzyme. Because of the extra bulk introduced by the G237A substitution, the carboxyl of serine shifts 0.3 Å toward motif 3 (Fig. 3e) and this shift, in turn, may explain why the kcat for serine activation is greatly reduced (Fig. 3b). However, the desirable outcome of reducing the kinetic activation of serine comes at a high price. The shrinking of the pocket (in hopes of excluding serine) sharply raised the Km for alanine, while at the same time did not greatly perturb the Km for serine, or did not provide a sufficient offsetting reduction in kcat for serine. The problem is further compounded with glycine (as expected, the kcat/Km ratio for glycine is least affected by the mutation), which is over 150-times more competitive with alanine, with G237A AlaRS441 than with wild-type AlaRS441.
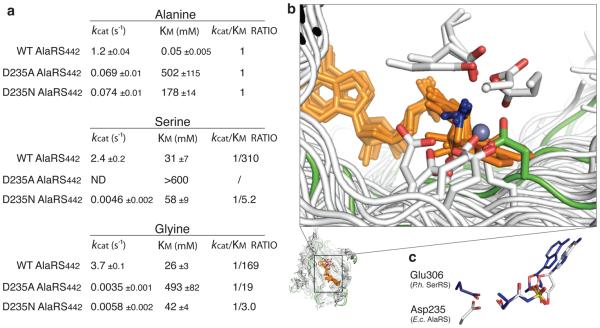
Because D235 is the major reason for the binding of serine in the pocket for alanine, we investigated whether this specificity problem could be solved by changing (D235E) or removing (D235N, D235Q) the acidic group or the side chain (D235A) of D235 (Fig. 4a, Supplementary Fig. 6, and Supplementary Material). None of these substitutions improved the Ala versus Ser discrimination. For example, misincorporating serine, relative to incorporating alanine, was much greater with the D235N and D235A mutants than with the wild type enzyme (Supplementary Fig. 6 and 7). Collectively, these results further highlight the dilemma that creates the serine paradox, and how difficult it is to bypass Asp235.
Figure 4. Critical role of Asp235 shows dilemma for AlaRSs.
a, Kinetic parameters for amino acid activation of D235A and D235N.
b, Various designs of acidic residues that recognize α-amino group of amino acids in Class II AARSs, which structures are aligned by their amino acid substrates (orange, with α-amino groups in blue). The central β-sheets are shown (AlaRS in green, others in white). The acidic residues are shown with carboxyl groups in red.
c, Ser-SA ligands in the active site of E. coli (E.c.) AlaRS and Pyrococcus horikoshii (P.h.) SerRS29. Both acidic residues form bifurcated interactions with the α-amino group and γ-OH of serine.
While they are continuously challenged by canonical and non-canonical amino acids, AARSs control genetic code fidelity on the protein synthesis production line. Only AlaRS and ThrRS are known to misactivate both smaller and larger non-cognate amino acids. However, the problematic larger amino acid (hydroxynorvaline) for ThrRS is not naturally occurring25. For that reason, ThrRS has no selective pressure for rejecting hydroxynorvaline. In contrast, AlaRSs are challenged by canonical amino acids, both the smaller glycine and the larger serine.
The work described above shows that this ‘serine paradox’ is rooted in a dilemma that flows out of the ancient design of AlaRSs. For both class I and class II enzymes, the ancient scaffold simultaneously captures the α-amino and α-carboxyl groups of the amino acid substrate. A natural consequence of this circumstance is the need for an acidic residue to pin down the α-NH3+. In contrast to previous understanding26, the way this is done is idiosyncratic to the synthetase, and many synthetases found alternative residues during evolution (Fig. 4b and Supplementary Fig. 8). Asp235 is universally conserved in AlaRSs across all three kingdoms of life. This deeply rooted architecture causes the serine-interaction paradox for AlaRSs, and the interaction of Asp235 with the serine OH is so robust that it was retained even after creation of a shrunken pocket, with the G237A substitution (Fig. 3). Interestingly, similar to AlaRS, the groups that bind the α-NH3+ in SerRS and ThrRS (glutamate or zinc) are also used to recognize the γ-OH of their cognate substrates25,27 (Fig. 4c).
The structural architecture of class II (and class I) tRNA synthetases goes back to the last universal common ancestor ~ 3.5 billion years ago. Asp235 has been in place in AlaRSs for that entire time. Despite billions of years of evolution, an alternative to Asp235, within the context of the structural scaffold of class II enzymes, was not found. Apparently unable to find a better architecture for recognition of alanine, the problem of serine misactivation was solved through the free-standing AlaXps. (Indeed, in a bacterial model, AlaXp rescued a cell from serine toxicity15) These proteins appeared contemporaneously with the early AlaRSs17. No doubt this solution removed much of the evolutionary pressure to find a variation of the structural architecture of AlaRS that would solve the serine paradox (Supplementary Fig. 1).
METHODS SUMMARY
To aid crystallization, a ‘leucine-zipper’ mutation28 (His104Leu, Gln108Leu and Glu112Leu) was created in an active 441 amino acid fragment of E. coli AlaRS. Wild-type and mutant E.coli AlaRS441-LZ proteins were overexpressed from Escherichia coli and purified by chromatography. The ligand complexes were generated by mixing at 4 °C the protein with Ala-SA, Ser-SA, Gly-SA, AMPPCP, or alanine and AMPPCP (together) with 10 mM MgCl2 and 10 mM β-mercaptoethanol. Crystals were obtained by the micro-sitting-drop vapor diffusion method. All wild-type and mutant AlaRS441-LZ complex structures were determined by molecular replacement using the apo A. aeolicus AlaRS structure (Protein Data Bank accession code 1RIQ) as the initial model. The ATP-pyrophosphate exchange assays were carried at 22 °C in a buffer composed of 100 mM Tris-HCl (pH 8.0), 1 mM ATP, 10 mM MgCl2, 10 mM KF, 0.1 mg/ml BSA, 5 mM β-mercaptoethanol, and 0.5 mM Na-32PPi. Linear initial rates were determined using measurements of incorporation of 32PPi into ATP at 4 sequential time intervals of 3 min each, and the data were fit to the Michaelis-Menten equation using KaleidaGraph. Details of all crystallographic and biochemical procedures are listed in Methods.
METHODS
Preparation of Proteins for Crystallization
A leucine-zipper mutant was introduced on N-terminus aminoacylation domain of E. coli AlaRS, based on rational mutagenesis designed to aid crystallization of the E. coli AlaRS protein (manuscript in preparation). Plasmids for expression of E. coli AlaRS fragments were constructed through PCR amplification of the desired region using primers containing NdeI-XhoI sites, and ligated into pET20b to generate pET20b-EcAlaRS0-441 which encodes the AlaRS fragment without a tag (as with endogenous E. coli AlaRS, the initial methionine, Met0, is cleaved in E. coli during expression and the mature sequence starts from the following serine, Ser1). The leucine-zipper mutant was created by mutating residues His104, Gln108 and Glu112 into leucines. This AlaRS441-LZ protein was expressed in E. coli at room temperature with 0.1 mM IPTG induction, and purified sequentially by a DEAE column, a Q high performance column, followed by a Phenol high performance column (GE Healthcare, Little Chalfont, Buckinghamshire, U.K.). Purified protein was dialyzed against 5 mM Tris-HCl buffer (pH 8.0), 50 mM NaCl and 1 mM β-mercaptoethanol. The G237A mutant AlaRS441-LZ was purified similarly.
Crystallization, Data Collection, Structure Determination and Refinement
A previous and limited analysis of a cocrystal of Aquifex aeolicus AlaRS with alanine resolved the interactions of the cognate amino acid with the protein at a resolution of 2.08 Å30. While the structure itself was accurate, it later came to our attention that the details of the proposed interactions with free serine were incorrect (see newly deposited coordinates pdb 3htz). In addition, we were concerned about the observation that, due to their small size and the absence of the AMP moiety, co-crystallized amino acids can bind to alternative sites within the active center31. Therefore we set out to obtain co-crystals of aminoacyl adenylates with E. coli AlaRS. Purified AlaRS441-LZ was then mixed with different ligands (40 mg/ml protein, with 2 mM Ala-SA/Ser-SA/Gly-SA/AMPPCP, or with 20 mM L-alanine and 2 mM AMPPCP), together with 10 mM MgCl2 and 10 mM β-mercaptoethanol, at 4 °C. Crystallization was done by the micro-sitting drop method. A drop was prepared by mixing 0.1 μl of protein solution with 0.1 μl of precipitant solution and was equilibrated against 70 μl precipitant solution. Ala-SA, Ser-SA and Gly-SA complex crystals were crystallized under 27-32% PEG400, 100 mM HEPES-Na (pH 7.5–8.0), at 4 °C. AMPPCP crystals were crystallized under 20% PEG400, 0.2 M Sodium Sulfate, 4 °C. Ala/AMPPCP crystals were crystallized under 20% PEG3000, 0.1 M Tris-HCl (pH 8.5), at 4 °C. Crystals were collected after incubation for 5-14 days and were flash-frozen at 100 K for data collection.
The data sets were obtained from SSRL beamline 11-1 and beamline 9-1 and were processed with HKL200032. The initial Ala-SA complex structure was solved by molecular replacement using the A. aeolicus AlaRS structure (pdb1riq). Iterative model building and refinement was performed using Coot and Refmac533,34, with Ala-SA ligand added only after the R-factor was lower than 25%. The current model has Rwork and Rfree of 15.3% and 19.0%, respectively. Inspection of this structure indicated that both the introduced leucine-zipper and the end residue (Phe441) formed an extensive interface for crystal packing and were crucial for crystallization. AlaRS441-LZ structures of other ligand complexes were solved using the Ala-SA complex as the initial model. Each structure has only one AlaRS441-LZ molecule per asymmetric unit (Table S1). Inspection of the difference density of the Ala/AMPPCP complex indicated these substrates partially reacted in the crystal, with clear density of dissociating βγ-PCP. The current model has an AMPPCP, an Ala-AMP and a βγ-PCP within the same active site, each with occupancy of 0.5.
G237A AlaRS441-LZ in complex with Ala-SA or Gly-SA was crystallized under similar conditions as wild-type AlaRS441-LZ by mixing with 2 mM Ala-SA or Gly-SA. G237A AlaRS441-LZ in complex with Ser-SA could only be crystallized by mixing with 10 mM Ser-SA, under 25% PEG550 MME, 0.1 M MES (pH 6.5), at room temperature. Data on the G237A AlaRS441-LZ complexes crystals was collected at SSRL beamline 7-1. In contrast to the wild-type AlaRS441-LZ crystals, each ligand induced the G237A mutant crystal into a distinct space group, with 2 molecules per asymmetric unit for Gly-SA (P212121), and 4 for Ala-SA (C2). The molecules within each asymmetric unit closely resembled each other by noncrystallographic symmetry. The Ser-SA G237A mutant crystal contained 2 different molecules in one asymmetric unit, including one in complex with Ser-SA and another in apo-form. This difference indicates that the G237A mutant has a lower binding affinity for Ser-SA analog, which is consistent with our observation that the G237A mutation caused a sharp decrease in the rate of Ser-AMP formation. All ligands were added only after the R-factor was lower than 25% (28% for Ser-SA G237A structure, 2.8 Å). All current models have good geometry and no residues in disallowed regions of the Ramachandran plot. Data collection and refinement statistics are given in Supplementary Table 1.
Preparation of Proteins for Kinetic Assays
The E. coli AlaRS fragment of interest was isolated through PCR amplification and inserted into pET20b to generate pET20b-His-EcAlaRS0-441, with a 6xHis tag at the N-terminus of the AlaRS fragment, but without the leucine-zipper mutations. The G237A, D235A, D235N, D235Q, D2325E, and D235AG237A mutations were introduced by the Quikchange method (Stratagene) and verified by DNA sequencing. The D235E mutant was insoluble. All of the other proteins were expressed and purified by a gravity column packed with Ni-NTA beads (QIAGEN), washed, and eluted with 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 250 mM imidazole. Proteins were then dialyzed against 5 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM β-mercaptoethanol.
ATP-Pyrophosphate Exchange Assays
The ATP-pyrophosphate exchange assays were carried at 22 °C in a buffer composed of 100 mM Tris-HCl (pH 8.0), 1 mM ATP, 10 mM MgCl2, 10 mM KF, 0.1 mg/ml BSA, 5 mM β-mercaptoethanol, and 0.5 mM Na-32PPi. Exchange assays contained 100 nM enzyme for the alanine, glycine and serine substrates for both native and G237A mutant, or up to 148 μM G237A mutant for serine substrates. Initial rates were determined over 3 min linear intervals using four points and fit to the Michaelis-Menten equation using KaleidaGraph. The values were determined from three independent experiments.
Supplementary Material
Acknowledgements
We are grateful for Randy J. Read pointing out the errors in the previous AlaRS-ligand structures and crystallographic discussions. We also thank Gerard J. Kleywegt, Zbyszek Otwinowski, Anastassis Perrakis for technical assistance. X-ray diffraction data was collected at Stanford Synchrotron Radiation Laboratory (SSRL) beamline 7-1, 9-1 and 11-1. This work was supported by grant GM 15539 from the National Institutes of Health and by a fellowship from the National Foundation for Cancer Research.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3hxu (WT/Ala-SA), 3hxv (WT/Gly-SA), 3hxw (WT/Ser-SA), 3hxx (WT/AMPPCP/Mg(II)), 3hxy (WT/Ala-AMP/PCP/AMPPCP/Mg(II)), 3hxz (G237A/Ala-SA), 3hy0 (G237A/Gly-SA), 3hy1 (G237A-apo and G237A/Ser-SA).
Reprints and permissions information are available at www.nature.com/reprints.
The authors declare no competing financial interests.
REFERENCES
- 1.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–75. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–5. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 3.Beebe K, Mock M, Merriman E, Schimmel P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–3. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- 4.Carter CW., Jr. Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–48. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 5.Giege R. The early history of tRNA recognition by aminoacyl-tRNA synthetases. J Biosci. 2006;31:477–88. doi: 10.1007/BF02705187. [DOI] [PubMed] [Google Scholar]
- 6.Norris AT, Berg P. Mechanism of Aminoacyl Rna Synthesis: Studies with Isolated Aminoacyl Adenylate Complexes of Isoleucyl Rna Synthetase. Proc. Natl. Acad. Sci. U.S.A. 1964;52:330–7. doi: 10.1073/pnas.52.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eldred EW, Schimmel PR. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 1972;247:2961–4. [PubMed] [Google Scholar]
- 8.Boniecki MT, Vu MT, Betha AK, Martinis SA. CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19223–8. doi: 10.1073/pnas.0809336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fersht AR. Enzyme Structure and Mechanism. Freeman; San Francisco: 1977. [Google Scholar]
- 10.Nureki O, et al. Structural basis for amino acid and tRNA recognition by class I aminoacyl-tRNA synthetases. Cold Spring Harb Symp Quant Biol. 2001;66:167–73. doi: 10.1101/sqb.2001.66.167. [DOI] [PubMed] [Google Scholar]
- 11.Fersht AR. Sieves in sequence. Science. 1998;280:541. doi: 10.1126/science.280.5363.541. [DOI] [PubMed] [Google Scholar]
- 12.Fukai S, et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 13.Tsui WC, Fersht AR. Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli. Nucleic Acids Res. 1981;9:4627–37. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15422–7. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong YE, Yang XL, Schimmel P. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J. Biol. Chem. 2008;283:30073–8. doi: 10.1074/jbc.M805943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling J, et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell. 2009;33:654–60. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo M, et al. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–7. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokabe M, Okada A, Yao M, Nakashima T, Tanaka I. Molecular basis of alanine discrimination in editing site. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11669–74. doi: 10.1073/pnas.0502119102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends. Biochem. Sci. 1997;22:211–6. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 21.Davis MW, Buechter DD, Schimmel P. Functional dissection of a predicted class-defining motif in a class II tRNA synthetase of unknown structure. Biochemistry. 1994;33:9904–11. doi: 10.1021/bi00199a012. [DOI] [PubMed] [Google Scholar]
- 22.Jakubowski H. Accuracy of Aminoacyl-tRNA Synthetases: Proofreading of Amino Acids. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Eurekah; Georgetown: 2005. pp. 384–396. [Google Scholar]
- 23.Shi JP, Musier-Forsyth K, Schimmel P. Region of a conserved sequence motif in a class II tRNA synthetase needed for transfer of an activated amino acid to an RNA substrate. Biochemistry. 1994;33:5312–8. doi: 10.1021/bi00183a039. [DOI] [PubMed] [Google Scholar]
- 24.Xin Y, Li W, First EA. Stabilization of the transition state for the transfer of tyrosine to tRNA(Tyr) by tyrosyl-tRNA synthetase. J. Mol. Biol. 2000;303:299–310. doi: 10.1006/jmbi.2000.4126. [DOI] [PubMed] [Google Scholar]
- 25.Sankaranarayanan R, et al. Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase. Nat. Struct. Biol. 2000;7:461–5. doi: 10.1038/75856. [DOI] [PubMed] [Google Scholar]
- 26.First EA. Catalysis of the tRNA Aminoacylation Reaction. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Eurekah; Georgetown: 2005. pp. 328–352. [Google Scholar]
- 27.Belrhali H, et al. Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science. 1994;263:1432–6. doi: 10.1126/science.8128224. [DOI] [PubMed] [Google Scholar]
- 28.Cieslik M, Derewenda ZS. The role of entropy and polarity in intermolecular contacts in protein crystals. Acta Crystallogr. D Biol. Crystallogr. 2009;65:500–9. doi: 10.1107/S0907444909009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh Y, et al. Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii. RNA Biol. 2008;5:169–77. doi: 10.4161/rna.5.3.6876. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 30.Swairjo MA, Schimmel PR. Breaking sieve for steric exclusion of a noncognate amino acid from active site of a tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:988–93. doi: 10.1073/pnas.0409024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malde AK, Mark AE. Binding and enantiomeric selectivity of threonyl-tRNA synthetase. J Am Chem Soc. 2009;131:3848–9. doi: 10.1021/ja9002124. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.