Abstract
The retinoblastoma tumor suppressor gene (Rb1) is centrally important in cancer research. Mutational inactivation of Rb1 causes the pediatric cancer retinoblastoma, while deregulation of the pathway in which it functions is common in most types of human cancer. The Rb1 encoded protein (pRb) is well known as a general cell cycle regulator, and this activity is centrally important for pRb-mediated tumor suppression. The main focus of this review, however, is on more recent evidence demonstrating the existence of additional, cell type specific pRb functions in cellular differentiation and survival. These additional functions are relevant to carcinogenesis suggesting that the net effect of Rb1 loss on the behavior of resulting tumors is highly dependent on biological context. The molecular mechanisms underlying pRb functions are based on the cellular proteins it interacts with and the functional consequences of those interactions. Better insight into pRb-mediated tumor suppression and clinical exploitation of pRb as a therapeutic target will require a global view of the complex, interdependent network of pocket protein complexes that function simultaneously within given tissues.
Keywords: tumor suppressor gene, cell cycle, differentiation, DNA damage repair, apoptosis
The retinoblastoma tumor suppressor gene (Rb1) has been a focal point for research into the molecular underpinnings of cancer since its identification and cloning nearly twenty years ago(Dryja et al. 1986; Friend et al. 1986; Lee et al. 1987; Fung et al. 1987). As foreshadowed by Knudson’s “two-hit hypothesis” for the origins of retinoblastoma(Knudson 1971), mutational inactivation of both Rb1 alleles is the primary molecular alteration causing the pediatric cancer retinoblastoma. In about 10% of retinoblastoma cases, the disease is inherited as a simple autosomal dominant trait with greater than 95% penetrance(Lohmann and Gallie 2004). Hereditary retinoblastoma is caused by germ line transmission of one mutationally inactivated Rb1 allele and loss of the remaining wild type allele in somatic retinal cells. Hence hereditary retinoblastoma typically has an earlier onset and a greater number of tumor foci than sporadic retinoblastoma where both Rb1 alleles must be inactivated in somatic retinal cells. To this day, Rb1 remains an exception among cancer-associated genes in that its mutation is apparently both necessary and sufficient, or at least rate limiting, for the genesis of a human cancer. The simple genetics of retinoblastoma has spawned the hope that a complete molecular understanding of the Rb1 encoded protein (pRb) would lead to deeper insight into the processes of neoplastic transformation and, ultimately, to therapeutic benefit for patients. This hope has been buoyed by the discovery that Rb1 mutation is observed in a number of other common human cancers. The expression of proteins that regulate pRb are also frequently altered in an even broader spectrum of cancers, indicating that deregulation of the normal pathways in which pRb functions may be very common in human cancer(Sherr 2000; Malumbres and Barbacid 2001). What is learned about pRb, therefore, is likely to be applicable to cancer in general.
Shortly after its identification, pRb was identified as a binding partner for mitogenic oncoproteins expressed by DNA tumor viruses including SV40 large T antigen (DeCaprio et al. 1988), adenovirus E1A(Whyte et al. 1988), and papillomavirus E7 proteins(Munger et al. 1989). In addition, pRb was discovered to be phosphorylated in synchrony with the cell cycle(Buchkovich et al. 1989; Chen et al. 1989; DeCaprio et al. 1989). This data suggested that pRb may be a general cell cycle regulator. This hypothesis was initially supported by the observations that exogenous unphosphorylated pRb could arrest cells in the G1 phase of the cell cycle(Goodrich et al. 1991; Connell-Crowley et al. 1997), and that depletion of pRb lead to an accelerated G1/S transition(Herrera et al. 1996). An important role for pRb in cell cycle regulation has been consistently validated by research over the ensuing decade. The reader is referred to a recent review appearing in the pages of this journal for a more in depth review of this topic(Cobrinik 2005).
The finding that pRb could bind viral proteins spurred the search for cellular proteins that might interact with pRb. The first cellular pRb binding partner discovered was the E2F1 transcription factor(Helin et al. 1992; Kaelin, Jr. et al. 1992; Shan et al. 1992). The active, unphosphorylated form of pRb preferentially bound E2F1, and disruption of the pRb/E2F1 complex caused cell cycle deregulation(Shan et al. 1996). It was soon realized that E2F1 is a member of a family of related sequence specific DNA binding transcription factors. Rb1 protein was able to bind several members of this family, particularly the transcriptional activators E2F1–3. When in complex with E2F1–3, pRb blocked their ability to activate transcription. In addition, pRb recruited a number of chromatin modifying factors, such as histone deacetylases, to facilitate active gene silencing. As E2F activity regulated many cell cycle genes(Ren et al. 2002; Ishida et al. 2001) and was required for a normal cell cycle(Wu et al. 2001; Humbert et al. 2000), repression of E2F-dependent transcription has generally been considered the principal mechanism underlying pRb-mediated cell cycle control. This hypothesis was supported by in vivo evidence indicating that loss of Rb1 lead to unscheduled cell proliferation in mice, and that compound loss of individual E2F family members partially rescued this cell cycle defect(Saavedra et al. 2002; Tsai et al. 1998; Ziebold et al. 2001). Dimova and Dyson(Dimova and Dyson 2005) recently reviewed the scientific literature relevant to E2F family of transcription factors.
The ability of pRb to bind E2Fs, silence E2F-dependent transcription, and restrain cell cycle progression is inhibited by phosphorylation. The activity of cyclin-dependent kinases (CDKs) correlates with the onset of pRb phosphorylation and the G1/S cell cycle transition. CDKs can phosphorylate pRb in vitro and this phosphorylation inhibits pRb/E2F complex formation(Dynlacht et al. 1994) as well as pRb-mediated cell cycle arrest activity(Connell-Crowley et al. 1997). Regulation of pRb by phosphorylation is complex. Up to sixteen possible CDK phosphorylation sites exist on pRb, and multiple CDKs can phosphorylate pRb with some site specificity(Connell-Crowley et al. 1997; Takaki et al. 2005; Zarkowska and Mittnacht 1997). Other proline directed kinases like p38 stress activated kinase 1(Nath et al. 2003; Hou et al. 2002), ERK1/2(Guo et al. 2005), and possibly Raf-1(Dasgupta et al. 2004) can also phosphorylate pRb. While some phosphorylation sites appear to be critical for negative regulation of pRb (Connell-Crowley et al. 1997; Knudsen and Wang 1996), the effects of phosphorylation at individual sites varies depending on the phosphorylation status of other sites(Barrientes et al. 2000; Knudsen and Wang 1997; Brown et al. 1999). How phosphorylation at individual sites or combinations of sites regulates pRb function has not been clearly defined. A model emerges wherein pRb functions as a general cell cycle inhibitor that restrains the G1/S transition by binding E2F and repressing E2F-dependent transcription. Phosphorylation of pRb, as a result of appropriate mitogenic stimulation and activation of CDKs, blocks pRb/E2F complex formation resulting in derepression and activation of E2F-dependent gene expression. This E2F-dependent gene expression facilitates continued cell cycle progression.
Rb1 protein has also been implicated in other aspects of cell cycle control. For example, pRb can inhibit S phase progression by attenuating cyclin A/Cdk2 activity, resulting in disruption of PCNA function and DNA replication(Knudsen et al. 1998; Sever-Chroneos et al. 2001). Rb1 protein is required for efficient G1/S cell cycle checkpoint activation in response to DNA damage(Harrington et al. 1998), and is required to maintain cell cycle exit in quiescent and senescent cells(Sage et al. 2003). Loss of pRb can cause G2/M cell cycle defects through deregulation of the E2F target gene Mad2, a mitotic checkpoint gene(Hernando et al. 2004). Rb1 protein can also regulate the cell cycle through mechanisms that are independent of E2F. For example, pRb can interact with Skp2, the substrate-recognition subunit of the ubiquitin protein ligase SCFSKP2. Binding of pRb to Skp2 prevents degradation of the CDK inhibitor p27KIP1, a normal target for SCFSKP2 mediated protein degradation(Ji et al. 2004). Stabilization of p27KIP1 inhibits the CDK activity that is required for normal cell cycle progression. Likewise, pRb can bind the transcription factor Mitf and cooperate with Mitf in activating transcription of the CDK inhibitor p21CIP1, at least in some cell types(Carreira et al. 2005).
Despite the significant progress that has been made in elucidating the general role of pRb in cell cycle control, it is still not entirely clear what contribution these mechanisms make to pRb-mediated tumor suppression. While it makes intuitive sense that pRb suppresses tumorigenesis by restraining the cell cycle, some have argued that cancer is fundamentally a disease of defective differentiation(Harris 2004). Perhaps pRb integrates the processes of cell cycle exit and cellular differentiation. Indeed, recent evidence indicates pRb has additional functions that may have a direct role in the regulation of cellular differentiation and cell survival. These additional functions are likely to influence the behavior of cells that lose pRb activity.
The pocket protein family
One of the factors that complicates attempts to unravel the molecular mechanisms responsible for pRb-mediated tumor suppression is the presence of two cellular proteins, p107 and p130 that are structurally related to pRb. All three proteins contain a protein interaction domain called the “pocket” that is required to mediate their interactions with other viral and cellular proteins. All of the pocket proteins share the ability to bind E2Fs, restrain cell cycle progression, and serve as substrates for CDKs (Cobrinik 2005). This functional overlap is reflected in the observation that cells lacking p107 or p130 have cell cycle defects similar to those lacking pRb(Hurford et al. 1997; Classon et al. 2000b), and loss of all three pocket proteins causes more pronounced cell cycle defects than loss of individual members(Dannenberg et al. 2000; Sage et al. 2000). Functional compensation is also observed in vivo as developmental defects associated with Rb1 loss are exacerbated by compound loss of p107(Lee et al. 1996a; MacPherson et al. 2004; Zhang et al. 2004b; Chen et al. 2004) or p130(MacLellan et al. 2005).
Despite the existence of functional compensation between the pocket proteins, it is clear that pRb has unique functions. Only pRb among the pocket proteins is required for embryonic development (Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992) while mice lacking p107 or p130 develop normally(Cobrinik et al. 1996; Lee et al. 1996b). The molecular bases for these differences are not entirely clear. For one, the pocket proteins are expressed at different times in the cell cycle and are likely to have different, but overlapping, expression patterns in vivo. Moreover, pRb preferentially binds E2F1–4 while p107 and p130 primarily interact with E2F4–5. This probably accounts for the observation that while loss of individual pocket proteins has similar effects on the cell cycle, the E2F target genes deregulated in each case are different(Hurford et al. 1997; Wells et al. 2000). However, it should be noted that even p107 and p130 can bind and regulate E2F1–3 under certain conditions(Lee et al. 2002).
Perhaps the most important difference between the pocket proteins is that only mutation of Rb1 is consistently associated with tumorigenesis in both mice and humans. Heterozygous Rb1 null mice develop normally, but spontaneously develop pituitary tumors of the intermediate and anterior lobes as well as medullary thryroid carcinomas with nearly complete penetrance(Hu et al. 1994; Zhou et al. 2005). Conditional ablation of Rb1 in mouse skin(Balsitis et al. 2003; Ruiz et al. 2004) or prostate(Maddison et al. 2004) also leads to the appearance of hyperplastic lesions. Conditional ablation of pRb in the mouse cerebellum in the absence of p53 can cause medulloblastoma(Marino et al. 2000), and loss of pRb and p107 is sufficient for the genesis of murine retinoblastoma(Chen et al. 2004; MacPherson et al. 2004; Zhang et al. 2004a). In contrast, mice completely lacking p107 or p130 are not tumor prone. In humans, mutational inactivation of Rb1 is a prerequisite for retinoblastoma. Rb1 mutation also occurs in a large fraction of osteosarcomas and soft tissue sarcomas and these diseases have been epidemiologically linked to retinoblastoma(Bookstein and Lee 1991). Rb1 mutation is also detected in almost all small cell lung carcinomas and, to a lesser extent, in carcinoma of the breast, prostate, and bladder to name a few. Consistent with the data in mice, mutational inactivation of p107 or p130 is rarely observed in human cancer. Although arguments have been made in support of potential tumor suppressor activity for p130 and p107 in human cancer(Paggi and Giordano 2001), it is clear that among pocket proteins, pRb plays a special role in tumor suppression. Thus the identification of molecular mechanisms unique to pRb are likely to be particularly relevant to carcinogenesis. However, in assessing the possible consequences of pRb inactivation, it is important to consider the potentially overlapping functions of the other pocket proteins.
Rb1 and cellular differentiation
The advent of mice with genetically engineered deletions of the Rb1 gene has led to a growing appreciation of the important role it plays in cell fate specification and differentiation. Rb1 null mice die in midgestation(Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992). Defects are observed in a number of tissues including the eye lens, brain, peripheral nervous system, muscle, liver, placenta, and the hematopoietic system among others(Wu et al. 2003; Iavarone et al. 2004; Spike et al. 2004; Tsai et al. 1998; Zacksenhaus et al. 1996; Novitch et al. 1996). Conditional, tissue specific gene ablation has also been used to examine the effects of Rb1 loss in adult tissue. Such studies have implicated Rb1 in the normal development of the epidermis(Balsitis et al. 2003; Ruiz et al. 2004), melanocytes(Yu et al. 2003), hair cells(Sage et al. 2005), liver(Mayhew et al. 2005), prostate(Maddison et al. 2004), lung(Wikenheiser-Brokamp 2004), cerebellum(Marino et al. 2003), pituitary(Vooijs et al. 1998), and retina(Chen et al. 2004; MacPherson et al. 2004; Zhang et al. 2004a). Affected tissues typically show signs of unscheduled cell proliferation, incomplete differentiation, and apoptotic cell death. Although some of these tissues initiate a differentiation program in the absence of pRb, as indicated by the expression of early lineage specific markers, they generally fail to reach a completely differentiated state.
The developmental defects observed upon loss of Rb1 have been attributed to both cell autonomous and non-cell autonomous mechanisms. For example, the failure of proper placental development(Wu et al. 2003) and/or fetal liver macrophage differentiation (Iavarone et al. 2004) contributes to the failure of erythrocyte maturation in the absence of Rb1. Rb1−/− erythroblasts also fail to differentiate normally in vitro or when reconstituting the hematopoietic system in irradiated wild type mice(Spike et al. 2004; Clark et al. 2004). Thus the pRb-associated hematopoiesis defects are due to both non-cell autonomous and cell autonomous mechanisms. In turn, the hematopoiesis defect contributes to developmental defects observed in the central nervous system (MacPherson et al. 2003). Conditional ablation of Rb1 in the central nervous system yields only a subset of the brain phenotypes observed in Rb1 nullizygous mice because hematopoiesis is not compromised in the conditionally mutant mice. Rb1 loss, therefore, elicits a number of direct and indirect effects on cellular differentiation and survival that contribute to the developmental phenotypes observed.
In vitro differentiation model systems have also provided support for a direct role for pRb in cellular differentiation. Loss of pRb compromises, and exogenous pRb expression restores muscle differentiation in vitro(Gu et al. 1993; Schneider et al. 1994). Initial growth arrest and expression of early differentiation markers does not require pRb, but permanent cell cycle withdrawal and expression of late differentiation markers does (Novitch et al. 1996; Schneider et al. 1994). In contrast cells lacking p107 and/or p130 are fully capable of muscle differentiation in vitro. Rb1 is also required for adipocyte differentiation (Chen et al. 1996a) and lineage commitment(Hansen et al. 2004) in vitro. Emphasizing the unique role pRb plays in this system, loss of p107 and p130 actually facilitates adipogenesis (Classon et al. 2000a). Consistent with the frequent mutation of Rb1 observed in osteosarcoma, pRb also potentiates osteogenic differentiation in vitro(Thomas et al. 2001).
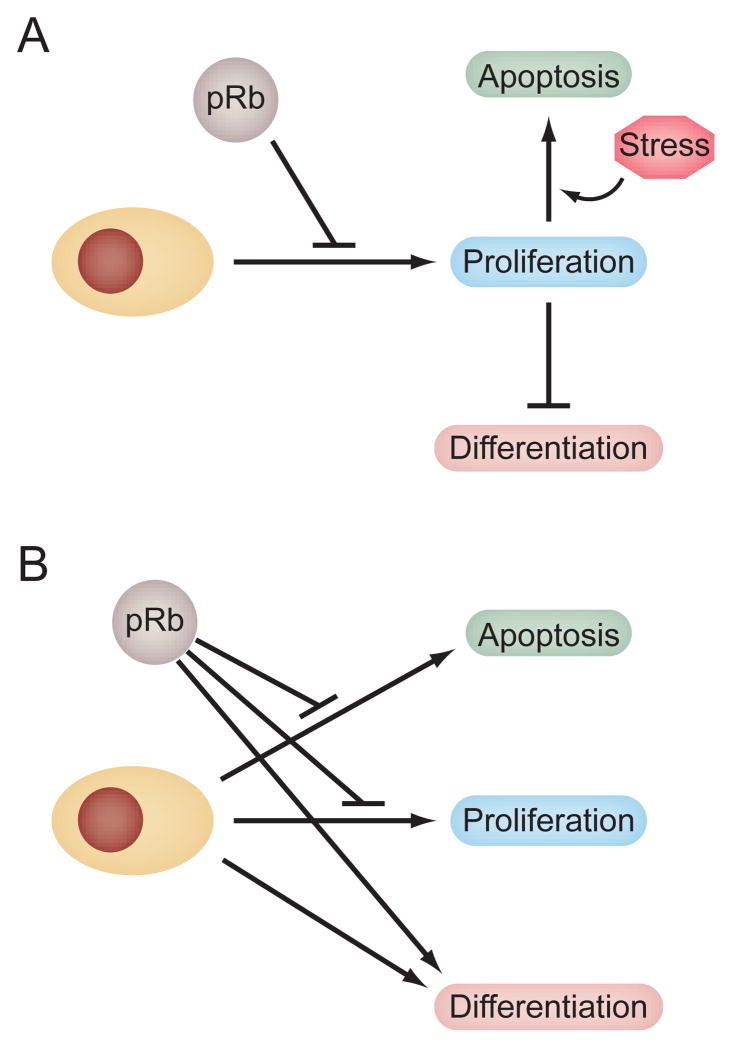
Since cellular differentiation is tightly coupled to cell cycle exit, a major unanswered question is whether the effects of pRb on differentiation reflect additional, cell type specific mechanisms, or whether they are an indirect consequence of pRb-mediated cell cycle control (Figure 1). In other words, does pRb facilitate differentiation simply by enforcing cell cycle exit, or does it use additional mechanisms to directly facilitate the execution of lineage specific gene expression programs. The ability of pRb to bind and augment the transcriptional activity of tissue specific transcription factors like MyoD (Gu et al. 1993), C/EBPs(Chen et al. 1996a), NF-IL6(Chen et al. 1996b), Pax8(Miccadei et al. 2005), CBFA1(Thomas et al. 2001), and nuclear hormone receptors(Batsche et al. 2005; Lu and Danielsen 1998; Singh et al. 1995) to name a few supports the latter hypothesis. However, there is some debate whether a pRb/MyoD complex is directly responsible for augmentation of muscle specific gene expression(Li et al. 2000). An alternative explanation is that pRb can bind and block the activity of differentiation inhibitors like EID-1(MacLellan et al. 2000; Miyake et al. 2000), RBP2(Benevolenskaya et al. 2005), or Id2(Iavarone et al. 1994; Iavarone et al. 2004). These inhibitors block the ability of lineage specific transcription factors to activate gene expression. Hence pRb may augment lineage specific gene expression indirectly by blocking the activity of inhibitors, or directly by augmenting the activity of tissue specific transcriptional activators.
Figure 1. Models for explaining the effects of pRb on cellular differentiation and survival.
During development, pRb is a wide spread regulator of the cell cycle. However, loss of pRb elicits tissue specific defects in cellular differentiation and survival as well as in cellular proliferation. The effects of pRb loss on differentiation and survival may be an indirect consequence of the inability to exit the cell cycle (A). This model is based on the frequent observation that cellular differentiation and apoptotis is coupled to the cell cycle. The model predicts that the molecular mechanisms underlying Rb1 associated differentiation and survival defects will be similar to those involved in pRb-mediated cell cycle control, primarily negative regulation of E2F transcription factors. Rb1-dependent differentiation and survival may also be mediated by direct mechanisms (B). In this model, pRb is proposed to utilize additional, cell cycle independent mechanisms to directly influence cell differentiation and survival. These mechanisms may or may not involve E2F transcription factors. This model predicts that the effects of pRb on proliferation, differentiation, and apoptosis should be genetically separable, at least in some cases. The models are not mutually exclusive. It is conceivable that both models are operable, and the relative contribution of each to the phenotypes associated with pRb loss may be dependent on biological context.
There is accumulating evidence that pRb-mediated mechanisms involved in cellular differentiation are separable from the ability of pRb to restrain the cell cycle. For example, increased expression of EID-1 blocks the ability of pRb to activate lineage specific gene expression, but does not block the ability of pRb to repress transcription and constrain the cell cycle(MacLellan et al. 2000; Krutzfeldt et al. 2005). Compound loss of Rb1 and N- or K-Ras in mice rescues a subset of phenotypes characteristic of Rb1 nullizygous mice. In particular, defective skeletal muscle differentiation is improved upon compound loss of a Ras gene despite evidence that the deregulated cell proliferation typical of Rb1 nullizygous embryos is unaffected(Takahashi et al. 2004; Takahashi et al. 2003). Evidence from Rb1 null mouse retinae indicates there is only mild deregulation of retinal progenitor cell proliferation but a dramatic reduction in mature rod photoreceptors(Donovan and Dyer 2004; Dyer and Bremner 2005; MacPherson et al. 2004; Sage et al. 2003; Schweers and Dyer 2005; Zhang et al. 2004a). Lineage and gene expression analysis suggests that the role of pRb in rod photoreceptor differentiation is distinct from its role in retinal progenitor cell proliferation(Zhang et al. 2004a).
Perhaps the most compelling evidence that pRb utilizes cell cycle independent mechanisms to regulate cellular differentiation is the identification of pRb mutants that genetically separate its effects on differentiation from its effects on the cell cycle. Several Rb1 mutations have been identified that fail to stably bind E2F transcription factors, fail to repress E2F-dependent transcription, and fail to regulate the cell cycle. Yet these mutants are still able to activate lineage specific gene expression and facilitate cellular differentiation in vitro (Sellers et al. 1998). In some cases, the ability of these mutants to promote differentiation correlates with their ability to bind proteins like RBP2(Benevolenskaya et al. 2005), thus underscoring the potential biological relevance of these protein interactions. One of these pRb mutants, an arginine for tryptophan substitution at codon 661, is naturally occurring and is associated with partially penetrant hereditary retinoblastoma(Lohmann et al. 1994; Onadim et al. 1992). This mutation belongs to a small class of naturally occurring, partially penetrant Rb1 mutations that affect the primary structure of pRb, but do not affect Rb1 expression. Presumably the residual differentiation promoting functions of the mutant protein account for the reduced penetrance and expressivity of retinoblastoma observed in families carrying the mutant allele(Harbour 2001).
Our laboratory has recently generated the analogous mutation (R654W) in mice in order to assess the relative contribution of pRb/E2F-depedent and –independent mechanisms to embryonic development. Like Rb1 nullizygous mice, mice homozygous for this mutant allele exhibit widespread deregulation of the cell cycle as might be expected given its inability to stably bind and regulate the transcriptional activators E2F-1, −2, and −3. Despite this, homozygous mutant embryos live longer than Rb1 nullizygous embryos, and exhibit improved cellular differentiation in some tissues(Sun et al. 2005). For example, improved erythrocyte maturation and fetal liver macrophage differentiation is observed compared to Rb1 nullizygous tissue. Improved differentiation correlates with the ability of the R654W pRb to retain protein interactions with E2F4 and Id2, two transcription factors known to influence the differentiation of these tissues. Thus the mechanisms that pRb uses to facilitate cellular differentiation in these tissues is genetically separable from its general ability to stably bind activating E2F transcription factors and regulate the cell cycle. It is likely that the mechanisms involve interactions with other proteins that are more directly involved in cell fate specification and differentiation.
Rb1 and cell survival
A prominent phenotype observed in Rb1 null embryos is excessive apoptosis in several tissues including the nervous system, eye lens, and skeletal muscle(Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992). Ectopic expression of pRb has also been demonstrated to confer resistance to a number of apoptotic triggers in vitro (McConkey et al. 1996). As with cellular differentiation, the analogous question arises whether the ability to regulate apoptosis is an indirect consequence of pRb-mediated cell cycle regulation, or whether the apoptotic defects reflect distinct pRb mechanisms that directly regulate apoptosis (Figure 1). Consistent with the former explanation, excessive apoptosis is typically observed in the same tissues that exhibit loss of cell cycle control in the absence of Rb1. Further, compound loss of E2F1 or E2F3, important targets for pRb-mediated cell cycle control, can partially rescue both the unscheduled cell proliferation and excessive apoptosis phenotypes(Saavedra et al. 2002; Tsai et al. 1998; Ziebold et al. 2001). Ectopic expression of E2F1 has long been known to sensitize cells to apoptotic cell death(Shan et al. 1996; Agah et al. 1997; DeGregori et al. 1997), and cells lacking E2F1 are resistant to some apoptotic triggers(Leone et al. 2001; Field et al. 1996). Ectopic expression of E2F3 can also induce apoptosis, although this apoptosis is mediated through E2F1 because the E2F1 promoter is itself a target for regulation by activating E2Fs(Denchi and Helin 2005). Gene expression profiling has suggested that E2F1 regulates genes important for apoptosis(Muller et al. 2001), including the apoptosis protease activating factor-1 gene (Apaf1) and procaspases(Moroni et al. 2001; Nahle et al. 2002). Apaf1 is a key downstream effector of the mitochondrial pathway of apoptosis and is upregulated in the absence of pRb, presumably due to the effects of E2F1 deregulation. Procaspase activation is the rate-limiting step in apoptotic cell death, so higher levels of procaspases presumably sensitize cells to apoptosis. Compound loss of Apaf1 or procaspases-3 diminishes the extent of apoptosis observed in the central nervous system of Rb1 nullizygous embryos(Guo et al. 2001; Simpson et al. 2001).
While pRb may influence both the cell cycle and cell survival through interaction with E2F transcription factors, it is not clear how expression of the cell cycle and apoptotic transcriptional targets of E2F are coordinated to determine the appropriate cell fate. A paradox arises wherein a cell that is fated to survive and proliferate will contain phosphorylated pRb in order to disrupt pRb/E2F complexes and relieve restraint on the cell cycle. However, disruption of pRb/E2F complexes is expected to upregulate both cell cycle and apoptotic E2F target genes. Conversely, it is paradoxical for a tumor suppressor gene to inhibit apoptosis given that this is a well-known oncogenic mechanism. This functional property predicts that pRb expression may be oncogenic, a prediction that has actually been verified under certain experimental conditions(Borges et al. 2005; Jiang and Zacksenhaus 2002; Yamamoto et al. 1999). Hence the cell must have some way to suppress the pro-apoptotic effects of pRb inactivation as it attempts to proliferate. One possibility is the presence of a distinct, E2F1-specific binding site on the carboxy-terminus of pRb that may be important for regulation of apoptosis, but not cell proliferation(Dick and Dyson 2003). Presumably, a distinct pRb/E2F1 complex that suppresses apoptosis would be retained even after pRb phosphorylation to relieve the cell cycle restraint. Similarly, phosphorylated pRb has been proposed to interact with and inhibit the pro-apoptotic nuclear protein pp32(Adegbola and Pasternack 2005). Alternatively, the cell may utilize independent anti-apoptotic pathways, perhaps mediated by survival factors, to dampen the expected pro-apoptotic effects of pRb/E2F complex disruption.
There is evidence that the effects of pRb on cell proliferation and survival are separable in some biological contexts. For example, compound loss of Rb1 and E2F1 partially rescues the proliferative defects in both the central and peripheral nervous systems, but only causes a significant reduction of apoptosis in the central nervous system(Tsai et al. 1998). In contrast, compound loss of E2F3 significantly diminishes apoptosis observed in the peripheral nervous system in the absence of Rb1(Ziebold et al. 2001). Conditional ablation of Rb1 in the nervous system elicits unscheduled cell proliferation as expected, but does not cause elevated apoptosis(Ferguson et al. 2002; MacPherson et al. 2003). Hence cell proliferation and apoptosis are not necessarily coupled upon pRb loss. Another hypothesis proposed to reconcile these results with the previously demonstrated involvement of pRb/E2F complexes is that apoptosis requires a cellular stress in addition to pRb loss. Hypoxia caused by defective placental development and hematopoiesis may provide the required cellular stress. The stress response gene p53 responds to hypoxia and is activated in the central nervous system of Rb1 nullizygous embryos. Compound loss of p53 diminishes apoptosis in the central nervous system caused by Rb1 loss(Macleod et al. 1996). Hence any condition that diminishes cellular stress or the stress response is expected to reduce apoptosis in the absence of pRb.
Another piece of circumstantial evidence that supports the hypothesis that pRb is more directly involved in the regulation of apoptosis is that pRb is a substrate for caspases(Boutillier et al. 2000; Fattman et al. 2001; Fattman et al. 1997; Tan et al. 1997). Caspases are activated to transduce death signals and execute cell killing during apoptosis. Cleavage of pRb by caspases can either inactivate the protein, or possibly activate novel pRb regulatory functions(Lemaire et al. 2005). Engineered mutations that block cleavage at particular caspase recognition sites within pRb have been shown to attenuate apoptosis in vitro induced by tumor necrosis factor-α(Tan et al. 1997) or potassium deprivation(Boutillier et al. 2000). Inhibition of apoptosis is unrelated to the cell cycle since the cell cycle response to tumor necrosis factor-α is the same in cells expressing wild type or the mutant pRb. The physiological relevance of pRb caspase cleavage has been demonstrated by introduction of an analogous mutation into the mouse(Chau et al. 2002; Borges et al. 2005). The caspase cleavage resistant Rb1 allele supports normal growth and development, but mice containing this mutation exhibit tissue specific resistance to apoptosis when challenged with bacterial endotoxin. The caspase resistant Rb1 allele also promotes colorectal cancer. It is of interest that the caspase cleavage resistant pRb does not inhibit apoptosis triggered by other apoptotic stresses like DNA damage.
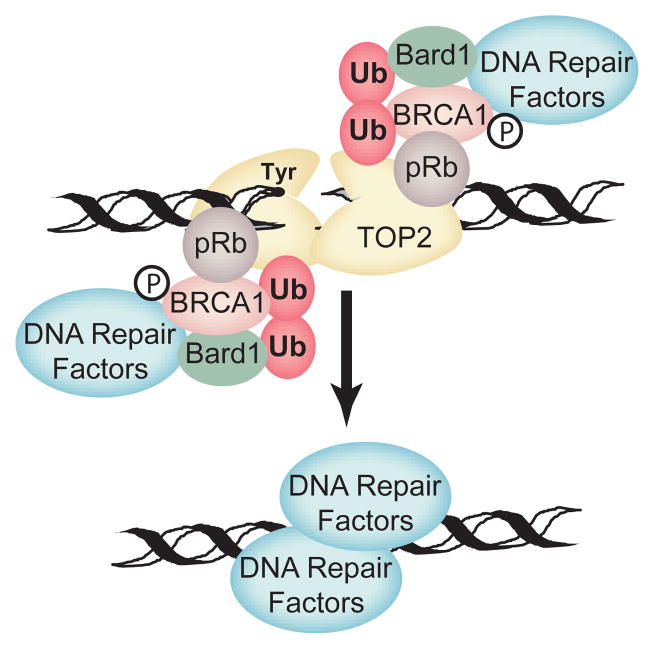
Rb1 has long been known to influence sensitivity to DNA damaging agents as pRb deficient cells have increased sensitivity to cisplatin, methotrexate, etoposide, and camptothecin among others(Almasan et al. 1995; Harrington et al. 1998; Knudsen et al. 2000). In particular, pRb is required to efficiently trigger cell cycle checkpoints in response to DNA damage. In the absence of checkpoint activation, cells usually undergo apoptotic cell death. This function of pRb can be accounted for by the pRb/E2F mechanism, but other evidence has suggested that more direct mechanisms may also be involved. Topoisomerase II (TopII) poisons like etoposide create a unique double stranded DNA break where the ends of the DNA are trapped in covalent linkage with TopII. In order for this damage to be repaired, the covalently linked TopII must be degraded to liberate free DNA ends. TopII and pRb are capable of binding one another, and loss of pRb slows the degradation of etoposide trapped TopII and the repair of the resulting DNA break(Xiao and Goodrich 2005). As a result, cells lacking pRb are more sensitive to the cytotoxic effects of TopII than cells expressing pRb. Efficient degradation and repair of trapped TopII lesions correlates with BRCA1/TopII interaction, and BRCA1/TopII interaction requires the presence of pRb. BRCA1 complexes not only have ubiquitin ligase activity, but also contain DNA repair factors. This suggests a model wherein pRb recruits factors like BRCA1 complexes to trapped TopII lesions (Figure 2). These factors facilitate the degradation of trapped TopII and the repair of the resulting DNA break. While pRb is also necessary for cell cycle checkpoint activation in response to etoposide, processing and repair of trapped TopII complexes is independent of cell cycle checkpoint activation. The E2F1–3 binding deficient R654W pRb mutant is still capable of facilitating processing and repair of trapped TopII lesions despite its inability to trigger the normal cell cycle checkpoint response(Xiao and Goodrich 2005). The above analysis suggests pRb may use multiple mechanisms to directly regulate cell survival in response to cellular stress, including stress induced by cancer chemotherapeutics. These mechanisms may be dependent or independent of E2F and the cell cycle.
Figure 2. A model for a direct role for pRb in repair of trapped TopII DNA lesions.
Rb1 protein is suggested to serve as an adaptor protein to recruit processing and repair factors to trapped TOP2 cleavage complexes. BRCA1/BARD1 E3 ubiquitin ligase activity facilitates degradation of TOP2 to reveal free DSBs that are then repaired by BRCA1 associated double stranded DNA break repair factors. The model is consistent with the observed requirement for an E2F- and cell cycle-independent pRb activity in the efficient processing and repair of etoposide induced DNA lesions.
The pRb-associated proteome
At the molecular level, pRb function can be understood through the proteins it interacts with and the functional consequences of those interactions. Our current perception of Rb1 function is based on an E2F-centric view of the underlying molecular mechanisms. This is understandable considering that the E2F transcription factors were the first cellular pRb associated proteins identified. Further, the functional consequences of the pRb/E2F interactions are the most thoroughly elaborated. The biological importance of this mechanism, particularly for pRb-mediated cell cycle control, has consistently been validated in numerous in vitro and in vivo studies over the last decade. Yet by one important criterion, this view of Rb1 is lacking. With the possible exception of the retina, we are still unable to reasonably predict what effect Rb1 pathway deregulation will have on tumorigenesis in a given tissue in mice or in man. This limits our ability to rationally target this important tumor suppressor gene pathway for therapeutic benefit.
How best to realize a molecular understanding of pRb sufficient to achieve this goal? There are still a sufficiently large number of gaps in our understanding of pRb/E2F-dependent mechanisms and their regulation to suggest that a continued focus on E2F may yield the desired result. But what of the potentially important E2F-indpendent mechanisms that are being identified? How many additional pRb-mediated mechanisms are yet to be discovered? A previous survey of the published literature has identified some 110 cellular proteins that have been implicated to physically interact with pRb (Morris and Dyson 2001). Roughly 15% of these proteins are kinases or phosphatases that could potentially regulate pRb activity, or the activity of pRb-associated proteins. About two-thirds of the proteins are involved in transcription. The remaining 20% have varied functions that are not obviously related to transcription. In the ensuing four years since this study appeared, more than 40 additional cellular proteins have been proposed in the published literature to physically interact with pRb, with the grand total now exceeding 150 different cellular proteins. This represents an average discovery rate of more than 10 new pRb-associated proteins per year. At least eleven new pRb-associated proteins have been newly described in 2005, so the rate of pRb-associated protein discovery is not diminishing. It is unclear how many of these pRb protein interactions form the basis for biologically relevant molecular mechanisms. Yet even by conservative estimate, dozens of distinct pRb protein complexes may be operable at any one time in a given cell. This is plausible given that pRb is generally in significant stoichiometric excess over most if its protein binding partners.
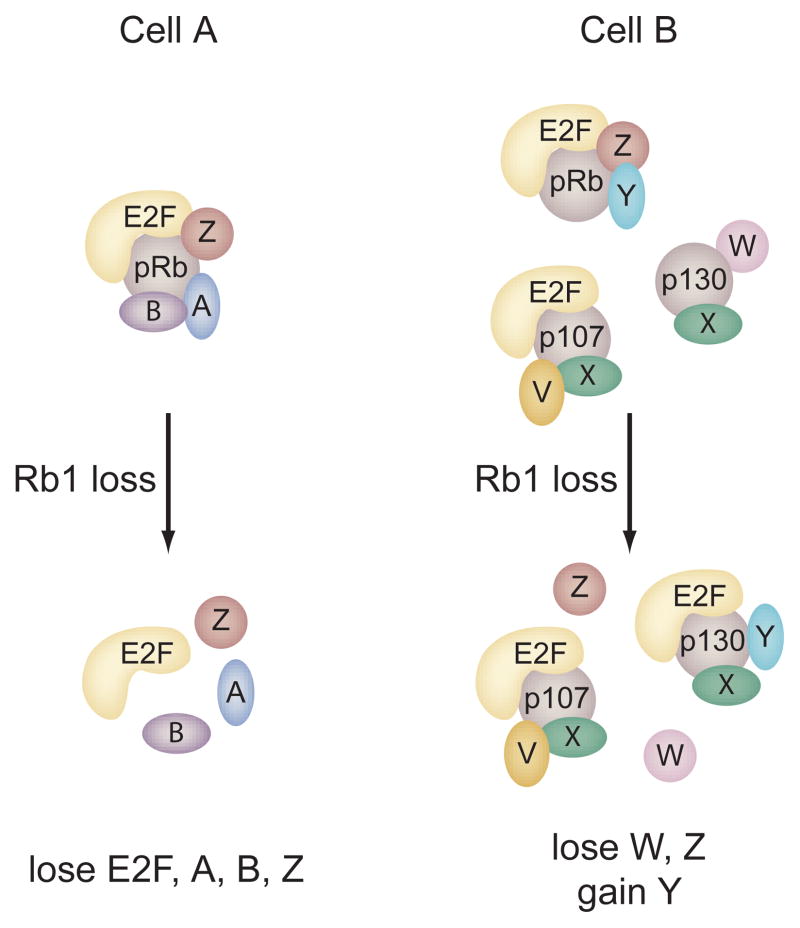
Another consideration is the existence of the structurally and functionally related pocket proteins p107 and p130. The cellular proteins that interact with p107 or p130 are largely undefined, but it is clear that they can bind some of the same proteins that associate with pRb. Hence the pocket proteins may be in competition for the binding of an overlapping subset of cellular proteins. Given this, any perturbation of the pocket proteins will have a substantial ripple effect based on a reshuffling of pocket protein complexes due to the altered stoichiometry of unbound protein partners. Evidence exists in the literature that supports such a reshuffling of pocket protein complexes upon experimental perturbation. For example, loss of E2F4, the preferred E2F binding partner for p107 and p130, results in the formation of novel p107/E2F1–3 and p130/E2F1–3 complexes that are not detected under normal conditions. These novel complexes have functional consequences as they can partially suppress tumorigenesis initiated by Rb1 loss(Lee et al. 2002). Further, specific disruption of pRb/E2F1–3 interactions by mutation can augment the formation of other pRb complexes, presumably because E2F1–3 no longer compete for pRb binding(Sun et al. 2005). It can be inferred from these observations that disruption of particular pocket protein interactions will not only result in loss of function, but may also yield gain of function as novel complexes form due to altered stoichiometric conditions within the cell. Viewing pocket protein function from the more global perspective of a highly interconnected protein interaction network may be required in order to accurately predict the consequences of pRb inactivation in a particular biological context (Figure 3).
Figure 3. The pocket protein interaction network.
While the pRb/E2F complex is central to pRb mediated cell cycle control and contributes to pRb-dependent differentiation and cell survival, more than 150 additional pRb protein binding partners have been described in the literature. The other pocket proteins, p107 and p130, interact with an overlapping subset of these cellular proteins. The model pictured attempts to illustrate some of the functional implications of this interdependent protein interaction network. Cell A and cell B represent two different cell types that express different constellations pocket protein complexes. Hence the function of pRb in these two cells is different. In cell A, p107 and p130 are either absent or they are unable to interact with the pRb-associated proteins shown. In this simple case, Rb1 loss results in the loss of all functions mediated by the indicated protein interactions. Cell B expresses a unique set of cellular proteins that interact with each of the pocket proteins with differing affinities. Based on these differing affinities and simple stoichiometric considerations, loss of Rb1 causes a reshuffling of pocket protein complexes resulting in loss of some functions (W,Z), gain of other functions (Y), or no significant change in function (E2F-functional compensation). Even more complicated scenarios can be envisioned upon partial inactivation of pocket proteins by phosphorylation.
Perspectives
Consideration of pRb function as a component of such a complex and interdependent protein interaction network has both theoretical and practical implications for our understanding of cancer and its treatment. Many pocket protein binding partners are restricted to particular tissues and cell types. Hence the distribution of distinct pocket protein complexes will vary from one cell type to another. The biological consequence of pRb loss in a particular cell will be determined by the protein interactions that are lost, and how the liberation of these pRb-associated proteins affects the distribution of the remaining pocket protein complexes. In other words, pRb may suppress tumorigenesis in different ways that vary depending on the tissue and cell type. The net effect of pRb loss in a given cell will also be determined by the status of independent regulatory pathways. For example, it has been proposed that the cell of origin for retinoblastoma is naturally resistant to apoptosis(Chen et al. 2004; Zhang et al. 2004b). This may explain why complete pRb loss in this cell initiates retinoblastoma rather than cell death. A practical consequence of this is that pRb inactivation may have different prognostic implications for different cancers. This could occur, for example, if pRb inactivation has a different effect on the response to cancer therapy in one tissue than it does in another.
How the Rb1 pathway or network is deregulated during carcinogenesis, either direct mutation of Rb1 or mutation of genes that regulate pRb, is not randomly distributed in human cancer (Figure 4). Retinoblastoma, small cell lung cancer, and osteosarcoma are commonly associated with Rb1 mutation. In contrast, Rb1 mutation is rare in melanoma. Melanoma is commonly associated with perturbations of cyclin D or p16Ink4a that lead to unscheduled pocket protein phosphorylation. This clustering suggests that different types of cancers select for different mechanisms of pRb inactivation. The implication is that the different mechanisms of pRb inactivation yield different functional consequences. A relevant example may be the ability of pRb to negatively regulate apoptotic cell death. As discussed above, the cell of origin for retinoblastoma may be naturally resistant to apoptotic cell death. Carcinogenesis in such a cell may benefit from complete loss of pRb since the potential pro-apoptotic consequences are irrelevant. In contrast, the cell of origin for melanoma may possess robust apoptotic responses. Carcinogenesis in such a cell would select against complete loss of pRb since the ensuing apoptotic response would preclude tumor progression. In this case, deregulated phosphorylation of pRb may partially inactivate critical tumor suppression functions, but not the anti-apoptotic functions. Alternatively, other pocket proteins may also need to be partially or completely inactivated. In this case deregulation of the kinases that regulate multiple pocket proteins would be selected for. Hence the consequences of pRb incativation will not only be dependent on cell type, but also on the mechanism of inactivation. Thus the behavior of cancers of a given type may vary depending on the nature the perturbation in the pRb network. For example, retinoblastomas with complete loss of pRb behave differently than partially penetrant cases that express mutant pRb or low levels of wild type pRb. This concept may apply more broadly to the common adult cancers suggesting that sub-classification by the nature of the lesion in the pocket protein interaction network may be informative.
Figure 4. Mechanisms of pRb inactivation in human cancer.
Three main mechanisms of pRb inactivation are observed in human cancer, and these mechanisms are not randomly distributed. Rb1 mutation is common in some cancers, but rare in others. Likewise, inactivation by deregulated pRb phosphorylation is common in a distinct subset of human cancers, but Rb1 mutation is rarely observed in this subset. The model proposes that each mechanism of inactivation has different molecular and functional consequences. Deregulated pocket protein phosphorylation is proposed to abrogate some protein interactions but not others. Coupled with considerations of the pocket protein interaction network outlined in Figure 3, different cell types may select for different mechanisms of pRb inactivation because the molecular consequences are more favorable for carcinogenesis.
The simple genetics of retinoblastoma is a rare exception in human cancer. Most common adult cancers arise through alterations in multiple combinations of many genes. The molecular cloning of the Rb1 gene has been met with great fanfare since it provides, in a sense, a simple model system in which to study the molecular basis for neoplastic transformation. Mutational inactivation of a single gene is apparently necessary and rate limiting for the genesis of a human cancer. In the ensuing twenty years, significant progress has been made in the characterization of pRb as a general cell cycle regulator, elucidating the molecular mechanisms responsible for this regulation, and accounting for how this activity may be regulated by phosphorylation. Our current level of understanding, however, does not explain a number of nagging questions. Why is Rb1 a tissue specific tumor suppressor given its more general role in cell cycle regulation? Why among the functionally related pocket proteins is mutation of Rb1 uniquely associated with human cancer? Why does the mechanism of pRb inactivation cluster in particular types of cancer? If pRb is fundamentally a cell cycle regulator, why does it bind so many cellular proteins with seemingly divergent functions? What do these other protein interactions contribute to pRb-mediated tumor suppression? A more complete consideration of cell cycle independent pRb functions may be necessary to answer these questions.
Ultimately, it seems, pRb function is considerably more complex than is currently understood. Rb1 is seemingly involved in many fundamentally important cellular processes including the cell division cycle, cellular stress responses, differentiation, cellular senescence, and programmed cell death, among others. The Rb1 encoded protein utilizes a large and complex array of protein interactions to influence these processes. This complexity is reminiscent of the genetic complexity typical of most human cancer. In a sense, Rb1 is the exception that proves the rule that a large number of interdependent molecular alterations are required for neoplastic transformation. Yet, pRb remains a convenient starting point for addressing this complexity. Applying the experimental and computational approaches being rapidly developed in the field of protein interactome mapping(Vidal 2005) to the pocket protein network may yet help us justify the scientific attention paid this important tumor suppressor gene over the two decades since its discovery.
Acknowledgments
I am grateful to the Goodrich lab members and colleagues at Roswell Park Cancer Institute for stimulating discussions. This work is supported by a grant from the National Cancer Institute (CA70292).
References
- Adegbola O, Pasternack GR. J Biol Chem. 2005;280:15497–15502. doi: 10.1074/jbc.M411382200. [DOI] [PubMed] [Google Scholar]
- Agah R, Kirshenbaum LA, Abdellatif M, Truong LD, Chakraborty S, Michael LH, Schneider MD. J Clin Invest. 1997;100:2722–2728. doi: 10.1172/JCI119817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasan A, Yin Y, Kelly RE, Lee EY, Bradley A, Li W, Bertino JR, Wahl GM. Proc Natl Acad Sci U S A. 1995;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsitis SJ, Sage J, Duensing S, Munger K, Jacks T, Lambert PF. Mol Cell Biol. 2003;23:9094–9103. doi: 10.1128/MCB.23.24.9094-9103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientes S, Cooke C, Goodrich DW. Oncogene. 2000;19:562–570. doi: 10.1038/sj.onc.1203332. [DOI] [PubMed] [Google Scholar]
- Batsche E, Desroches J, Bilodeau S, Gauthier Y, Drouin J. J Biol Chem. 2005;280:19746–19756. doi: 10.1074/jbc.M413428200. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Bookstein R, Lee WH. Critical Reviews In Oncogenesis. 1991;2:211–227. [PubMed] [Google Scholar]
- Borges HL, Bird J, Wasson K, Cardiff RD, Varki N, Eckmann L, Wang JY. Proc Natl Acad Sci USA. 2005;102:15587–15592. doi: 10.1073/pnas.0503925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP. Oncogene. 2000;19:2171–2178. doi: 10.1038/sj.onc.1203532. [DOI] [PubMed] [Google Scholar]
- Brown VD, Phillips RA, Gallie BL. Mol Cell Biol. 1999;19:3246–3256. doi: 10.1128/mcb.19.5.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich K, Duffy LA, Harlow E. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- Chau BN, Borges HL, Chen TT, Masselli A, Hunton IC, Wang JY. Nature Cell Biology. 2002;4:757–765. doi: 10.1038/ncb853. [DOI] [PubMed] [Google Scholar]
- Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Chen PL, Riley DJ, Chen Y, Lee WH. Gene Dev. 1996a;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- Chen P-L, Riley DJ, Chen-Kiang S, Lee W-H. Proc Natl Acad Sci USA. 1996b;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-L, Scully P, Shew J-Y, Wang JYJ, Lee W-H. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Doyle KM, Humbert PO. Blood. 2004;104:1324–1326. doi: 10.1182/blood-2004-02-0618. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Classon M, Kennedy BK, Mulloy R, Harlow E. Proc Natl Acad Sci USA. 2000a;97:10826–10831. doi: 10.1073/pnas.190343597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M, Salama S, Gorka C, Mulloy R, Braun P, Harlow E. Proc Natl Acad Sci U S A. 2000b;97:10820–10825. doi: 10.1073/pnas.190343497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L, Harper JW, Goodrich DW. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van RA, Schuijff L, te RH. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta P, Sun J, Wang S, Fusaro G, Betts V, Padmanabhan J, Sebti SM, Chellappan SP. Mol Cell Biol. 2004;24:9527–9541. doi: 10.1128/MCB.24.21.9527-9541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston DM. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C-M, Livingston DM. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, Helin K. EMBO Rep. 2005;6:661–668. doi: 10.1038/sj.embor.7400452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Dyson N. Mol Cell. 2003;12:639–649. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Dyer MA. Vision Res. 2004;44:3323–3333. doi: 10.1016/j.visres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Dryja TP, Rapaport JM, Joyce JM, Petersen RA. Proc Natl Acad Sci U S A. 1986;83:7391–7394. doi: 10.1073/pnas.83.19.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Bremner R. Nat Rev Cancer. 2005;5:91–101. doi: 10.1038/nrc1545. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Flores O, Lees JA, Harlow E. Gene Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- Fattman CL, An B, Dou QP. J Cell Biochem. 1997;67:399–408. [PubMed] [Google Scholar]
- Fattman CL, Delach SM, Dou QP, Johnson DE. Oncogene. 2001;20:2918–2926. doi: 10.1038/sj.onc.1204414. [DOI] [PubMed] [Google Scholar]
- Ferguson KL, Vanderluit JL, Hebert JM, McIntosh WC, Tibbo E, MacLaurin JG, Park DS, Wallace VA, Vooijs M, McConnell SK, Slack RS. EMBO J. 2002;21:3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. Nature (London) 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- Fung YKT, Murphree AL, T’Ang A, Qian J, Hinrichs SH, Benedict WF. Science. 1987;236:1657–1661. doi: 10.1126/science.2885916. [DOI] [PubMed] [Google Scholar]
- Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- Gu W, Schneider JW, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- Guo J, Sheng G, Warner BW. J Biol Chem. 2005;280:35992–35998. doi: 10.1074/jbc.M504583200. [DOI] [PubMed] [Google Scholar]
- Guo Z, Yikang S, Yoshida H, Mak TW, Zacksenhaus E. Cancer Res. 2001;61:8395–8400. [PubMed] [Google Scholar]
- Hansen JB, Jorgensen C, Petersen RK, Hallenborg P, De Matteis R, Boye HA, Petrovic N, Enerback S, Nedergaard J, Cinti S, te Riele H, Kristiansen K. Proc Natl Acad Sci USA. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW. Arch Ophthalmol. 2001;119:1699–1704. doi: 10.1001/archopht.119.11.1699. [DOI] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, Dyson N. Proc Natl Acad Sci USA. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. Nature. 2004;427:201. doi: 10.1038/427201a. [DOI] [PubMed] [Google Scholar]
- Helin K, Lees JA, Vidal M, Dyson N, Harlow E, Fattaey A. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou ST, Xie X, Baggley A, Park DS, Chen G, Walker T. J Biol Chem. 2002;277:48764–48770. doi: 10.1074/jbc.M206336200. [DOI] [PubMed] [Google Scholar]
- Hu N, Gutsmann A, Herbert DC, Bradley A, Lee WH, Lee EY. Oncogene. 1994;9:1021–1027. [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. Gene Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- Hurford RK, Cobrinik D, Lee MH, Dyson N. Gene Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. Gene Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- Iavarone A, King ER, Dai XM, Leone G, Stanley ER, Lasorella A. Nature. 2004;432:1040–1045. doi: 10.1038/nature03068. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, Zhu L. Mol Cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Zacksenhaus E. J Cell Biol. 2002;156:185–198. doi: 10.1083/jcb.200106084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Krek W, Sellers WR, DeCaprio JA, Ajchenbaum F, Fuchs CS, Chittenden T, Li Y, Farnham PJ, Blanar MA, et al. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Buckmaster C, Chen TT, Feramisco JR, Wang JY. Gene Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Wang JYJ. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Wang JYJ. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES. Molecular & Cellular Biology. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt M, Ellis M, Weekes DB, Bull JJ, Eilers M, Vivanco MD, Sellers WR, Mittnacht S. Mol Cell. 2005;18:213–224. doi: 10.1016/j.molcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Lee EY, Cam H, Ziebold U, Rayman JB, Lees JA, Dynlacht BD. Cancer Cell. 2002;2:463–472. doi: 10.1016/s1535-6108(02)00207-6. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Genes Dev. 1996b;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Genes Dev. 1996a;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- Lee W-H, Bookstein R, Hong F, Young L-J, Shew J-Y, Lee EYHP. Science. 1987;235:1394–1399. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- Lemaire C, Godefroy N, Costina-Parvu I, Rincheval V, Renaud F, Trotot P, Bouleau S, Mignotte B, Vayssiere JL. Oncogene. 2005;24:3297–3308. doi: 10.1038/sj.onc.1208493. [DOI] [PubMed] [Google Scholar]
- Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, Thompson MA, Yang H, Fujiwara Y, Greenberg ME, Orkin S, Smith C, Nevins JR. Mol Cell. 2001;8:105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- Li FQ, Coonrod A, Horwitz M. Mol Cell Biol. 2000;20:5129–5139. doi: 10.1128/mcb.20.14.5129-5139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann DR, Brandt B, Hopping W, Passarge E, Horsthemke B. Hum Genet. 1994;94:349–354. doi: 10.1007/BF00201591. [DOI] [PubMed] [Google Scholar]
- Lohmann DR, Gallie BL. Am J Med Genet. 2004;C129:23–28. doi: 10.1002/ajmg.c.30024. [DOI] [PubMed] [Google Scholar]
- Lu J, Danielsen M. J Biol Chem. 1998;273:31528–31533. doi: 10.1074/jbc.273.47.31528. [DOI] [PubMed] [Google Scholar]
- MacLellan WR, Garcia A, Oh H, Frenkel P, Jordan MC, Roos KP, Schneider MD. Mol Cell Biol. 2005;25:2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan WR, Xiao G, Abdellatif M, Schneider MD. Mol Cell Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod KF, Hu Y, Jacks T. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Mol Cell Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison LA, Sutherland BW, Barrios RJ, Greenberg NM. Cancer Res. 2004;64:6018–6025. doi: 10.1158/0008-5472.CAN-03-2509. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Marino S, Hoogervoorst D, Brandner S, Berns A. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der GH, Jonkers J, Berns A. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- McConkey DJ, Goodrich D, Bucana C, Klostergaard J. Oncogene. 1996;13:1693–1700. [PubMed] [Google Scholar]
- Miccadei S, Provenzano C, Mojzisek M, Giorgio Natali P, Civitareale D. Oncogene. 2005;24:6993–7001. doi: 10.1038/sj.onc.1208861. [DOI] [PubMed] [Google Scholar]
- Miyake S, Sellers WR, Safran M, Li X, Zhao W, Grossman SR, Gan J, DeCaprio JA, Adams PD, Kaelin WG., Jr Mol Cell Biol. 2000;20:8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Denchi EL, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Nature Cell Biology. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, Lazebnik Y, Bar-Sagi D, Lowe SW. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- Nath N, Wang S, Betts V, Knudsen E, Chellappan S. Oncogene. 2003;22:5986–5994. doi: 10.1038/sj.onc.1206843. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Mulligan GJ, Jacks T, Lassar AB. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onadim Z, Hogg A, Baird PN, Cowell JK. Proc Natl Acad Sci U S A. 1992;89:6177–6181. doi: 10.1073/pnas.89.13.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paggi MG, Giordano A. Cancer Res. 2001;61:4651–4654. [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. Gene Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, Paramio JM, Vooijs M. Development. 2004;131:2737–2748. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- Saavedra HI, Wu L, de Bruin A, Timmers C, Rosol TJ, Weinstein M, Robinson ML, Leone G. Cell Growth & Differ. 2002;13:215–225. [PubMed] [Google Scholar]
- Sage C, Huang M, Karimi K, Gutierrez G, Vollrath MA, Zhang DS, Garcia-Anoveros J, Hinds PW, Corwin JT, Corey DP, Chen ZY. Science. 2005;307:1114–1118. doi: 10.1126/science.1106642. [DOI] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- Schweers BA, Dyer MA. Visual Neuorsci. 2005 doi: 10.1017/S0952523805225026. in press. [DOI] [PubMed] [Google Scholar]
- Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WG., Jr Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever-Chroneos Z, Angus SP, Fribourg AF, Wan H, Todorov I, Knudsen KE, Knudsen ES. Molecular & Cellular Biology. 2001;21:4032–4045. doi: 10.1128/MCB.21.12.4032-4045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Durfee T, Lee WH. Proc Natl Acad Sci U S A. 1996;93:679–684. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Zhu X, Chen PL, Durfee T, Yang Y, Sharp D, Lee WH. Mol Cell Biol. 1992;12:5620–5631. doi: 10.1128/mcb.12.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- Simpson MT, MacLaurin JG, Xu D, Ferguson KL, Vanderluit JL, Davoli MA, Roy S, Nicholson DW, Robertson GS, Park DS, Slack RS. J Neurosci. 2001;21:7089–7098. doi: 10.1523/JNEUROSCI.21-18-07089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Coe J, Hong W. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- Spike BT, Dirlam A, Dibling BC, Marvin J, Williams BO, Jacks T, Macleod KF. EMBO J. 2004;23:4319–4329. doi: 10.1038/sj.emboj.7600432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Chang Y, Schweers B, Dyer MA, Zhang X, Hayward SW, Goodrich DW. An E2F1–3 binding deficient Rb1 protein partially rescues developmental defects associated with Rb1 nullizygosity. Molecular and Cellular Biology. 2005;26:1527–1537. doi: 10.1128/MCB.26.4.1527-1537.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi C, Bronson RT, Socolovsky M, Contreras B, Lee KY, Jacks T, Noda M, Kucherlapati R, Ewen ME. Mol Cell Biol. 2003;23:5256–5268. doi: 10.1128/MCB.23.15.5256-5268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi C, Contreras B, Bronson RT, Loda M, Ewen ME. Mol Cell Biol. 2004;24:10406–10415. doi: 10.1128/MCB.24.23.10406-10415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki T, Fukasawa K, Suzuki-Takahashi I, Semba K, Kitagawa M, Taya Y, Hirai H. J Biochem. 2005;137:381–386. doi: 10.1093/jb/mvi050. [DOI] [PubMed] [Google Scholar]
- Tan X, Martin SJ, Green DR, Wang JYJ. J Biol Chem. 1997;272:9613–9616. doi: 10.1074/jbc.272.15.9613. [DOI] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- Vidal M. FEBS Lett. 2005;579:1834–1838. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Vooijs M, van d V, te RH, Berns A. Oncogene. 1998;17:1–12. doi: 10.1038/sj.onc.1202169. [DOI] [PubMed] [Google Scholar]
- Wells J, Boyd KE, Fry CJ, Bartley SM, Farnham PJ. Molecular & Cellular Biology. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp KA. Development. 2004;131:4299–4310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- Xiao H, Goodrich DW. Oncogene. 2005;24:8105–8113. doi: 10.1038/sj.onc.1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Soh JW, Monden T, Klein MG, Zhang LM, Shirin H, Arber N, Tomita N, Schieren I, Stein CA, Weinstein IB. Clin Cancer Res. 1999;5:1805–1815. [PubMed] [Google Scholar]
- Yu BD, Becker-Hapak M, Snyder EL, Vooijs M, Denicourt C, Dowdy SF. Proc Natl Acad Sci USA. 2003;100:14881–14886. doi: 10.1073/pnas.2431391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. Gene Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- Zarkowska T, Mittnacht S. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gray J, Wu L, Leone G, Rowan S, Cepko CL, Zhu X, Craft CM, Dyer MA. Nat Genet. 2004a;36:351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schweers B, Dyer MA. Cell Cycle. 2004b;3:952–959. [PubMed] [Google Scholar]
- Zhou Z, Flesken-Nikitin A, Levine CG, Shmidt EN, Eng JP, Nikitina EY, Spencer DM, Nikitin AY. Cancer Res. 2005;65:787–796. [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA. Gene Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]