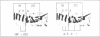Abstract
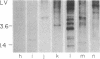
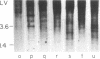
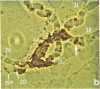
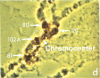
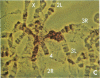
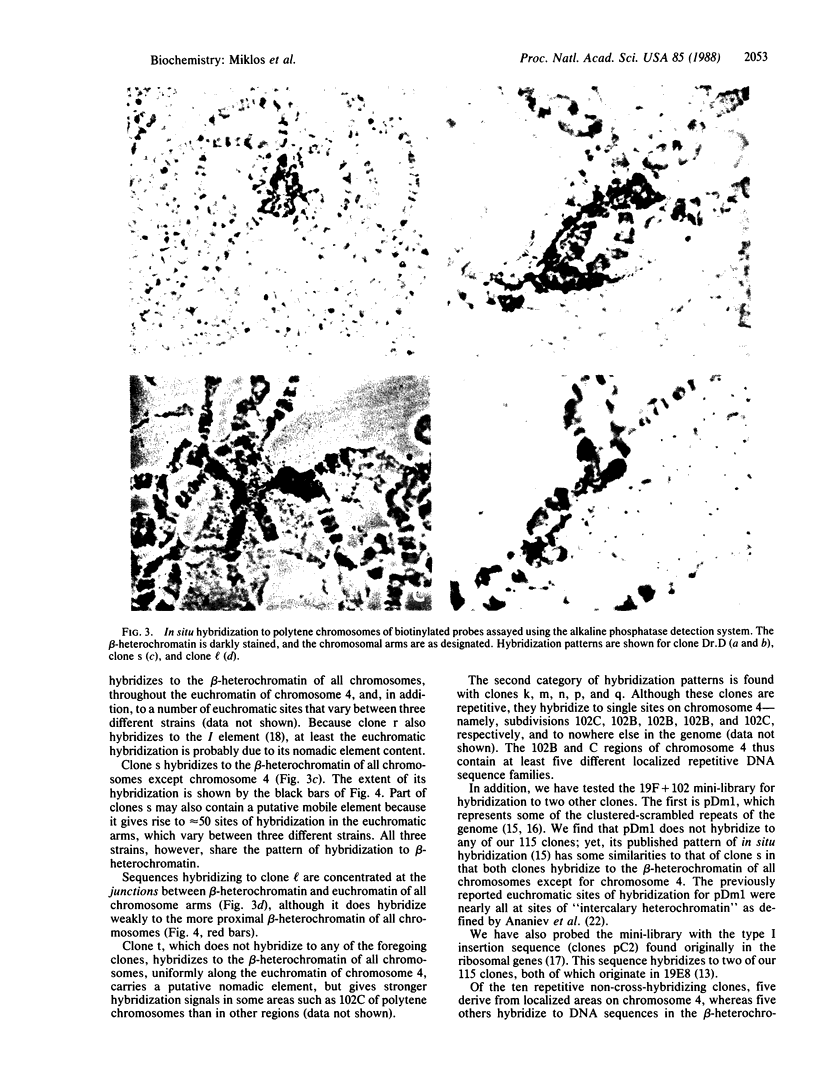
Microdissection and microcloning of the euchromatin-heterochromatin transition region of the Drosophila melanogaster polytene X chromosome and part of the euchromatin of chromosome 4 reveals that they share certain features characteristic of beta-heterochromatin, which is morphologically defined as the loosely textured material at the bases of some polytene chromosome arms. Both are mosaics of many different middle-repetitive DNA sequences interspersed with single-copy DNA sequences. Sixty percent of cloned inserts derived from division 20 and about 40 percent from subdivisions 19EF of the X chromosome harbor at least one repetitive DNA sequence in an average insert of 4.5 kilobases. No repeats have significant cross-hybridization to any of the eleven satellite DNAs, or to the clustered-scrambled sequences present in pDm1. The repetitive elements are, in general, confined to the beta-heterochromatic regions of polytene chromosomes, but some are adjacent to nomadic elements. Chromosome 4, however, has some repeats spread throughout its entire euchromatin. These data have implications for the structure of transition zones between euchromatin and heterochromatin of mitotic chromosomes and also provide a molecular basis for reexamining some of the unusual classical properties of chromosome 4.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ananiev E. V., Gvozdev V. A., Ilyin Yu V., Tchurikov N. A., Georgiev G. P. Reiterated genes with varying location in intercalary heterochromatin regions of Drosophila melanogaster polytene chromosomes. Chromosoma. 1978 Dec 21;70(1):1–17. doi: 10.1007/BF00292211. [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Bucheton A., Paro R., Sang H. M., Pelisson A., Finnegan D. J. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984 Aug;38(1):153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., Stephan W. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics. 1986 Apr;112(4):947–962. doi: 10.1093/genetics/112.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Long E. O., DiNocera P. P., Pardue M. L. Ribosomal insertion-like elements in Drosophila melanogaster are interspersed with mobile sequences. Cell. 1981 Aug;25(2):399–408. doi: 10.1016/0092-8674(81)90058-1. [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P., Dawid I. B. Interdigitated arrangement of two oligo(A)-terminated DNA sequences in Drosophila. Nucleic Acids Res. 1983 Aug 25;11(16):5475–5482. doi: 10.1093/nar/11.16.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly R. J., Kiefer B. I. DNA sequence adjacent to and specific for the 1.672 g/cm3 satellite DNA in the Drosophila genome. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7172–7176. doi: 10.1073/pnas.83.19.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H., Polan M. L. Reptitive DNA sequences in drosophila. Chromosoma. 1971;33(3):319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Green M. M., Yamamoto M. T., Miklos G. L. Genetic instability in Drosophila melanogaster: cytogenetic analysis of MR-induced X-chromosome deficiencies. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4533–4537. doi: 10.1073/pnas.84.13.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Mason P. J., Spierer P. Transcripts, genes and bands in 315,000 base-pairs of Drosophila DNA. J Mol Biol. 1983 Sep 5;169(1):83–96. doi: 10.1016/s0022-2836(83)80176-4. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Appels R. Pleiotropic effects associated with the deletion of heterochromatin surrounding rDNA on the X chromosome of Drosophila. Chromosoma. 1982;86(4):469–490. doi: 10.1007/BF00330122. [DOI] [PubMed] [Google Scholar]
- Kidd S. J., Glover D. M. A DNA segment from D. melanogaster which contains five tandemly repeating units homologous to the major rDNA insertion. Cell. 1980 Jan;19(1):103–119. doi: 10.1016/0092-8674(80)90392-x. [DOI] [PubMed] [Google Scholar]
- Lakhotia S. C., Jacob J. EM autoradiographic studies on polytene nuclei of Drosophila melanogaster. II. Organization and transcriptive activity of the chromocentre. Exp Cell Res. 1974 Jun;86(2):253–263. doi: 10.1016/0014-4827(74)90711-3. [DOI] [PubMed] [Google Scholar]
- Lefevre G. The distribution of randomly recovered X-ray-induced sex-linked genetic effects in Drosophila melanogaster. Genetics. 1981 Nov-Dec;99(3-4):461–480. doi: 10.1093/genetics/99.3-4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre G., Watkins W. The question of the total gene number in Drosophila melanogaster. Genetics. 1986 Aug;113(4):869–895. doi: 10.1093/genetics/113.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E. Fine-Structure Analysis and Genetic Organization at the Base of the X Chromosome in DROSOPHILA MELANOGASTER. Genetics. 1978 Mar;88(3):457–467. doi: 10.1093/genetics/88.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., Brutlag D. L. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1986 Feb;83(3):696–700. doi: 10.1073/pnas.83.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Beckendorf S. K. A transposable element inserted just 5' to a Drosophila glue protein gene alters gene expression and chromatin structure. Cell. 1983 Aug;34(1):75–84. doi: 10.1016/0092-8674(83)90137-x. [DOI] [PubMed] [Google Scholar]
- Miklos G. L., Healy M. J., Pain P., Howells A. J., Russell R. J. Molecular and genetic studies on the euchromatin-heterochromatin transition region of the X chromosome of Drosophila melanogaster. 1. A cloned entry point near to the uncoordinated (unc) locus. Chromosoma. 1984;89(3):218–227. doi: 10.1007/BF00295003. [DOI] [PubMed] [Google Scholar]
- Miklos G. L., Kelly L. E., Coombe P. E., Leeds C., Lefevre G. Localization of the genes shaking-B, small optic lobes, sluggish-A, stoned and stress-sensitive-C to a well-defined region on the X-chromosome of Drosophila melanogaster. J Neurogenet. 1987 Jan;4(1):1–19. doi: 10.3109/01677068709102329. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Appels R., Endow S., Glover D. Chromosomal distribution of the major insert in Drosophila melanogaster 28S rRNA genes. Genet Res. 1981 Apr;37(2):209–214. doi: 10.1017/s0016672300020176. [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Hadfield C., Pretorius G. H. Microdissection and cloning of the white locus and the 3B1-3C2 region of the Drosophila X chromosome. EMBO J. 1983;2(6):927–934. doi: 10.1002/j.1460-2075.1983.tb01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Tabata S., Pachl C. The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell. 1979 Dec;18(4):1231–1246. doi: 10.1016/0092-8674(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Young B. S., Pession A., Traverse K. L., French C., Pardue M. L. Telomere regions in Drosophila share complex DNA sequences with pericentric heterochromatin. Cell. 1983 Aug;34(1):85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]