Abstract
The novel yeast protein Tar1p is encoded on the anti-sense strand of the multi-copy nuclear 25S rRNA gene, localizes to mitochondria, and partially suppresses the mitochondrial RNA polymerase mutant, rpo41-R129D. However, the function of Tar1p in mitochondria and how its expression is regulated are currently unknown. Here we report that Tar1p is subject to glucose repression and is up-regulated during post-diauxic shift in glucose medium and in glycerol medium, conditions requiring elevated mitochondrial respiration. However, Tar1p expression is down-regulated in response to mitochondrial dysfunction caused by the rpo41-R129D mutation or in strains lacking respiration. Furthermore, in contrast to the previously reported beneficial effects of moderate over-expression of Tar1p in the rpo41-R129D strain, higher-level over-expression exacerbates the ROS-derived phenotypes of this mutant, including decreased respiration and life span. Finally, two-hybrid screening and in vitro-binding studies revealed a physical interaction between Tar1p and Coq5p, an enzyme involved in synthesizing the mitochondrial electron carrier and anti-oxidant, coenzyme Q. We propose that Tar1p expression is induced under respiratory conditions to maintain oxidative phosphorylation capacity, but that its levels in mitochondria are typically low and stringently controlled. Furthermore, we speculate that Tar1p is down-regulated when respiration is defective to prevent deleterious ROS-dependent consequences of mitochondrial dysfunction.
Keywords: TAR1, RPO41, Mitochondria, COQ5, Respiration, rDNA
Introduction
In the budding yeast Saccharomyces cerevisiae, ribosomal RNA (rRNA) is transcribed from a tandem array of 100–200 copies of a 9.8 kb sequence on chromosome XII referred to as the rDNA locus (Petes and Botstein 1977). We previously reported that, in addition to the four rRNA genes, at least one protein is also encoded by the rDNA locus (Coelho et al. 2002). This protein, Tar1p, was identified in a transposon-based screen for expressed ORFs missed in the initial annotation of the yeast genome and is expressed from a transcript derived from the anti-sense strand of the 25S rRNA gene. Epitope-tagged Tar1p localizes to mitochondria and moderate over-expression of Tar1p partially rescues the growth phenotype resulting from the R129D point mutation in the amino-terminal domain of mitochondrial RNA polymerase, Rpo41p (Coelho et al. 2002). The biogenesis of ribosomes is a major energetic expenditure for eukaryotic cells and is regulated at multiple levels in response to a complex set of inputs such as growth rate and prevailing nutrient conditions (Warner 1989, 1999). Thus, based on the unique location of the TAR1 gene and its mitochondrial localization, we speculated previously that one aspect of this regulation could involve a novel communication pathway mediated by Tar1p, linking the energetically expensive process of ribosome biogenesis in the nucleus to the energy-producing function mitochondria (Coelho et al. 2002). However, the control of TAR1 expression from its endogenous promoter in the rDNA locus and the function of Tar1p in mitochondria have remained uncharacterized.
In eukaryotes, the mitochondrion occupies a central position in cellular metabolic and signaling pathways and contains ~1,500 proteins. Mitochondria also harbor their own DNA genome (mtDNA), which is essential for the production of ATP via oxidative phosphorylation (OXPHOS), as it encodes critical subunits of the electron transport and ATP synthase complexes of the mitochondrial inner membrane (Bonawitz et al. 2006a). In yeast and mammals, mtDNA also encodes the two rRNA subunits of mitochondrial ribosomes and a unique set of tRNAs dedicated to translation of the mtDNA-encoded mRNAs the mitochondrial matrix. However, the vast majority of the mitochondrial proteome is encoded in the nucleus and imported into the organelle, including the remaining OXPHOS complex subunits and the factors required for expression and maintenance of mtDNA (e.g. mitochondrial ribosomal proteins and the aforementioned mitochondrial RNA polymerase) (Bonawitz et al. 2006a). Thus, the biogenesis and function of mitochondria requires the coordination of nuclear and mitochondrial gene expression via complex signaling networks that are only beginning to be understood (Kelly and Scarpulla 2004).
Mitochondrial OXPHOS is an intricate process involving movement of electrons through the electron transport chain (or respiratory chain) (Saraste 1999). As already mentioned, these multi-protein complexes are composed of subunits encoded by genes in both the nucleus and mtDNA. In addition to the complexes themselves, a large number of additional proteins and co-factors are required for OXPHOS. For example, cytochrome c and CoQ are mobile electron carriers that transport electrons between complexes in the chain. CoQ synthesis occurs on the matrix side of the inner mitochondrial membrane via the actions of multiple enzymes encoded by COQ genes in the nucleus (COQ1-COQ10) (Tran and Clarke 2007), representing a salient example of the commitment of a large number of nuclear gene products to a single mitochondrial component. Coq5p is a C-methytransferase that carries out one of several modifications to the benzoquinone backbone of CoQ required for its synthesis and also stabilizes other enzymes in the CoQ biosynthesis pathway (Barkovich et al. 1997; Baba et al. 2004). Interestingly, expression of the COQ5 gene is regulated by energy source via transcription factors known to be involved in regulation of mitochondrial function (Hagerman et al. 2002; Hagerman and Willis 2002), suggesting this protein may be a uniquely modulated member of the CoQ pathway. CoQ is also found elsewhere in the cell and exhibits antioxidant function (James et al. 2004).
In addition to its role in ATP production, mitochondrial respiration is also a major source of reactive oxygen species (ROS), which can react with and damage critical biological molecules. ROS production may be exacerbated by dysfunctional mitochondria, and is thought to contribute substantially to both disease states and normal aging (Wallace 2005; Bonawitz and Shadel 2007). For example, we have shown that disrupted mitochondrial translation in the rpo41-R129D mutant yeast strain causes ROS-mediated reductions in respiration and severely decreased life span (Bonawitz et al. 2006b). In contrast, increasing mitochondrial respiration via reduced TOR signaling extends yeast life span by reducing ROS (Bonawitz et al. 2007). Antioxidants like CoQ are needed to defend against damage from endogenous and exogenous ROS and can influence aging and life span (Rodriguez-Aguilera et al. 2005).
The biogenesis and function of mitochondria has been extensively studied in budding yeast, and regulatory changes in mitochondrial respiration as a function of growth state and carbon source are well documented in this organism (Johnston and Carlson 1991; Pon and Schatz 1991). For example, during log-phase growth in glucose medium, mitochondrial respiration and biogenesis are repressed, and ATP is generated primarily by glycolysis. As the glucose in the medium is depleted, a shift in metabolism occurs (the diauxic shift) that involves up-regulation of mitochondrial biogenesis and respiration in order to sustain ATP production as glucose becomes limiting (Pon and Schatz 1991). After diauxic shift, yeast growth slows, and in stationary phase cell division eventually ceases altogether. Furthermore, yeast can be grown in non-fermentable medium (e.g. glycerol) in which mitochondrial respiration is absolutely required and mitochondrial biogenesis is up-regulated (Schuller 2003). Finally, in respiration-deficient, petite strains (e.g. ρ− or ρ0 strains that lack functional mtDNA), retrograde signaling pathways are activated (Liu and Butow 2006), thereby altering nuclear gene expression and cellular metabolism. Interestingly, the same growth conditions that influence mitochondrial biogenesis (e.g. diauxic shift and glycerol medium) involve reductions in growth rate, which as noted above is an important control point for cytoplasmic ribosome biogenesis (Kief and Warner 1981; Warner 1989). Consequently, this is also when major transcriptional changes occur at the rDNA locus (Kief and Warner 1981; Warner 1989), where the TAR1 gene is located. In the present study, the expression dynamics of the TAR1 gene from its endogenous locus within the rDNA array was examined and a yeast 2-hybrid screen was performed to identify potential interacting factors in mitochondria. We discovered (1) that the TAR1 gene is stringently regulated, subject to glucose repression, and responsive to mitochondrial cues, and (2) that Tar1p interacts with the Coq5p enzyme involved in the biosynthesis of CoQ. We speculate that these properties are important in a role for Tar1p in modulating mitochondrial respiration, as well as managing cellular oxidative stress when respiration is defective.
Materials and methods
Yeast strains, media, and plasmids
Standard media and techniques for culturing and transforming yeast were used as described (Sherman 2002). Rich glucose medium (YPD), rich glyerol medium (YPG), and synthetic glucose medium (SD) supplemented with the appropriate nutrients were used as indicated in the text. Construction of the rpo41-R129D mitochondrial RNA polymerase mutant (also called GS129) and its isogenic wild type control strain (GS122) have been described previously (Rodeheffer et al. 2001). Strain 155F6 contains a genomic fusion of Tar1p to LacZ as a result of transformation with a Tn3-based transposon as described (Ross-Macdonald et al. 1997; Coelho et al. 2002; Kumar et al. 2002), and is a derivative of strain Y800 (Kumar et al. 2002). Petite isolates of Y800 and 155F6 were obtained by streaking each strain onto YPD medium and screening for white colonies (strain Y800 and its derivatives lack the ADE2 gene, and are thus red if respiratory competent and white if petite) (Shadel 1999). Inability to respire was confirmed by streaking onto glycerol medium. Strains in which Tar1p-LacZ reporter expression was analyzed were constructed by sporulation of strain 155F6 to obtain haploids, followed by disruption of the endogenous RPO41 gene and transformation with a plasmid encoding either the wild type RPO41 gene (positive control) or the rpo41-R129D allele as described previously (Rodeheffer et al. 2001). The yeast 2-hybrid assay was carried out in strain Y190, which contains GAL4-driven genomic copies of HIS3 and lacZ (Rodeheffer et al. 2001).
The pTEF-Tar1p over-expression plasmid was constructed using the following forward and reverse PCR primers: 5′-AATTTCTAGAATGCGAGATTCCCCTACC-3′ and 5′-AATTCTCGAGTCAAATTTGAAATCTGGTAC-3′, respectively, which amplify the TAR1 ORF from start codon to stop codon and append a XbaI site at the 5′ end and a XhoI site at the 3′ end. The resulting PCR product was cloned using the TA cloning vector pGEM-T EZ (Promega), and transferred from this vector into the XbaI and XhoI sites in the multiple cloning site of plasmid p427-TEF.
The TAR1 yeast two-hybrid bait plasmid was constructed by amplifying the TAR1 ORF by PCR using the following forward and reverse primers: 5′-AACATATGC GAGATTCCCT-3′ and 5′-TTCTCGAGAGTGAAGCGG CAAAAGC-3′, respectively. The resulting ~500 bp PCR product, which had the TAR1 ORF flanked by an NdeI restriction site that overlapped the ATG start codon and an XhoI site 17 nucleotides downstream of the predicted stop codon, was cloned using the TA-cloning vector pGEM-T (Promega). The resulting plasmid was then digested with NdeI and XhoI and the TAR1 fragment was cloned into the yeast two-hybrid plasmid, pAS1 that was digested with NdeI and SalI.
To generate a bacterial expression plasmid encoding GST-tagged Tar1p, the TAR1 ORF was first amplified by PCR using primers that contained a BamH1 and a Not1 site overlapping the start and stop codon, respectively. The resulting PCR product was then cut with BamHI and NotI and cloned directly into the plasmid pGEX-4T-3 (Amersham Biosciences), resulting in an in-frame fusion of GST to the N-terminus of Tar1p under control of an IPTG-regulated Ptac promoter. The bacterial Coq5p expression plasmid was a gift from Dr. F.W. Studier (Brookhaven National Laboratory, Upton, NY) and encodes an untagged Coq5p ORF inserted into the plasmid PET-13a under control of the bacteriophage T7 promoter as well as a kanamycin resistance marker.
Reverse transcriptase-PCR
RNA was extracted from 10 mL of stationary phase cultures using the following protocol. (Standard precautions for working with RNA were used, and all water used was first treated with DEPC). Cells were pelleted and resuspended in 400 μL of AE buffer (50 mM sodium acetate pH 5.2, 10 mM EDTA). A total of 40 μL of 10% SDS and samples were vortexed, followed by addition of 500 μL of AE-buffered phenol. Samples were incubated at 65°C for 4 min, then transferred to dry ice for 2 min, then spun at full speed in a microcentrifuge for 5 min at room temperature. Supernatant was transferred to a new tube, re-extracted with phenol, and RNA was precipitated using a standard ethanol precipitation protocol. RNase treatment was carried out by resuspending in TE containing 25 μg/mL RNase A. DNase treatment was performed using RQ1 DNase (Promega) according to the manufacturer’s instructions. Reverse transcriptase reactions were carried out using AMV reverse transcriptase (Promega) according to manufacturer’s instructions and the TAR1 reverse primer 5′-TTAAGAATC CTCAAATTTGAAATCTGGT-3′. After first-strand cDNA synthesis, PCR was performed using standard conditions after adding a TAR1 forward primer for PCR, 5′-TTAAGG ATCCATGCGAGATTCCCCTACC-3′.
Detection of Tar1p by western blot
Total protein was isolated from ~4 × 107 cells using an alkaline lysis and trichloroacetic acid precipitation protocol (Yaffe and Schatz 1984). Proteins (20 μg) were resolved by electrophoresis on 15% SDS-PAGE gels and transferred to a PVDF membrane (Millipore Immobilon). Blocking and antibody incubations were done in TBS-T (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20) with 5% nonfat dry milk. The primary antibody was a rabbit polyclonal antibody raised against a synthesized epitope of Tar1p and was used at a dilution factor of 1:1,000. The secondary antibody was HRP-conjugated donkey anti-rabbit IgG (Santa Cruz) and was used at a dilution factor of 1:5,000. Detection of the secondary antibody was performed using Western Lighting Chemiluminescent Reagent Plus (PerkinElmer) following the manufacturer’s instructions.
β-Galactosidase assays of Tar1p-LacZ translational fusion strains
β-Galactosidase (LacZ) activity was determined essentially as described (Burke et al. 2000) using ~0.5 − 1 × 108 cells. Samples were taken from cultures at the time points indicated and cells were collected by centrifugation and then resuspended in 1 mL of LacZ buffer (Burke et al. 2000). A total of 50 μL of 0.1% SDS and 75 μL of chloroform were added, samples were vortexed for 10 s, and 200 μL of 0.4 mg/mL o-nitrophenyl-β-D-galactoside (Sigma) were added. After 24 h at 30°C, samples were spun for 5 min at full speed in a microcentrifuge. The absorbance of 1 mL of the resulting supernatant at 420 nm was measured using a spectrophotometer and units of β-galactosidase activity were expressed as OD420/OD600 units of cells assayed.
Oxygen consumption assays
Oxygen consumption was measured with a Clark-type electrode exactly as described in (Bonawitz et al. 2006b).
Chronological lifespan assays
Determination of chronological lifespan was performed using a Trypan blue staining assay as previously described (Bonawitz et al. 2006b). Briefly, 50-mL cultures were inoculated to an OD600 of 0.05 in SD medium and incubated with shaking at 30°C. Viability was assessed at the given time points by removing a 100 μL sample of the culture and incubating it with an equal volume of 0.4% Trypan blue for 5 min at 30°C, then dispensing 10 μL of this mix onto a standard hemocytometer and counting live (clear) versus dead (blue) cells. Assays were carried out in triplicate and at least 100 cells were counted for each point.
Yeast 2-hybrid screening
The yeast 2-hybrid system we employed was essentially the same as that we described previously (Rodeheffer et al. 2001) except the Y190 2-hybrid yeast strain was transformed with the pAS1-TAR1 plasmid described above to generate the reporter strain for Tar1p-interacting partners. This strain was then transformed with a yeast genomic 2-hybrid library in the plasmid pACT II and Trp+ Leu+ transformants were plated on SD plates that also contained 40 mM 3 aminotriazole (3-AT) to select for those expressing high levels of HIS3 reporter gene activity (i.e. putative 2-hybrid positives). From a screen of ~50,000 Leu + Trp + colonies, 34 potential positives were isolated. These were re-screened for their ability to grow in the presence 40 mM 3-AT and for LacZ activity as described previously. Nineteen of these were classified as false positives based on one or both of these secondary screens, leaving 15 true positives for analysis. The library plasmids were liberated from each of these yeast strains and transformed into E. coli using a standard “smash and grab” protocol (Hoffman and Winston 1987). These were then subjected to restriction analysis to determine if the pACT plasmids had inserts. Seven of these had indistinguishable restriction patterns using multiple diagnostic enzymes and were considered the same and therefore only one was analyzed further as representative of this group along with the remaining eight. These nine plasmids were then transformed back into a fresh Y190 strains containing either pAS1-TAR1 or a negative control bait plasmid pAS-SNF1 and again screened for positive LacZ activity in the TAR1 bait strain and lack of LacZ activity in the control strain. Only five of the nine survived this selectivity screen, but one of these was the representative of the group of seven that were identical based on restriction mapping. Thus, a total of 11 true positives were identified. The inserts in these eleven plasmids were sequenced to determine the identity of Tar1p-interacting proteins they encoded.
Protein binding assays in vitro
Cultures of bacterial strain BL21 (DE3) were transformed separately with plasmids encoding GST alone (pGEX-4T-3), GST-Tar1p (pGEX-4T-Tar1), and Coq5p (PET-13-Coq5). Overnight cultures of the resulting strains were inoculated into 100 mL of selective media (LB ampicillin or LB kanamycin) to an OD600 of 0.01 and grown at 37°C with shaking. After cultures had reached an OD600 of 0.8, protein expression was induced by addition of 0.4 mM IPTG. Cultures were incubated for an additional 4 h at 37°C with shaking and then harvested by centrifugation. The resulting bacterial cell pellets from the GST alone and GST-Tar1 expressing strains were resuspended in 10 mL of NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris pH 8.0, 0.5% NP-40) with 1 mM PMSF, and lysed by sonication. Centrifugation, lysis, and all subsequent steps were carried out at 4°C. Following sonication, insoluble protein and unlysed cells were removed by centrifuging the lysate at maximum speed in an SS-34 rotor for 10 minutes. Next, 200 μL of a 50% slurry of Glutathione-Sepharose 4B beads (Amersham Biosciences) were added to soluble lysates obtained from bacteria expressing GST and GST-Tar1p. The resulting suspension was incubated for 1 h at 4°C with gentle rocking, then spun down and washed three times with 10 mL of ice-cold NETN buffer. SDS-PAGE gels and western blots were conducted to confirm significant affinity purification of GST and GST-Tar1p proteins (data not shown). Whole cell lysate was obtained from the Coq5p-expressing bacteria by the same procedure as described above. This lysate was pre-cleared of proteins that non-specifically bind to glutathione sepharose beads by adding 200 μL of 50% bead slurry and incubating for 1 h at 4°C with gentle rocking and then removing the beads by centrifugation.
To assay for an interaction between Tar1p and Coq5p, 10 mL of pre-cleared Coq5 lysate was added to glutathione sepharose beads containing either GST or GST-Tar1p, followed by incubation for 1 h at 4°C with gentle rocking. Next, each sample was spun down, washed 3 times with NETN buffer, and then split in into two equal volumes, each containing a bead bed volume of 50 μL. To one aliquot of beads from each sample (GST/Coq5p or GST-Tar1p/Coq5p) was added 50 μL of SDS gel loading buffer to harvest all proteins bound to the beads. To the other aliquot of each sample was added 50 μL of NETN buffer containing 40 mM reduced glutathione, to harvest only those proteins that specifically elute with GST or GST-Tar1p. Proteins in all samples were resolved by electrophoresis on a 10% SDS-PAGE gel and transferred to a PVDF membrane (Millipore Immobilon). Blocking and antibody binding were performed in TBS-T (see above) containing 5% nonfat dry milk. The primary antibody used for detection of GST was goat anti-GST (GE Healthcare) at a dilution factor of 1:1,000, and the secondary antibody used was HRP-conjugated swine anti-goat (BioRad) at a dilution factor of 1:5,000. The primary antibody used for detection of Coq5p was a rabbit anti-Coq5p polyclonal antibody (a gift from Dr. C.F. Clark, University of California, Los Angeles) at a dilution factor of 1:5,000 and the secondary antibody used was HRP-conjugated donkey anti-rabbit IgG (Santa Cruz), also at a dilution factor of 1:5,000. Detection of the secondary antibody was performed using Western Lightning Chemiluminescent Reagent Plus (PerkinElmer) according to the manufacturer’s instructions. X-ray film was exposed for 1–4 min to obtain the western signals in Fig. 5b.
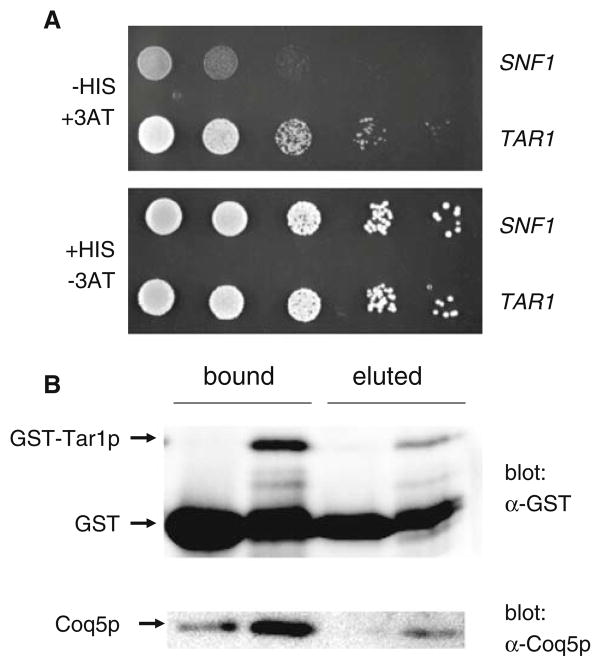
Fig. 5.
Tar1p interacts with Coq5p. a Tar1p exhibits a yeast two-hybrid interaction with Coq5p. Shown is the growth of a representative experimental strain (TAR1) and a negative control strain (SNF1) on media that is either selective (−HIS, +3AT; top panel) or non-selective (+HIS, −3AT; bottom panel) for a bait–prey interaction. The experimental strain contains plasmids expressing a Coq5p-Gal4p DNA binding domain fusion protein as well as a Tar1p-Gal4p transcriptional activation domain fusion protein. The negative control strain expresses the Gal4p activation domain fused to Snf1p (a putative non-interactor with Coq5p) instead of Tar1p. The top panel shows growth on media lacking histidine and containing 40 mM 3AT (a toxic histidine analog), which selects for a bait–prey interaction, whereas the bottom panel shows growth on media containing histidine without 3AT, and thus non-selective for a bait–prey interaction. b Tar1p binds to Coq5p in vitro. GST (lanes 1 and 3) and GST-Tar1p (lanes 2 and 4) were bound to glutathione sepharose beads and used to test for an interaction for Coq5p as described in the text. Lanes 1 and 2 contains all proteins that were able to be stripped from the beads by boiling in SDS gel-loading buffer, while lanes 3 and 4 contains only those proteins that eluted from the beads following the addition of reduced glutathione (i.e. only those interacting with the GST or GST-Tar1p fusion protein). In the top panel, all samples were analyzed by western blotting using an anti-GST antibody. The bottom panel shows the same samples probed with an anti-Coq5p antibody
Results
Detection of endogenous TAR1 expression
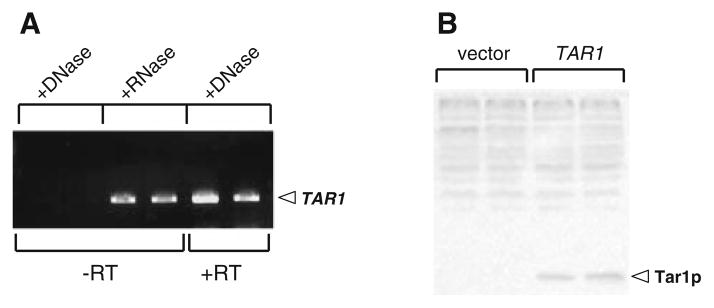
It was previously concluded, based on dot-blot analysis, that that the endogenous TAR1 mRNA is transcribed in vivo (Coelho et al. 2002). However, the TAR1 transcript cannot be unequivocally identified by Northern blot (data not shown). Given the potential for non-sequence-specific hybridization or hybridization to DNA, we tested whether expression of endogenous TAR1 could be unambiguously determined by reverse-transcriptase PCR (RT-PCR). Using primers specific to TAR1 we were able to amplify a cDNA fragment by RT-PCR in a sample treated with DNase I to remove genomic DNA (Fig. 1a; two right-hand lanes, +DNase, +RT). This product was not the result of amplification of contaminating (undigested) genomic DNA since no product was observed in the same sample in absence of reverse transcripase under identical PCR conditions (Fig. 1a; left-hand two lanes, +DNase, −RT). Finally, that the RT-PCR product is of the predicted size for TAR1 (and not affected by the presence of RNA) was confirmed by performing the same PCR reaction in an identical sample in which the genomic DNA was not digested by DNase I, but treated with RNase (Fig. 1a; middle two lanes, +RNase, −RT). This latter control amounts to a normal TAR1 PCR reaction from genomic DNA (in the absence of competing rRNA) to confirm the size of the expected product from the RT-PCR reaction. We next attempted to detect endogenous Tar1p protein by western blot analysis, using a rabbit polyclonal antibody raised against a synthesized peptide corresponding to a portion of Tar1p. We were unable to detect endogenous Tar1p in whole cell lysates or in purified mitochondria from strains grown under a variety of standard growth conditions (data not shown). However, we were able to detect untagged Tar1p expressed from the strong constitutive TEF1 promoter in whole cell lysates using this antibody (Fig. 1b).
Fig. 1.
Expression of the endogenous TAR1 gene can be detected at the transcript level, but not the protein level. a Detection of the endogenous TAR1 transcript by reverse transcriptase PCR (RT-PCR). All reactions were carried out in duplicate using two different RNA preparations. Lanes 1 and 2 show the results of RT-PCR on samples treated with DNase and with no RT enzyme included in the cDNA synthesis step (control for contaminating genomic DNA); lanes 3 and 4 show PCR products generated from genomic DNA in samples treated with RNase (control for size of expected RT-PCR product); and lanes 5 and 6 show a product amplified from the endogenous TAR1 transcript by RT-PCR in the complete absence of genomic DNA. b Western blot using a Tar1p peptide antibody. Lanes 1 and 2 are duplicate protein samples from a wild type strain transformed with empty vector; lanes 3 and 4 are duplicate protein samples from a strain transformed with a vector expressing Tar1p from the constitutive TEF1 promoter
Tar1p is up-regulated in response to pro-respiratory growth conditions and in the absence of glucose repression
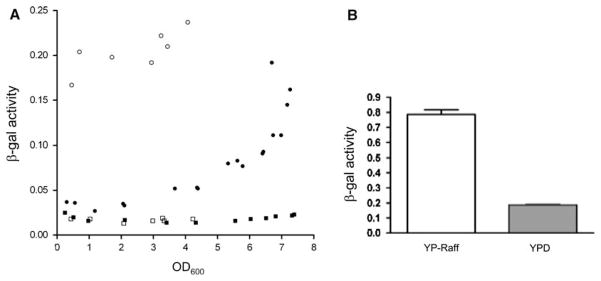
In the absence of an ability to detect endogenous Tar1p by Western blot, we investigated its expression under different growth conditions using a translational reporter, consisting of the E. coli LacZ gene fused in-frame with a TAR1 ORF in the chromosomal rDNA locus (Kumar et al. 2002). Interestingly, when grown to mid-log phase in rich glucose medium, cells harboring the Tar1p-LacZ translational fusion showed levels of β-galactosidase activity only marginally higher than the background observed in the isogenic control strain lacking LacZ (Fig. 2a). However, β-galactosidase activity rose sharply after post-diauxic shift, when metabolism is shifted from anaerobic fermentation to aerobic respiration, with highest reporter activity observed in stationary phase (Fig. 2a). Saccharomyces cerevisiae is well known for its preference for anaerobic fermentation as its main energy metabolism pathway, and many genes involved in respiration and mitochondrial biogenesis are inhibited in the presence of glucose. Since Tar1p is a mitochondrial protein, we hypothesized that its expression could be stimulated by depletion of glucose from the growth medium in stationary phase. To test this hypothesis, we measured expression of the Tar1p β-galactosidase reporter in cells grown in YPG medium containing the non-fermentable carbon source glycerol. Under these growth conditions, which require respiration, Tar1p-LacZ activity was much higher at all stages of growth than in YPD (rich glucose) medium (Fig. 2a). Interestingly, although there may be a slight increase in LacZ activity at higher culture densities during YPG growth, the magnitude of this increase is much less than that exhibited in YPD medium, suggesting that Tar1p is up-regulated under respiratory growth conditions specifically and not simply during stationary phase. It should be pointed out that the maximum achievable culture density is substantially lower for YPG cultures than for YPD cultures. Finally, in log-phase cultures, TAR1-LacZ activity was also substantially higher when raffinose was substituted for glucose in rich medium (Fig. 2b), indicating that the repression of TAR1 expression in log-phase glucose cultures (and its induction in glycerol medium) is largely due to the effects of glucose repression. Consistent with the results described above (Fig. 1b), we were unable to detect the presence of the Tar1p-LacZ fusion protein by western blot (data not shown), even though its existence could be unambiguously confirmed by the detectable β-galactosidase activity (i.e. the fusion protein activity must be the product of transcription and translation of the chromosomal TAR1 gene since LacZ is fused in-frame with the TAR1 ORF).
Fig. 2.
Expression of Tar1p increases during growth in conditions requiring respiration. a Plotted are measurements of β-galactosidase activity (y-axis; see “Materials and methods”) as a function of growth state (OD600, x-axis) in the LacZ reporter strain 155F6 (circles) and its isogenic parental strain Y800 (squares) in either YPD (solid symbols) or YPG (open symbols). b Shown are β-galactosidase activity measurements (y-axis) of the 155F6 strain during log-phase growth (OD600 between 0.4 and 0.6) in rich raffinose (YP-Raff) or glucose (YPD) medium. The bars represent the mean obtained from triplicate samples of each culture with the standard deviation represented by the brackets
Tar1p is down-regulated in cells exhibiting mitochondrial dysfunction
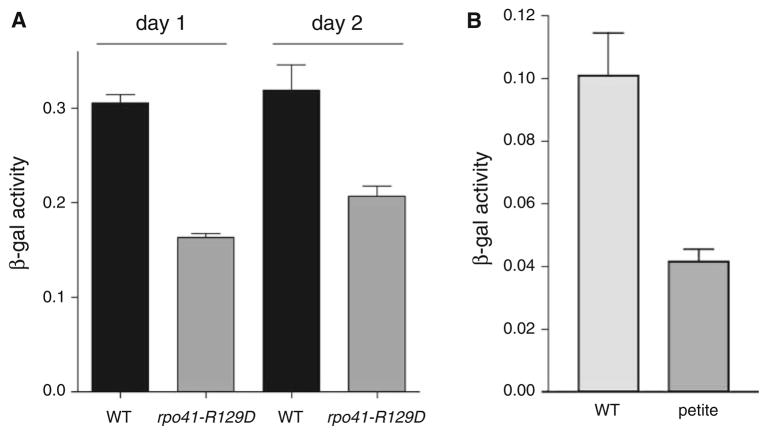
Given its unique position in the nuclear genome, imbedded within the ribosomal RNA array, we sought to investigate the possibility that Tar1p could represent a novel pathway of communication between the nucleus and the mitochondrion. Previous work has shown that in addition to coordinating nuclear and mitochondrial gene expression in response to changing nutrient conditions, pathways such as the retrograde response can also serve to signal mitochondrial dysfunction to the nucleus, enacting a program of compensatory gene expression changes. To test the possibility that Tar1p could be involved in such a pathway we measured expression of our Tar1p reporter in the previously described rpo41-R129D mitochondrial RNA polymerase mutant. This mutant shows an array of phenotypes indicative of mitochondrial dysfunction, including increased reactive oxygen species (ROS), slow growth in glycerol medium, and decreased chronological lifespan (Bonawitz et al. 2006b). Interestingly, this mutant exhibited levels of Tar1p approximately half of wild type levels in stationary phase (Fig. 3a). This effect was not due to a simple lag in growth or entry into stationary phase in the mutant, as Tar1p levels were still deficient in rpo41-R129D after an additional 24 h in stationary phase (Fig. 3a). Furthermore, the decreased expression of Tar1p exhibited by the rpo41-R129D mutant is not the result of a general down-regulation of mitochondrial biogenesis, since levels of the mitochondrial-encoded OXPHOS subunit Cox2p were identical in wild type and rpo41-R129D stationary phase cultures (data not shown). We next tested whether the down-regulation of Tar1p described above is peculiar to the rpo41-R129D mutant, or occurs as a general response in strains exhibiting defective respiration. To obtain this information, we took advantage of the fact that most strains of yeast spontaneously generate clones unable to respire (petite) typically due to mutation or loss of mtDNA. We isolated three independent petite clones of a strain harboring the Tar1p-LacZ genomic fusion described above, and tested β-galactosidase activity in stationary phase rich media. The results of this experiment show that, like the rpo41-R129D mutant, these petite strains also exhibited down-regulation of Tar1p (Fig. 3b). We conclude from these data that Tar1p expression is down-regulated in response to generalized mitochondrial respiratory dysfunction.
Fig. 3.
Tar1p expression is down-regulated in strains exhibiting mitochondrial dysfunction. a β-galactosidase activity at day 1 and day 2 (post-diauxic shift) from strains 155F6 expressing either wild type RPO41 gene (WT) or the rpo41-R129D allele (rpo41-R129D). b Tar1-LacZ activity at day 1 post-diauxic shift of strain 155F6 containing functional mitochondria (WT) or respiratory deficient mitochondria (petite). Strains shown in (b) were grown in rich glucose medium (YPD), but those in (a) were grown in synthetic glucose medium (SD) to select for plasmids. Error bars in both panels represent standard deviation
Over-expression of Tar1p exacerbates the phenotype of a mitochondrial RNA polymerase point mutant
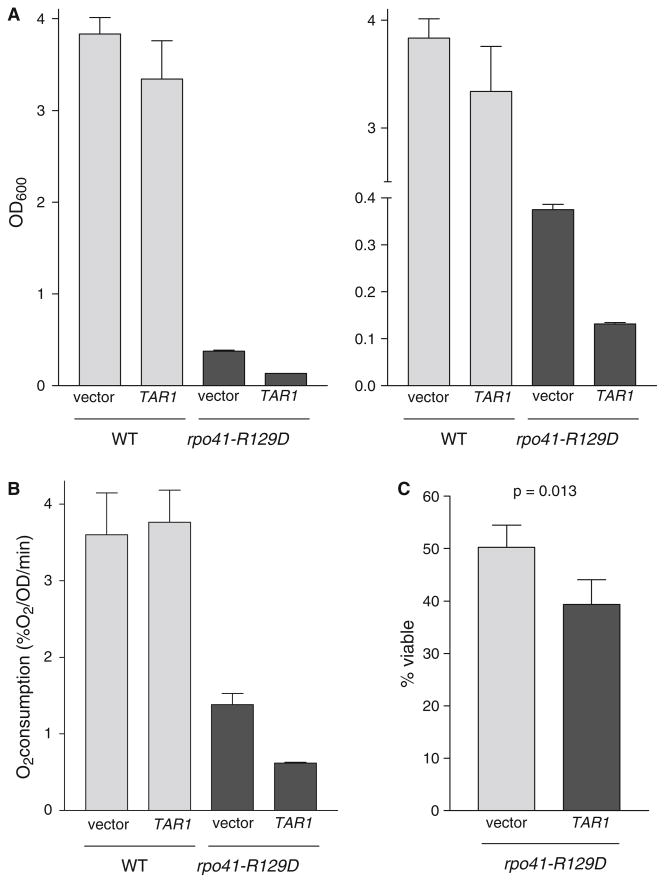
One of the best-understood pathways of nuclear-mitochondrial communication in yeast is the retrograde response, which prevents the failure of amino acid biosynthesis in petite cells by increasing the expression of several tricarboxylic acid cycle enzymes (Liu and Butow 2006). By analogy, we hypothesized that down-regulation of Tar1p in cells with mitochondrial dysfunction could potentially serve as a protective or compensatory mechanism undertaken to mitigate the negative effects associated with the inhibition of respiration. To test this hypothesis, we over-expressed Tar1p from the strong, constitutive TEF1 promoter in the rpo41-R129D mutant. This promoter led to a substantial increase in the steady-state level of Tar1p (Fig. 1b). Interestingly, this high level over-expression of Tar1p exacerbated the previously described phenotypes of the rpo41-R129D mitochondrial RNA polymerase mutant, leading to slower growth in glycerol, lower levels of respiration, and shortened lifespan (Fig. 4). In contrast, over-expression of Tar1p from the same promoter had no effect on any of these phenotypes in the isogenic wild type strain (Fig. 4). These data are consistent with a model wherein Tar1p down-regulation in response to mitochondrial dysfunction serves a protective function, and thus forced expression from an exogenous promoter circumvents this function, exacerbating the negative downstream impact of defective respiration.
Fig. 4.
High-level over-expression of Tar1p exacerbates the phenotype of the mitochondrial RNA polymerase rpo41-R129D mutant. a OD600 of wild type and rpo41-R129D strains containing empty vector or TAR1 over-expression vector grown for 24 h in glycerol medium (YPG) after inoculation to an OD600 of 0.01. The left and the right panels show the same information, but for clarity the y-axis scale has been broken in the right panel to better illustrate the effect of TAR1 over-expression on YPG growth in the rpo41-R129D mutant. b Oxygen consumption at day 1 (post-diauxic shift) in synthetic glucose medium (SD) of the strains described in (a), normalized to number of viable cells. c Viability (i.e. chronological life span) of rpo41-R129D strains grown in synthetic glucose medium (SD) and containing either empty vector or over-expressing TAR1 at day 1 (post diauxic shift). Corresponding wild type cultures were greater than 99% viable at this same time point (data not shown)
Identification of Coq5p as a binding partner for Tar1p
To gain insight into the mechanism by which Tar1p impacts mitochondrial function, we performed a yeast 2-hybrid screen to identify interacting protein partners. From a screen of ~50,000 library transformants, we identified 34 positives (strains that expressed high levels of a Gal4p-driven HIS3 reporter; see “Materials and methods” for details). These were carried through additional tests to confirm histidine prototrophy, expression of an interaction-dependent LacZ reporter, and the specificity of the interaction for Tar1p. Of the eleven positives that survived these secondary screens, nine were shown (via sequencing of the library plasmid they contained) to encode Coq5p, a mitochondrial C-methyltransferase involved in the synthesis of CoQ. Characterization of a representative from this group is shown in Fig. 5a. One requirement for the yeast 2-hybrid system to work properly is that the Gal4p-activation-domain (Gal4-AD) fusion protein (prey) must localize to the nucleus in order to interact with the Gal4-DNA-binding domain fusion protein (bait) and activate the GAL4-driven reporters. Sequencing of the fusion junctions between the COQ5 gene and the Gal4-AD in the isolated prey plasmids revealed not only that the COQ5 ORF was in-frame with the Gal4-AD, but also that a significant portion of the predicted N-terminal mitochondrial targeting sequence was deleted in each case (data not shown). This presumably was a requirement to prevent the fusion prey protein from localizing to the mitochondrion instead of the nucleus, in which case a 2-hybrid interaction would be prevented from occurring. The other two 2-hybrid positives contained a portion of the DJP1 gene, which is a peroxisomal chaperone protein.
Although the yeast two-hybrid result described above is strong evidence of an interaction between Tar1p and Coq5p, we sought to confirm this interaction biochemically via an in vitro-binding experiment using recombinant protein. We expressed N-terminal GST-tagged Tar1p and native Coq5p separately in E. coli, as well as unmodified GST peptide for use as a negative control. As described in detail in the “Materials and methods”, we incubated glutathione sepharose beads bound with either GST-Tar1p or GST with a pre-cleared soluble lysate from Coq5p-expressing bacteria. The beads were then split into two equal samples, one of which was boiled in SDS gel-loading buffer (to remove all bound proteins), and the other of which was treated with buffer containing reduced glutathione (to elute GST and GST-Tar1p while leaving behind any proteins that non-specifically bind to the beads). As shown in the top panel of Fig. 5b, western blotting using an anti-GST antibody revealed that both GST and GST-Tar1p were able to be bound to and eluted from the glutathione sepharose beads. The bound and eluted fractions were then probed for the presence of Coq5p by western analysis. While some Coq5p did bind to the negative control (GST) beads under these conditions, substantially more Coq5p was bound by beads containing GST-Tar1p (despite the fact the quantity of GST peptide bound was much greater than that of GST-Tar1p). More importantly, elution with reduced glutathione released Coq5p in the case of GST-Tar1p-bound beads, but not in the case of GST-bound beads (Fig. 5b, bottom panel, lanes 3 and 4), indicating that the Coq5p associated with the negative control, GST-containing beads (Fig. 5b, bottom panel, lane 1) was the result of binding non-specifically to either the beads or a non-GST protein in the lysate that itself was non-specifically bound to the beads. These results confirmed the yeast two-hybrid interaction described above by showing these proteins can also interact in vitro.
Discussion
This study represents the first detailed examination of Tar1p, a very unusual protein that we and our collaborators found is encoded by a previously non-annotated gene located on the anti-sense strand of the nuclear 25S ribosomal RNA gene (Coelho et al. 2002). We speculated previously that Tar1p may function to modulate mitochondrial gene expression and/or respiration and may also serve to coordinate nuclear rRNA expression with mitochondrial metabolism (Coelho et al. 2002). However, since the TAR1 gene is located in the repetitive rDNA array and completely within the 25S rRNA coding sequence, a traditional gene knock-out approach to gain insight into its function has been problematic. Thus, as an alternative approach, we have instead examined its expression from the rDNA locus under a variety of conditions and have performed a 2-hybrid screen to identify potential binding partners. We draw the following primary conclusions from this study: (1) Tar1p expression is stringently controlled (i.e. maintained at very low steady-state levels), but is up-regulated in the absence of glucose repression when mitochondrial respiration and biogenesis are elevated; (2) Tar1p expression is down-regulated in response to mitochondrial dysfunction in order to avoid or mitigate the deleterious consequences of this form of cellular stress; and (3) Tar1p interacts with Coq5p, an mitochondrial enzyme involved in the synthesis of the electron carrier/antioxidant CoQ, possibly in order to modulate respiration and/or ROS homeostasis. The basis for these conclusions and other salient points from this study are discussed below.
Consistent with its previously determined mitochondrial localization (Coelho et al. 2002), we found that the expression of Tar1p from its endogenous promoter in the rDNA locus increases in rich glucose medium after diauxic shift (Fig. 2a). At this stage of growth, glucose is largely exhausted from the culture medium, forcing yeast to switch from the preferred mode of anaerobic, fermentative metabolism to aerobic mitochondrial respiration. Furthermore, the expression of Tar1p increases in medium containing the non-fermentable carbon source glycerol (Fig. 2a) or the fermentable, but non-repressing sugar raffinose (Fig. 2b). These data, in conjunction with additional supporting evidence described below, reinforce the original conclusion that Tar1p is indeed a mitochondrial protein involved in some aspect of respiratory metabolism and indicate that its expression is subject to canonical glucose repression.
One potential key to understanding the function of TAR1 is our observation that the Tar1p protein accumulates to only very low levels in S. cerevisiae. Although we were able to specifically detect endogenous TAR1 transcript for the first time (Fig. 1), we were unable to detect its protein product by western blot. We are confident that our inability to detect Tar1p is not due to a defective antibody, since we are easily able to detect Tar1p over-expressed from the constitutive TEF1 promoter. We also conclude that the failure to detect endogenous Tar1p is not due to the complete lack of expression of Tar1p, as we cannot detect by western blot the Tar1p-LacZ fusion protein under the control of the endogenous TAR1 promoter, even when its presence can be unambiguously confirmed via β-galactosidase activity (Fig. 2). It is noted that endogenous levels of a number of other mitochondrial regulatory proteins, such as Pif1p or Pet309p, also cannot be detected by western blot despite their playing critical roles in mitochondria (Lahaye et al. 1991; Manthey et al. 1998).
Another important insight into the function of Tar1p is its down-regulation in response to mitochondrial dysfunction. We found that Tar1p expression is substantially decreased in petite strains lacking respiration or experiencing mitochondrial respiratory dysfunction as a result of the rpo41-R129D mutant (Fig. 3). In contrast to the previously reported beneficial effect of moderate over-expression of Tar1p (Coelho et al. 2002), we found that high-level over-expression of Tar1p from the strong TEF promoter greatly exacerbated all of the rpo41-R129D mutant phenotypes, including slow growth on glycerol, decreased chronological life span, and progressive failure of respiration (Fig. 4). Together, we interpret these observations to indicate that the level of Tar1p expression that is optimal for growth is stringently regulated, and that amounts that are either too low or too high are detrimental to fitness. Interestingly, this characteristic of a tight optimum range of expression is common among gene products involved in mitochondrial genome maintenance such as Mip1p and Abf2p (Lecrenier and Foury 1995; Zelenaya-Troitskaya et al. 1998). Finally, we also emphasize that although the ability of Tar1p to rescue the glycerol growth defect of rpo41-R129D colonies on solid medium is, in part, how Tar1p was identified originally (Coelho et al. 2002), this effect is variable and difficult to reproduce in liquid glycerol medium (data not shown). Thus, it is possible that the previously reported effects of Tar1p on the rpo41-R129D phenotype are due to a stochastic suppression event that occurred during selection on sold medium, whereas its high level over-expression reported here elicits a straightforward and consistent exacerbation of multiple ROS-mediated phenotypes of the rpo41-R129D mutant in liquid cultures.
The results discussed thus far strongly suggest that Tar1p expression, when optimal, plays a role in promoting respiration (either directly or indirectly) or in alleviating the production or harmful effects of mitochondrial-derived ROS. This conclusion is bolstered by the final result from this study, that Tar1p interacts with Coq5p (Fig. 5). Such an interaction makes a great deal of biological sense, since both Tar1p and Coq5p are involved in respiration-related processes. For example, Tar1p may influence the rate of synthesis (or methylation status) of CoQ as a means to regulate respiration or the production of ROS from the electron transport chain. Here, it is worth noting that the dwell time of reduced CoQ has been implicated directly in the production of superoxide from the mitochondrial respiratory chain (James et al. 2004). However, CoQ also has been shown to be an antioxidant and to be present in other cellular membranes (James et al. 2004). Thus, an alternative (or additional) possibility is that Tar1p (via Coq5p-mediated effects on CoQ synthesis) impacts the ability of cells to combat ROS-mediated damage. Finally, it is noteworthy that the expression of the COQ5 gene is regulated by energy source (Hagerman et al. 2002; Hagerman and Willis 2002) and, like TAR1 (Fig. 2a), its transcription is up-regulated after the post-diauxic shift (DeRisi et al. 1997) and in stationary phase (Gasch et al. 2000) glucose cultures. It is under specific conditions such as these when the interaction between these two proteins is most likely to be functionally relevant. Determining the precise functional ramifications of the Tar1p-Coq5p interaction and the transcription factors and signaling pathways that mediate their specific expression profiles in response to different environmental and growth conditions represents an intriguing area of future investigation.
It is tempting to expand on our idea that the unique genomic location of TAR1 is relevant to its function. Indeed, it is easy to imagine that transcription of TAR1 would be antagonized by robust rRNA transcription proceeding in the opposite direction. The observation that Tar1p expression increases in post-diauxic and stationary phases, when transcription of rRNA is strongly down-regulated (Fig. 2) is consistent with this idea, as is the up-regulation of Tar1p in glycerol medium (Fig. 2), where rDNA transcription and ribosome biogenesis are significantly down-regulated to match the slower growth rate (Kief and Warner 1981; Warner 1989). Furthermore, in addition to Pol I transcriptional regulation, the rDNA locus is subject to complex gene silencing mechanisms (Rusche et al. 2003). Thus, it will be of great interest to understand how such factors reciprocally impact rRNA and Tar1p expression from this dynamic locus, as well as how and why these changes are linked to mitochondrial metabolism, perhaps as one means to link the expensive process of ribosome biogenesis to the primary energy generator, the mitochondrion. Interestingly, the mammalian ribin protein (Kermekchiev and Ivanova 2001), the mRNA for which is in part complementary to rRNA and hence analogous to Tar1p, is involved in the response to growth factors, suggesting that mechanisms that link rDNA-related transcripts to cellular metabolism may be conserved. Finally, both respiration and extrachromosomal rDNA circles (ERCs), which contain TAR1, have profound effects on yeast replicative life span (Sinclair and Guarente 1997; Lin et al. 2002). Thus, it is tempting to speculate that altered expression of Tar1p from ERCs may play a role in yeast aging by influencing respiration or exacerbating the effects of mitochondrial ROS.
Acknowledgments
This work was supported by grant DAAD19-00-1-0650 from the Army Research Office. M.C-L. was supported by NIH Training grant GM07499-30. C.M.W. was a visiting student from the University of Bath when this work was performed. We wish to thank Dr. Michael Snyder for providing the Tar1p::LacZ reporter strain and the anti-Tar1p peptide antibody, Dr. Catherine Clarke for the Coq5p antibody, and Dr. William Studier for the bacterial Coq5p expression plasmid used in this study.
Footnotes
Communicated by R. Jensen.
Contributor Information
Nicholas D. Bonawitz, Department of Pathology and Genetics, Yale University School of Medicine, 310 Cedar Street, P.O. Box 208023, New Haven, CT 06520-8023, USA. Graduate Program in Genetics and Molecular Biology, Emory University School of Medicine, Altanta, GA, USA
Marc Chatenay-Lapointe, Department of Pathology and Genetics, Yale University School of Medicine, 310 Cedar Street, P.O. Box 208023, New Haven, CT 06520-8023, USA. Graduate Program in Genetics, Yale University School of Medicine, New Haven, CT, USA.
Christopher M. Wearn, Department of Biochemistry, Emory University School of Medicine, Rollins Research Center, Atlanta, GA 30322, USA
Gerald S. Shadel, Email: gerald.shadel@yale.edu, Department of Pathology and Genetics, Yale University School of Medicine, 310 Cedar Street, P.O. Box 208023, New Haven, CT 06520-8023, USA
References
- Baba SW, Belogrudov GI, Lee JC, Lee PT, Strahan J, Shepherd JN, Clarke CF. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J Biol Chem. 2004;279:10052–10059. doi: 10.1074/jbc.M313712200. [DOI] [PubMed] [Google Scholar]
- Barkovich RJ, Shtanko A, Shepherd JA, Lee PT, Myles DC, Tzagoloff A, Clarke CF. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J Biol Chem. 1997;272:9182–9188. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Shadel GS. Rethinking the mitochondrial theory of aging: the role of mitochondrial gene expression in lifespan determination. Cell Cycle. 2007;6:1574–1578. doi: 10.4161/cc.6.13.4457. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006a;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol Cell Biol. 2006b;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Steerns T. A Cold Springs Harbor Laboratory course manual. Cold Spring Harbor Press; Cold Spring Harbor: 2000. Methods in yeast genetics; p. 205. [Google Scholar]
- Coelho PS, Bryan AC, Kumar A, Shadel GS, Snyder M. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 2002;16:2755–2760. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RA, Willis RA. The yeast gene COQ5 is differentially regulated by Mig1p, Rtg3p and Hap2p. Biochim Biophys Acta. 2002;1578:51–58. doi: 10.1016/s0167-4781(02)00496-7. [DOI] [PubMed] [Google Scholar]
- Hagerman RA, Trotter PJ, Willis RA. The regulation of COQ5 gene expression by energy source. Free Radic Res. 2002;36:485–490. doi: 10.1080/10715760290021360. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- James AM, Smith RA, Murphy MP. Antioxidant and prooxidant properties of mitochondrial Coenzyme Q. Arch Biochem Biophys. 2004;423:47–56. doi: 10.1016/j.abb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Johnston M, Carlson M. Regulation of carbon and phosphate utilization. Cold Springs Harbor Press; Plainview: 1991. [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kermekchiev M, Ivanova L. Ribin, a protein encoded by a message complementary to rRNA, modulates ribosomal transcription and cell proliferation. Mol Cell Biol. 2001;21:8255–8263. doi: 10.1128/MCB.21.24.8255-8263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kief DR, Warner JR. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Harrison PM, Cheung KH, Lan N, Echols N, Bertone P, Miller P, Gerstein MB, Snyder M. An integrated approach for finding overlooked genes in yeast. Nat Biotechnol. 2002;20:58–63. doi: 10.1038/nbt0102-58. [DOI] [PubMed] [Google Scholar]
- Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. Embo J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecrenier N, Foury F. Overexpression of the RNR1 gene rescues Saccharomyces cerevisiae mutants in the mitochondrial DNA polymerase-encoding MIP1 gene. Mol Gen Genet. 1995;249:1–7. doi: 10.1007/BF00290229. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Manthey GM, Przybyla-Zawislak BD, McEwen JE. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur J Biochem. 1998;255:156–161. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- Petes TD, Botstein D. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci USA. 1977;74:5091–5095. doi: 10.1073/pnas.74.11.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon L, Schatz G. Biogenesis of yeast mitochondria. 1. Cold Springs Harbor Press; Plainview: 1991. [Google Scholar]
- Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Aguilera JC, Gavilan A, Asencio C, Navas P. The role of ubiquinone in Caenorhabditis elegans longevity. Ageing Res Rev. 2005;4:41–53. doi: 10.1016/j.arr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Sheehan A, Roeder GS, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:190–195. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- Shadel GS. Yeast as a model for human mtDNA replication. Am J Hum Genet. 1999;65:1230–1237. doi: 10.1086/302630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Suppl):S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregationrecombination and copy number in Saccharomyces cerevisiae. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]