Abstract
E. coli lipoic acid ligase (LplA) catalyzes ATP-dependent covalent ligation of lipoic acid onto specific lysine sidechains of three acceptor proteins involved in oxidative metabolism. Our lab has shown that LplA and engineered mutants can ligate useful small-molecule probes such as alkyl azides (Nat. Biotechnol. 2007, 25, 1483–1487) and photocrosslinkers (Angew. Chem Int. Ed Engl. 2008, 47, 7018–7021) in place of lipoic acid, facilitating imaging and proteomic studies. Both to further our understanding of lipoic acid metabolism, and to improve LplA’s utility as a biotechnological platform, we have engineered a novel 13-amino acid peptide substrate for LplA. LplA’s natural protein substrates have a conserved β-hairpin structure, a conformation that is difficult to recapitulate in a peptide, and thus we performed in vitro evolution to engineer the LplA peptide substrate, called “LplA Acceptor Peptide” (LAP). A ~107 library of LAP variants was displayed on the surface of yeast cells, labeled by LplA with either lipoic acid or bromoalkanoic acid, and the most efficiently labeled LAP clones were isolated by fluorescence activated cell sorting. Four rounds of evolution followed by additional rational mutagenesis produced a “LAP2” sequence with a kcat/Km of 0.99 μM−1min−1, >70-fold better than our previous rationally-designed 22-amino acid LAP1 sequence (Nat. Biotechnol. 2007, 25, 1483–1487), and only 8-fold worse than the kcat/Km values of natural lipoate and biotin acceptor proteins. The kinetic improvement over LAP1 allowed us to rapidly label cell surface peptide-fused receptors with quantum dots.
Introduction
Most proteins are evolved to interact with a multitude of cellular molecules and thus contain a number of distinct domains, binding sites, and activities. Often, it is useful to the biochemist to reduce a specific aspect of a protein’s function to just a peptide fragment. This can help to determine the minimal features of a protein required for a specific function such as binding, recognition by an enzyme, translocation, or folding.1–4 It may also be desirable to create a consensus peptide substrate for assay purposes,5;6 or to use a peptide in place of a protein to facilitate crystallography of multi-protein complexes.7;8 For therapeutic applications, replacement of protein drugs with peptides having similar activity can improve tissue penetration and reduce immunogenicity.9;10 Our lab is interested in protein minimization to peptides for the purpose of developing new protein labeling technologies. Size minimization of protein tags that direct the targeting of fluorescent probes11 can greatly reduce problems of tag interference with protein trafficking, folding, and interactions.
Conversion of proteins to peptides without loss of the function of interest, however, is challenging for a number of reasons. First, the function may require secondary structure that is difficult to recapitulate in a peptide. Second, the function may require contributions from multiple, non-contiguous regions of a protein. Third, structural information is not available for many proteins, and in some cases, even the regions that contribute to a protein’s relevant activity are not known. Fourth, due to their more flexible structure, peptide binding is often associated with a greater entropic penalty than is protein binding12, making it more difficult to engineer high-affinity interactions.
Numerous methods have been used to reduce proteins to peptides. Simple truncation and/or rational design can be successful,13–15 but is usually associated with at least a partial loss of activity and/or specificity. Peptide scanning16 or high-throughput screening17–19 approaches are more exhaustive, but library sizes are limited (typically 102 – 105), so it is difficult to identify optimal sequences. Peptide selections, on the other hand, can process libraries up to 109 in size, dramatically increasing the probability of identifying a successful sequence. Accordingly, selections on phage,20–23 inside bacteria,24 and on the surface of bacteria,25 yeast,26 and mammalian cells,27 have been used to evolve peptides with novel functions.
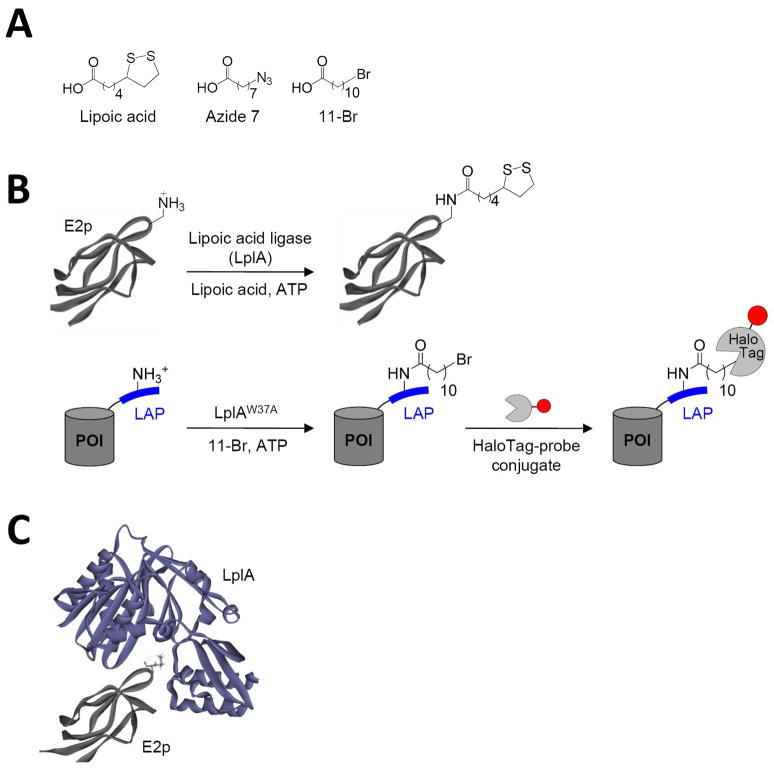
In this study, our goal was to identify novel, kinetically efficient peptide substrates for E. coli lipoic acid ligase (LplA) (Figure 1). LplA is a cofactor ligase that our lab has harnessed for fluorescent protein labeling applications.13;28 The natural function of LplA is to catalyze ATP-dependent, covalent ligation of lipoic acid (Figure 1A) onto specific lysine sidechains of three E. coli proteins involved in oxidative metabolism: pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, and the glycine cleavage system.29 Previously, we showed that LplA and engineered variants could ligate unnatural probes such as an alkyl azide (a functional group handle for fluorophore introduction; Figure 1A),13 a fluorinated aryl azide photocrosslinker,28 bromoalkanoic acid (a ligand for HaloTag30; Figure 1A),31 and a coumarin fluorophore32 in place of lipoic acid. To utilize these ligation reactions for protein imaging applications, we prepared recombinant fusions of proteins of interest (POIs) to the 9 kD E2p domain of pyruvate dehydrogenase (Figure 1B top).13 Such fusions could be labeled with high efficiency and specificity by our unnatural probes on the surface and in the cytosol of living mammalian cells.13;28;31;32
Figure 1.
LplA-catalyzed protein and peptide labeling reactions. (A) Natural (lipoic acid) and unnatural (Azide 7 and 11-Br) small-molecule substrates of LplA and its W37 mutant. (B) Natural and engineered ligation reactions. Top: LplA-catalyzed lipoylation of the 9 kD E2p domain of E. coli pyruvate dehydrogenase (structure from PDB 1QJO). Bottom: LplA-catalyzed 11-Br ligation onto an engineered LAP (“LplA Acceptor Peptide”), which is genetically fused to any protein of interest (POI). Ligated alkyl bromide can be specifically and covalently modified by HaloTag-fluorophore conjugates.31 The red circle represents any probe. (C) Model for interaction between LplA (purple, from PDB 1X2H)33 and E2p (grey, from PBD 1QJO).34 The lipoylation site on E2p, Lys41, is rendered in stick.
Even though 9 kD (85 amino acids) E2p is considerably smaller than Green Fluorescent Protein (27 kD) and other protein labeling tags such as HaloTag (33 kD)30 and SNAP tag (20 kD),35 we wanted to further reduce its size, to minimize steric interference with POI function. We previously attempted this by rational design of an “LplA acceptor peptide” (LAP1),13 based mostly on the sequence of LplA’s natural protein substrate 2-oxoglutarate dehydrogenase, with a few additional rational mutations. LAP1 is 17 amino acids long, or 22 amino acids with the recommended linker.13 We found that LAP1 fusion proteins could be ligated by LplA to some probes (lipoic acid,13 alkyl azide,13 and aryl azide28 in vitro and in cell lysate, but not on the cell surface except under conditions of high LAP1-POI over-expression.13;28 We could never detect LAP1 labeling in the cytosol.32 Other probes (bromoalkanoic acid and coumarin) could not be ligated to LAP1 fusions in any context.31;32 In contrast, E2p fusions could be labeled by all probes on the cell surface and in the cytosol.13;28;31;32 Since the measured kcat values for azide 7 ligation, for instance, are similar for LAP1 and E2p (0.048 ± 0.001 sec−1 and 0.111 ± 0.003 sec−1, respectively13), we attribute the difference in labeling outcomes to the gap in their Km values. H-protein of the glycine cleavage system has a Km of 1.2 μM,36 which is likely to be similar to E2p’s Km, due to their sequence and structural similarity.37 On the other hand, we estimate that the Km for LAP1 is >200 μM, as measured by HPLC (data not shown).
For this study, we selected yeast surface display38 as our platform to evolve a novel peptide substrate for LplA (called “LAP2”), with kinetic properties comparable to those of LplA’s natural protein substrates. We preferred yeast display to other evolution platforms for a number of reasons. Selections in bacterial cytosol24 do not allow fine adjustment of protein concentrations and selection conditions. Phage display has limited dynamic range, both due to displayed peptide copy number (3–5 on pIII or 2700 on pVIII39), and due to the all-or-nothing nature of affinity-based product capture. The limited dynamic range makes it very difficult to enrich kinetically efficient peptide substrates, as we discovered in our phage display evolution of yAP, a peptide substrate for yeast biotin ligase.21 Mammalian cell surface display is challenging due to the need for viral transfection to control the multiplicity of infection, and the low viability of cells after fluorescence activated cell sorting (FACS).40
By careful library design, tuning of selection conditions with the help of a model selection, four rounds of selection with decreasing LplA concentrations, and additional rational mutagenesis, we engineered a 13-amino acid LAP2 with a kcat of 0.22 ± 0.01 sec−1 and a Km of 13.32 ± 1.78 μM for lipoic acid ligation. The catalytic efficiency (kcat/Km = 0.99 μM−1min−1) is closer to that of LplA’s natural protein substrate H-protein (kcat/Km = 7.95 μM−1min−1)33 than that of LAP1 (est. kcat/Km < 0.0135 μM−1min−1 for azide ligation).13 As a consequence of this improvement, we could easily lipoylate cell surface LAP2 fusion proteins, even at low expression levels. We also performed LplA-mediated specific quantum dot targeting to LAP2-LDL receptor. In comparison, quantum dot labeling was undetectable when using the same receptor fused to LAP1.
Results
Model selections
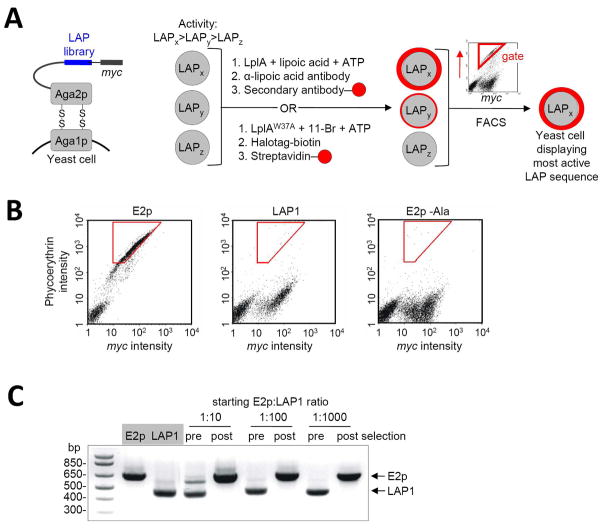
We designed the selection scheme shown in Figure 2A. A library of LAP variants is displayed on the C-terminus of Aga2p, a cell surface mating agglutinin protein commonly used for yeast display.38 A c-Myc epitope tag is also introduced to allow measurement of LAP expression levels by immunofluorescence staining. Each of 106–108 yeast cells expresses a single LAP mutant. Three hypothetical LAP mutants (LAPx, LAPy, and LAPz) with diminishing activity towards LplA are shown in Figure 2A. They are collectively labeled by LplA (e.g., with lipoic acid), and ligated probe is detected with a suitable fluorescent reagent (e.g., anti-lipoic acid antibody followed by phycoerythrin-conjugated secondary antibody). Since LAPx is the most active mutant in this scheme, yeast cells displaying this mutant should become brightly fluorescent. On the other hand, LAPy and LAPz-displaying yeast will be dimmer or unlabeled. To normalize for variations in expression level, the yeast pool is also collectively labeled with anti-c-Myc antibody, detected with a secondary antibody conjugated to Alexa Fluor 488, which is easily resolvable from phycoerythrin fluorescence. The double-labeled yeast cells are subjected to two-dimensional fluorescence activated cell sorting (FACS). Yeast cells displaying a high ratio of phycoerythrin intensity to Alexa Fluor 488 intensity (sorting gate shown in red in Figure 2A) represent the most efficiently labeled yeast, with the largest fraction of labeled LAPs, and are isolated by the FACS instrument. The captured yeast cells are amplified, sequenced, and subjected to further rounds of selection.
Figure 2.
Yeast display selection scheme and results of model selections. (A) Left: The LAP library (blue) is displayed on the yeast surface as a fusion to Aga2p protein. A C-terminal myc epitope is used to quantify LAP expression level. Right: Selection scheme. Yeast cells displaying three sample LAP sequences, with high (LAPx), moderate (LAPy), and low (LAPz) activity are shown. The yeast cells are collectively labeled with lipoic acid or 11-Br probe. The former is detected with anti-lipoic acid; the latter is detected with HaloTag-biotin,30 followed by streptavidin-fluorophore conjugate. The yeast pool is then sorted (sorting gate in red) on the basis of both ligation extent (red probe intensity) and LAP expression level (c-Myc staining intensity), to enrich the most kinetically efficient LAP peptides. (B) Determination of labeling and sorting conditions for model selections. FACS scatter plots are shown for yeast displaying E2p, LAP1,13 and E2p-Ala (E2p with a Lys41→Ala mutation), after lipoylation with 300 nM LplA for 30 minutes, and staining with anti-lipoic acid antibody. The plots show the distribution of single yeast cells as a function of phycoerthyrin staining intensity (reflecting extent of lipoylation) and c-Myc staining intensity (reflecting expression level of the Aga2p-LAP fusion). A cell population on the lower left is present in all three samples, and represents untransformed yeast. Optimized sorting gates, used for the model selections, are shown in red. (C) Results of model selections. E2p-displaying yeast and LAP1-displaying yeast were mixed at ratios of 1:10, 1:100, or 1:1000, labeled with 300 nM LplA for 30 minutes, and sorted. PCR analysis gives the ratio of yeast populations pre- and post-selection. E2p enrichment factor was >103-fold.
Before initiating selections on a LAP library, we tested and optimized our selection scheme using a model system consisting of mixtures of E2p-expressing yeast and LAP1-expressing yeast. Since LAP1 represents the best that we can achieve by rational design and E2p represents LplA’s natural substrate with evolutionarily optimized kcat/Km, we wished to design a selection that could maximally enrich E2p-yeast over LAP1-yeast. We performed lipoylation of E2p or LAP1 expressed on yeast surface by adding purified LplA, ATP, and lipoic acid to the media. FACS scanning showed that, for a 30 minute reaction time, we could obtain the largest difference in signal between E2p-yeast and LAP1-yeast using 300 nM LplA (Figure 2B). Higher LplA concentrations increased LAP1 intensity without increasing E2p intensity, diminishing the difference between them (data not shown). To check the site-specificity of LplA labeling on the yeast surface, we also performed a negative control using an E2p-Aga2p construct with a Lys→Ala mutation at the lipoylation site, and observed no phycoerythrin staining (Figure 2B).
Using 300 nM LplA, we performed 30-minute labeling on 1:10, 1:100, and 1:1000 mixtures of E2p-yeast and LAP1-yeast (E2p yeast in the minority). FACS was performed using the red gate shown in Figure 2B. We used a PCR assay to determine the ratio of yeast before and after a single round of selection, capitalizing on the different sizes of the E2p and LAP1 genes. Figure 2C shows that for all starting mixtures, the selection protocol enriched E2p yeast and depleted LAP1 yeast so completely that it could not be detected. We conclude that our selection can enrich kinetically efficient LplA substrates (e.g., E2p) over active but inefficient substrates (e.g., LAP1) by >1000-fold in a single round.
In addition to a selection based on lipoylation, we wished to develop a selection scheme based on ligation of an unnatural probe. This would serve two purposes. First, by using two different sets of probes and detection reagents in alternating rounds of selection, we could minimize the possibility of inadvertently isolating LAPs with affinity for one of our detection reagents. Second, we could increase the probability of isolating a LAP sequence that would be effective not just for lipoylation, but also for ligation of unnatural probes such as photocrosslinkers and fluorophores.
In separate work,31 we have identified mutants of LplA that catalyze ligation of bromoalkanoic acids. Once ligated to E2p or LAP, such probes can covalently react with the commercial protein HaloTag,30 which is derived from a microbial dehalogenase. Thus, we have used 11-bromoundecanoic acid (11-Br, Figure 1A) to target HaloTag-conjugated fluorophores to specific cell surface proteins (Figure 1B, bottom).31 For yeast display selections, we labeled cell surface E2p or LAP1 with the Trp37→Ala mutant of LplA mutant (LplAW37A), ATP, and the 11-Br probe. We then detected ligated bromoalkane with HaloTag protein, conjugated to biotin, and detected that in turn with streptavidin conjugated to phycoerythrin (Figure 2A). As with the lipoylation assay, we detected a large difference in phycoerythrin staining between E2p-yeast and LAP1-yeast, using 500 nM mutant LplA, and no labeling of E2p (Lys→Ala)-yeast (data not shown). Thus, 11-Br probe is also suitable for LAP selections on yeast cells.
Construction of LAP library and yeast display selections
We wished to shorten LAP, from LAP1’s 17–22 amino acids,13;28 and thus opted for a 12-mer peptide library. With complete randomization of the 11 residues flanking the central Lys, the theoretical diversity would be ~1014, far greater than the experimentally achievable library size, which is limited by yeast transformation efficiency to 107–108.41 Thus, we decided to create a partially randomized 12-mer library, guided by alignments of natural lipoate acceptor protein sequences, the NMR structure of E2p,34 and the structure of a functionally and structurally related biotin acceptor domain in complex with biotin ligase.42
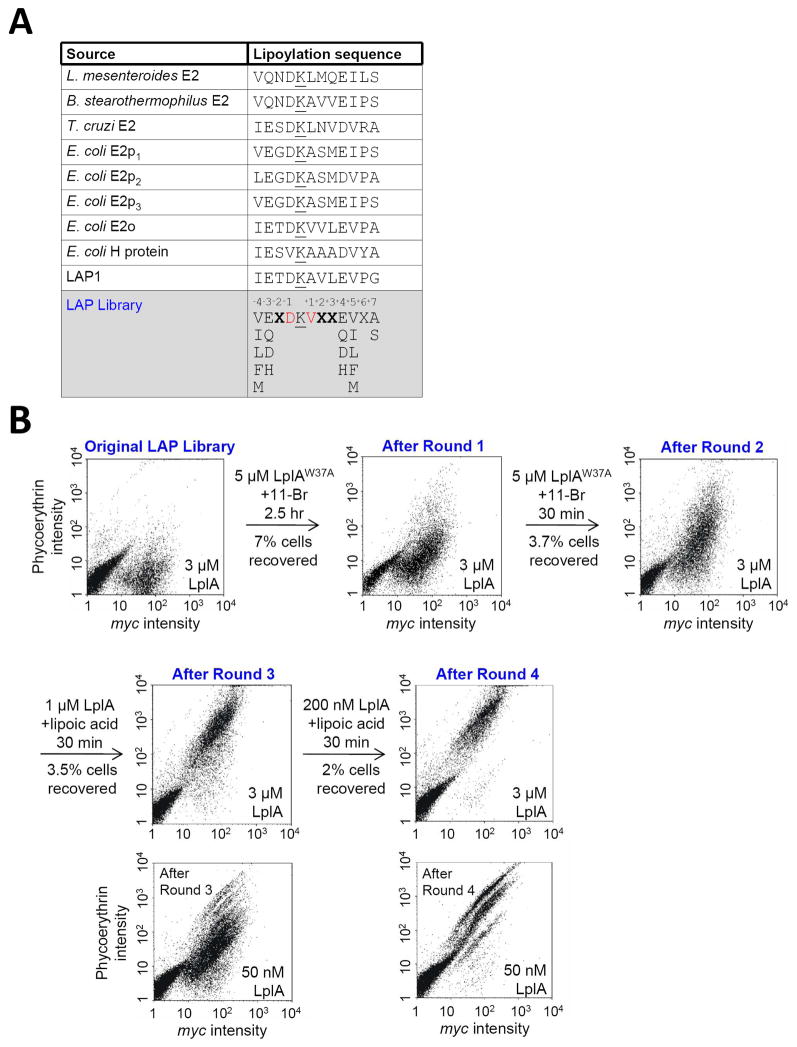
We aligned the sequences of 250 naturally lipoylated proteins (lipoate acceptor proteins) from >100 distinct species. The five lipoyl domains from E. coli (present in LplA acceptor proteins), along with lipoyl domains from three other species are shown in Figure 3A. Several trends were apparent from the alignment: (1) the −1 Asp is highly conserved; (2) positions +1, +5, and −4 are usually hydrophobic; (3) Glu and Asp are enriched at positions −3 and +4; and (4) position +6 is usually Ser or Ala. We introduced these preferences into our LAP library design, shown in Figure 3A.
Figure 3.
Library design and selection results. (A) Table showing sequences of natural LplA protein substrates, our previous rationally-designed LAP1,13 and our LAP library. Lysine modification sites are underlined. For the LAP library, we fixed positions −4 and +5 as hydrophobic amino acids (Val, Ileu, Leu, Phe, Met), positions −3 and +4 as polar amino acids (Glu, Asp, Gln, His), and position +7 as Ser or Ala. Red-colored positions are partially randomized (39% Asp or 49% Val). X represents any amino acid. (B) Results of four rounds of selection. Selection conditions, including small-molecule substrates used for labeling, are given above each arrow. To analyze amplified yeast pools following each round of selection, uniform lipoylation conditions were used (given in the lower right of each scatter plot). Yeast pools from rounds 3 and 4 were additionally analyzed under milder conditions, with 50 nM LplA.
In addition, we used structural data to inform our LAP library design. NMR structures are available for several lipoate acceptor domains.34;43–45 All of them show that the lipoylated lysine is presented at the tip of a β-hairpin turn. Though this is a challenging structure to recapitulate in a peptide, we took a cue from the structure of E. coli E2p, which shows that the −1 Asp sidechain hydrogen bonds with backbone amide N-H groups of both the central lysine and +1 Ala (Figure S1).34 To promote this loop-favoring interaction, we installed Asp at the −1 position with 39% frequency in our LAP library (Figure 3A).
There is no co-crystal structure of a lipoate acceptor domain with LplA, to indicate which residues might be important for interactions with the enzyme. However, lipoate domains are structurally similar to biotin acceptor domains,46;47 and LplA is structurally related to biotin ligase as well.48 The co-crystal structure of P. horikoshii biotin ligase with its biotin acceptor protein shows a hydrogen bond between the +4 Glu of the acceptor and Lys27 of the enzyme.42 In addition, the authors of the T. acidophilum LplA structure created a computationally docked model of their enzyme with E2p.37 The docked structure also predicts a hydrogen bond between the +4 Glu of E2p and Lys155 of the enzyme, which corresponds to Lys143 in E. coli LplA. Figure 1C shows our docked model of E. coli LplA with its E2p lipoate acceptor. Because these structures and models suggest that +4 Glu is important for interactions with LplA, we restricted the +4 position of our LAP library to polar residues (Glu, Asp, Gln, and His) to promote inter-molecular hydrogen bonding (Figure 3A).
The LAP library was cloned by Klenow-mediated fill-in of a synthetic oligonucleotide library. The insert was introduced into pCTCON2,41 containing Aga2p and the c-Myc tag, by homologous recombination. Our yeast transformation efficiency was ~107, 103-fold under our theoretical diversity of ~1010.
For reasons described above, we used both lipoic acid and bromoalkanoic acid (11-Br) probes for our selections. The latter was used for the first two rounds of selection, and lipoic acid was used for rounds 3 and 4 (Figure 3B). To successively increase selection stringency, we decreased LplA concentration throughout the selection, from 5 μM in rounds 1 and 2, to 1 μM in round 3, to 200 nM LplA in the final round. Reaction times were 2.5 hours for the first round, and 30 minutes for all subsequent rounds.
To compare the activities of recovered yeast from each round of selection, we re-amplified the yeast pools and labeled them with lipoic acid under identical conditions. Figure 3B shows that c-Myc intensities remain constant, while phycoerythrin intensities gradually increase. With 3 μM LplA, yeast recovered from rounds 3 and 4 looked identical; thus we also performed analysis under milder conditions (Figure 3B). With 50 nM LplA, it can be seen that yeast cells from round 4 are more extensively labeled by lipoic acid than yeast cells from round 3.
Characterization of selection results
The sequences of selected LAP clones from rounds 2, 3, and 4 are shown in Figures S2A and S2B. In addition, graphical representations of amino acid frequencies are shown in Figure S2C. We observed the following trends: (1) In general, selected LAP clones had interlaced hydrophobic and negatively-charged sidechains flanking the central lysine. (2) Position +2, which was fully randomized in the LAP library, became 100% Trp. This enrichment was apparent after just a single round of selection. (3) Position +3, which was also fully randomized, showed a preference for aromatic side chains. (4) Positions −3 and +4 were limited to one of 4 polar sidechains in the LAP library. Position −3 became 100% Glu. Position +4 became exclusively Glu or Asp, already by round 2. (5) Positions −4 and +5 were limited to hydrophobic residues in the LAP library. Position +5 did not converge, but position −4 became 100% Phe. (6) Position +1, which was 49% Val in the library, became 100% Val. We note that after round 4, we observed only 4 distinct clones, and further rounds of selection did not reveal any additional diversity.
A powerful feature of FACS-based selection is its dynamic range. For a single round of selection, different sorting gates can be used, and the sequences of clones obtained via different gates can be compared, to infer sequence-activity relationships. For round 4, in addition to our standard high phycoerthyrin gate (“Gate A”), we also collected yeast from a slightly lower gate (“Gate B”). Figure S2B shows that the major difference between Gate A clones and Gate B clones is the presence of Phe at the −4 position in Gate A clones. We surmised that the selection of −4 Phe may account for much of the jump in LAP activity between rounds 3 and 4. Indeed, when we mutated the −4 Phe of one of the Gate A clones, LAP4.1, to Val, its activity in a yeast surface lipoylation assay dropped to a level comparable to the Gate B clones (Figure S3).
We utilized the information from Gate A and Gate B clones (Figure S2C) to rationally design a new LAP sequence, called “LAP2”. Since Gate A clones showed clear amino acid preferences at positions −4, −3, −2, +1, +2, +4, +5, and +7, we introduced these preferred residues into our LAP2 sequence. Positions −1, +3, and +6 did not show consensus in Gate A clones, so we based these amino acids in LAP2 on preferences seen in the Gate B clones. We characterized this rationally designed LAP2 alongside the four evolved LAP clones from round 4, in cell-based and in vitro assays, described below.
Comparison of LAP sequences
To compare the round 4 LAP sequences and LAP2, we created genetic fusions to CFP-TM (cyan fluorescent protein fused to a transmembrane helix from PDGF receptor)13 for mammalian cell surface expression, and HP1 (heterochromatin protein 1)13 for bacterial expression. In all constructs, an N-terminal glycine from the Aga2p fusion was carried over, making the total LAP length 13 amino acids.
First, we compared the surface expression levels of the LAP fusions in HeLa mammalian cells. Whereas LAP4.1, LAP4.2, and LAP2 gave clear cell surface expression, both LAP4.3 and LAP4.4 showed poor expression (data not shown). We surmised that LAP4.3 expression might be hindered by its +6 Cys, due to intermolecular disulfide bond formation in the oxidizing secretory pathway. Since Gate B clones showed a preference for Asp at this position, we prepared a point mutant of LAP4.3 with a +6Cys→Asp mutation (LAP4.3D). Figure S4 shows that LAP4.3D gives improved cell surface expression compared to LAP4.3, as indicated by the pattern of CFP fluorescence. In addition, cell surface lipoylation with exogenous LplA gives a strong signal with LAP4.3D-CFP-TM, whereas little signal is detected under the same conditions with LAP4.3-CFP-TM. E. coli expression of the HP1 fusion protein also improved significantly upon introduction of the +6Cys→Asp mutation in LAP4.3. Based on these observations, we carried LAP4.3D into subsequent analyses, and we did not characterize LAP4.3 or LAP4.4 any further.
Second, we compared the LAPs in a cell surface lipoylation assay (Figure S5). CFP-TM fusion constructs were expressed in human embryonic kidney 293 (HEK) cells, and lipoylation was carried out by purified LplA enzyme added to the media. After 10 minutes of labeling, lipoylated cell surface proteins were imaged using anti-lipoic acid antibody. Figure S5A shows representative images of labeled E2p, LAP2, and LAP1.49 Whereas E2p and LAP2 are lipoylated to a similar degree, labeling is not detected under these conditions for LAP1. To quantitatively compare the labeling efficiencies of all the LAP sequences, we plotted lipoylation signal (as measured by antibody staining intensity) against CFP signal for single cells. Average signal ratios listed in Figure S5B indicate that LAP2 is labeled more efficiently than the other LAP sequences, and is comparable even to E2p.
Third, the LAP sequences were compared in an intracellular labeling assay. In separate work, we have engineered a coumarin fluorophore ligase for labeling of recombinant proteins in living mammalian cells.32 To compare the LAP sequences using this assay, we prepared fusions to nuclear-localized Yellow Fluorescent Protein (YFP), and labeled transfected cells with the coumarin probe for 10 minutes. Afterwards, images were analyzed by plotting mean single cell coumarin intensities against mean single cell YFP intensities. Figure S6 shows that LAP2 is labeled more efficiently than the other LAP sequences in the cytosol, and gives even higher signal intensities than E2p, at high expression levels.
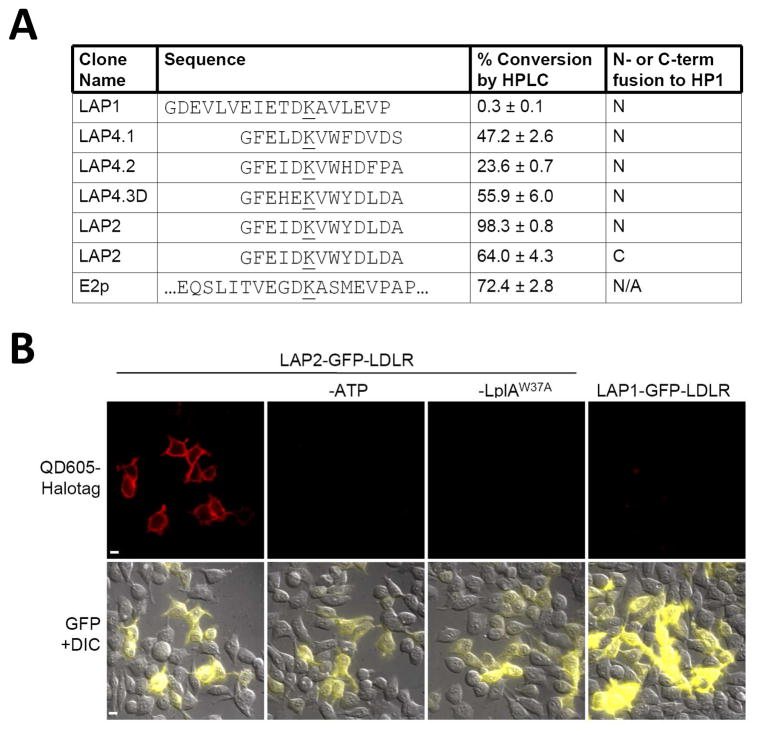
Fourth, we compared the LAP sequences in vitro in an HPLC assay,13 after expressing and purifying the HP1 fusion proteins13 from bacteria. Figure 4A shows the percent conversion to lipoylated product under identical reaction conditions. As in the cellular assays, LAP2 is the best sequence. When fused to the C- rather than N-terminus of HP1, the activity of LAP2 decreased somewhat, but was still higher than all other LAP sequences at the N-terminus. We also performed HPLC assays using other probes (azide 7, 11-Br, and coumarin) and found that LAP2 was the best substrate for these also (data not shown).
Figure 4.
Comparison of LAP clones and application to cell surface quantum dot tagging. (A) Various LAP sequences were compared to E2p protein, by lipoylation with 50 nM LplA for 1 hour. Product was detected by HPLC. All LAPs were tested as fusions to the N- or C-terminus of carrier protein HP1, as indicated.13 Error bars, ± 1 s.d. (B) HEK cells expressing LAP2 or LAP1-fused LDL receptor were labeled with LplAW37A and 11-Br for 5 minutes, followed by QD605-HaloTag31 for 5 minutes. QD605 emission is shown in the top row. Merged GFP (Green Fluorescent Protein) and DIC (differential interference contrast) images are shown in the bottom row. Negative controls are shown with ATP or LplA omitted. Scale bars, 10 μm.
Characterization of LAP2 and application to receptor labeling
Using HPLC to quantify product formation, we measured the kcat and Km values for LplA-catalyzed lipoylation of a synthetic LAP2 peptide (without an attached fusion protein). Figure S7 shows that the kcat is 0.22 ± 0.01 sec−1, slightly lower than that of E2p (kcat 0.253 ± 0.003 sec−113). The Km is 13.32 ± 1.78 μM, closer to that of LplA’s natural substrate H-protein (Km 1.2 μM33) than that of LAP1 (est. Km >200 μM; data not shown).
To utilize LAP2 for receptor imaging, we prepared a fusion to the low density lipoprotein (LDL) receptor. LAP2-LDL receptor expressed in HEK cells was labeled with LplAW37A and 11-Br probe. Ligated bromoalkane was derivatized with HaloTag-conjugated quantum dot 605 (QD605). Figure 4B shows specific QD605 labeling of LAP2-LDL receptor at the cell surface. Omission of ATP or LplA eliminates labeling. The same experiment performed with LAP1-fused LDL receptor did not produce any detectable QD605 signal.
Often, we use LplA labeling in conjunction with biotin ligase (BirA) labeling, for two-color imaging applications.13;31 We used HPLC to test the cross-reactivity of LAP2 with BirA and found no biotinylation after a 12 hour reaction with 5 μM BirA (data not shown).
Discussion
In summary, we have engineered a new peptide substrate for LplA using a novel selection platform based on yeast display. The peptide, LAP2, is lipoylated with a kcat similar to that of LplA’s protein substrate E2p, and has a Km much closer to that of LplA’s protein substrates than that of our previous rationally-designed LAP1.13 The consequence of this improvement in kinetic efficiency is the ability to label peptide-tagged cell surface receptors with unnatural probes, even at low or medium receptor expression levels. In other work, LAP2 also allows fluorophore tagging of intracellular proteins.32 In contrast, LAP1 fusions are difficult to label at the cell surface,13;28 and impossible to label inside of living cells.32 LAP2 is also shorter than LAP1 (13 amino acids instead of 17–22 amino acids) and can be recognized by LplA at the N-terminus, C-terminus, and internally.32
Comparing LAP2 to LplA’s natural protein substrates, the negatively charged residues at positions −1, −3, and +4, and the hydrophobic residues at positions −4 and +5 are shared. Since −1 Asp of E2p may promote loop formation (Figure S1), and +4 Glu in E2p may interact with Lys143 in LplA’s binding pocket (see above), LAP2 may interact with LplA in a manner similar to E2p. When overlaying the LAP2 sequence onto the E2p NMR structure (Figure S1),34 the −4 Phe and the +3 Tyr are positioned to interact in an intramolecular manner. We speculate that this interaction may help to stabilize LAP2 in a loop conformation that promotes high affinity binding to LplA. Interestingly, the engineered 15-amino acid acceptor peptide for biotin ligase50 also contains aromatic sidechains at these two positions. We also noticed that the +2 Trp that emerged in our selections may be positioned to interact with a hydrophobic patch on the LplA surface that includes Phe24.
Our study also introduces a new selection scheme for evolution of peptide substrates. Previously, yeast display has been used to evolve enzyme specificity,35;51 binding peptides,26 and binding proteins,38 but, to our knowledge, no enzymatic substrates have been evolved by this method. The appeal of yeast display for enzyme substrate evolution lies in its dynamic range: up to 104–105 copies of peptide can be displayed on the surface of each yeast cell,41 and FACS sorting allows fractionation of yeast into distinct pools based on the extent of surface peptide modification. In contrast, phage display has far more limited dynamic range due to the low copy number of displayed peptides, and the all-or-nothing nature of affinity-based product capture. As a consequence, we previously used two generations of phage display selections (as opposed to the single generation of selections used here) to produce a peptide substrate for yeast biotin ligase with a kcat/Km of only 0.00078 μM−1min−1,21 >1000-fold worse than the kcat/Km we obtained here for LAP2. Yin et al. have also used phage display to evolve peptide substrates for phosphopantetheinyl transferases, and obtained Km values in the 51–117 μM range, with kcat/Km in the range of 0.015–0.19 μM−1min−1.23 Again, these values are poorer than the corresponding values for LAP2. Our selection scheme should be generalizable to other classes of enzyme substrates, such as those for kinases and glycosyltransferases, as long as the enzymatic products can be detected by fluorescence.
Future work will involve the engineering of even shorter LAP sequences, performing biochemical assays and crystallography to determine the mode of LAP binding to LplA, and evolving orthogonal LAP/LplA pairs for multicolor imaging applications.
Materials and Methods
Cloning of Aga2p fusions to LAP1 and E2p for yeast display (Figure 2)
The E2p gene was amplified from E2p-CFP-TM13 using the primers E2p-NheI-PCR (5′GCATC GCTAGC ATG GCT ATC GAA ATC AAA GTA CCG G; incorporates an NheI site) and E2p-BamHI-PCR (5′GGTGA GGATCC CGC AGG AGC TGC CGC AG; incorporates a BamHI site). The resulting PCR product was digested with NheI and BamHI and ligated in-frame to NheI/BamHI-digested pCTCON2 vector.41 To clone the Aga2p fusion to LAP1, we hybridized the oligos LAP1-NheIBamHI-F (5′CTAGC GAC GAA GTA CTG GTT GAA ATC GAA ACC GAC AAA GCA GTT CTG GAA GTA CCG GGC GGT GAG GAG GAG G) and LAP1-NheIBamHI-R (5′GATCC CTC CTC CTC ACC GCC CGG TAC TTC CAG AAC TGC TTT GTC GGT TTC GAT TTC AAC CAG TAC TTC GTC G). The annealed oligos encode the 22-amino acid LAP1 sequence DEVLVEIETDKAVLEVPGGEEE.13 We then ligated the duplex DNA in-frame to NheI/BamHI-digested pCTCON2 vector. The E2p-Ala mutant was generated by Lys40→Ala mutagenesis using the QuikChange oligo 5′ GATCACCGTAGAAGGCGAC GCT GCTTCTATGGAAGTTCCGGC and its reverse complement.
Model selections on yeast with LAP1 and E2p (Figure 2)
Aga2p-E2p and Aga2p-LAP1 plasmids were transformed into S. cerevisiae EBY100 using the Frozen-EZ Yeast Transformation II kit (Zymo Research). After transformation, cells were grown in SDCAA media41 at 30 °C with shaking for 20 hours. The culture was then diluted to a cell density of 106 cells/mL in SGCAA media41 to induce protein expression for 20 hours with shaking at room temperature. Cells were harvested by centrifugation and washed with PBSB (Phosphate buffered saline, pH 7.4 + 0.5% BSA).
To lipoylate the yeast, 106–107 cells were pelleted at 14,000 g for 30 sec in a 1.5 mL eppendorf tube, then resuspended in 100 μL PBSB. To these cells, 750 μM (±)-α-lipoic acid, 300 nM LplA, 3 mM ATP, and 5 mM magnesium acetate were added. The cells were incubated on a rotator for 30 minutes at 30 °C. After washing the cells once with PBSB, cells were incubated with rabbit anti-lipoic acid antibody (1:300 dilution, Calbiochem) and mouse anti-c-Myc antibody (1:50 dilution, Calbiochem) for 40 minutes at 4 °C. The cells were washed again with PBSB followed by incubation with phycoerythrin-anti-rabbit antibody (1:100 dilution, Invitrogen) and Alexa Fluor 488-anti-mouse antibody (1:100 dilution, Invitrogen) for 40 minutes at 4 °C. Finally, cells were rinsed twice with PBSB and resuspended in 600 μL of PBSB for FACS analysis on a FACScan instrument, or FACS sorting on an Aria FACS instrument, both from BD Biosciences, and housed in the Koch Institute flow cytometry core facility.
We note that for c-Myc tag detection, we initially used a chicken anti-c-Myc antibody. However, we found that anti-chicken antibody cross-reacts with rabbit antibodies, and thus we switched to mouse anti-c-Myc antibody, which gives a lower signal, but does not bind to the rabbit anti-lipoic acid antibody.
To implement the model selections, E2p-displaying yeast and LAP1-displaying yeast were combined in various ratios. A total of 107 cells were lipoylated as described above in 100 μL PBSB. Following labeling, cells were sorted using a typical polygonal gate as shown in Figure 2B. We recovered ~5% of cells from the 1:10 mixture of E2p:LAP1, 0.5% of cells from the 1:100 mixture, and <0.1% of cells from the 1:1000 mixture. Collected cells were amplified in SDCAA media for 24–48 hours. Plasmids were isolated using Zymoprep II (Zymo Research). For PCR analysis of enrichment factors, the primers pctPCR.F (5′GCGGTTCTCACCCCTCAACAAC) and pctPCR.R (5′GTATGTGTAAAGTTGGTAACGGAACG) were used.
Cloning of LAP library (Figure 3A)
We ordered from IDT (Integrated DNA technologies) a partially randomized oligo with the following sequence: 5′A AAT AAG CTT TTG TTC GGA TCC NGM MNN NAN NTS MNN MNN AAC TTT ATC MNN NTS NAN TCC GCT AGC CGA CCC TCC. Underlined nucleotides were synthesized from mixtures containing 70% of the indicated base + 10% of each of the other bases. N designates an equimolar mixture of all bases. S designates a 1:1 mixture of G and C. M designates a 1:1 mixture of A and C.
This oligo was annealed with another oligo, Con2For.F (5′CT AGT GGT GGA GGA GGC TCT GGT GGA GGC GGT AGC GGA GGC GGA GGG TCG GCT AGC GGA), which overlaps with both pCTCON2 vector and the library oligo. The 5′ overhangs were filled in using Klenow polymerase. The resulting product was PCR-amplified using the primers Con2For.F and Con2Rev.R (5′TA TCA GAT CTC GAG CTA TTA CAAGTC CTC TTC AGA AAT AAG CTT TTG TTC GGA TCC). Meanwhile, pCTCON2 vector was prepared by digestion with NheI and BamHI, and gel-purified. PCR insert and pCTCON2 vector were transformed together into S. cerevisiae EBY100 (Invitrogen) by electroporation as described by Colby et al.52 Homologous recombination occurred inside the yeast. Serial dilutions of transformed yeast were plated on SDCAA plates and colonies were counted, to determine transformation efficiency.
Yeast display selection on LAP library (Figure 3B)
Yeast displaying the LAP library were prepared as described above (see “Model selections”). ~ 7×107 cells were washed and resuspended in 700 μL PBSB. For the first round, HaloTag labeling was performed. Cells were combined with 1 mM 11-Br, 5 μM LplA(W37A), 3 mM ATP, and 5 mM magnesium acetate for 2.5 hours at 30 °C. After washing with PBSB, 700 nM biotinylated-HaloTag protein31 was incubated with the cells in 50 μL PBSB for 30 minutes at 30 °C. Halotag protein was biotinylated by EZ-Link Sulfo-NHS-LC-Biotin (Sulfosuccinimidyl-6-(biotinamido) hexanoate) (Thermo Fisher Scientific) as described by the manufacturer. Then, cells were rinsed once with PBSB and labeled with streptavidin-phycoerythrin (1:100 dilution, Jackson ImmunoResearch) for 40 minutes at 4 °C. For detection of the c-Myc tag, chicken anti-c-Myc antibody (1:200 dilution, Invitrogen) and Alexa Fluor 488-anti-mouse antibody (1:100 dilution, Invitrogen) were used. Labeled cells were rinsed twice with PBSB and resuspended in 1 mL of PBSB for FACS sorting. After sorting, collected yeast were amplified in SDCAA media at 30 °C for 36–48 hr and induced with SGCAA media at 30 °C for 20 hr, for the next round.
Rounds 2–4 were implemented with 11-Br or lipoic acid labeling, under the conditions indicated in Figure 3B. Lipoylation was carried out as described above under “Model selections”.
Analysis of yeast pools after each round of selection (Figure 3B)
Yeast harvested from each round of selection were amplified and induced as described above. All pools were then treated identically with 3 μM LplA or 50 nM LplA, 750 μM (±)-α-lipoic acid, and 3 mM ATP for 30 minutes. To sequence individual clones, yeast were plated on SDCAA plates, single colonies were amplified in SDCAA media, and plasmid was isolated using the Zymoprep Yeast Plasmid Miniprep kit (Zymo Research). To increase DNA concentration, LAP genes were PCR-amplified from plasmid using the primers PctPCR.F and PctPCR.R (sequences under “Model selections”). Sequencing was completed using the primer PctSeq (5′GGCAGCCCCATAAACACAC).
Cloning and expression of LAP-HP1 fusion proteins (Figure 4A)
First, an MfeI restriction site was introduced into our previously described13 LAP1-HP1 expression plasmid, at the C-terminal end of the LAP1 sequence, using the QuikChange primer 5′ AAGCAGTTCTGGAAGTACCG CAATTG GGCGGTGAGGAGGAGTACGCC and its reverse complement. We then annealed the forward and reverse oligos shown below, and ligated the duplex DNA in-frame into NheI/MfeI-digested LAP1-(MfeI)-HP1 vector. The vector introduced a C-terminal His6 tag. Bacterial expression and purification were carried out as previously described.13
C-terminal fusion of LAP2 to HP1 was performed by annealing LAP2-C forward and reverse oligos (shown below), and ligating the duplex in-frame to NdeI/BamHI digested pET15b vector, which introduces an N-terminal His6 tag.
| Peptide | Reverse oligos |
| LAP4.1 | 5′AATTGTGAATCGACATCAAACCATACTTTATCAAGTTCAAATCCG |
| LAP4.2 | 5′AATTGTGCAGGGAAATCATGCCATACTTTATCAATCTCGAATCCG |
| LAP4.3D | 5′AATTGCGCATCGAGATCATACCAAACTTTCTCATGCTCAAATCCG |
| LAP2 | 5′AATTGGGCGTCCAGGTCGTACCACACCTTGTCGATCTCGAAGCCG |
| LAP2-C | 5′GATCCTCTTAGGCGTCCAGGTCGTACCACACCTTGTCGATCTCGAAGCCG |
| Peptide | Forward oligos |
| LAP4.1 | 5′CTAGCGGATTTGAACTTGATAAAGTATGGTTTGATGTCGATTCAC |
| LAP4.2 | 5′CTAGCGGATTCGAGATTGATAAAGTATGGCATGATTTCCCTGCAC |
| LAP4.3D | 5′CTAGCGGATTTGAGCATGAGAAAGTTTGGTATGATCTCGATGCGC |
| LAP2 | 5′CTAGCGGCTTCGAGATCGACAAGGTGTGGTACGACCTGGACGCCC |
| LAP2-C | 5′CTAGCGGCTTCGAGATCGACAAGGTGTGGTACGACCTGGACGCCTAAGAG |
Comparison of LAP clones by HPLC assay (Figure 4A)
To compare the labeling efficiencies of the different LAP-HP1 fusion proteins, we assembled labeling reactions as follows: 50 nM LplA, 60 μM LAP-HP1 or E2p, 750 μM (±)-α-lipoic acid, 3 mM ATP, and 5 mM magnesium acetate in Dulbecco’s Phosphate Buffered Saline (DPBS). Reactions were incubated at 30 °C for 1 hour, and then quenched with 180 mM EDTA (final concentration). The extent of conversion to lipoylated product was determined by HPLC as described in previous work.13;28
Cloning of LAP fusion proteins for mammalian expression
Three QuikChange mutations were made on the published pEGFP-LAP-LDLR construct.13 5′GAAGTACCATCAGCAGACGGC CAATTG ACTGTGAGCAAGGGCGAGG and its reverse complement were used to introduce MfeI site to 3′ end of LAP1. Subsequently, 5′GCACCTCGGTTCTATCGATA ACGCGT ACCATGGGGCCCTGGGGC and its reverse complement were used to mutate upstream (outside of the gene) NheI site to MluI. A new NheI site was then introduced to 5′ end of LAP1 using 5′CTGCAGTTGGCGACAGAAGT GCTAGC GACGAAGTACTGGTTGAAATC and its reverse complement. This expression vector was named LAP1-GFP-LDLR. LAP2-GFP-LDLR was obtained by annealing LAP2 forward and reverse oligos used for LAP2 HP1 fusion protein and ligating the duplex DNA in-frame into NheI/MfeI-digested LAP1-GFP-LDLR.
LAP2-CFP-TM was generated by annealing LAP2-BglIIAscI-F (5′GATCT GGC TTC GAG ATC GAC AAG GTG TGG TAC GAC CTG GAC GCC GG) and LAP2-BglIIAscI-R (5′CGCGCC GGC GTC CAG GTC GTA CCA CAC CTT GTC GAT CTC GAA GCC A) and ligating the duplex DNA in-frame into BglII/AscI digested LAP-CFP-TM (renamed as LAP1-CFP-TM).13 E2p-CFP-TM has previously been described.13
Cell surface quantum dot labeling of LAP2 with LplA (Figure 4B)
HEK 293T cells were transfected with LAP2-GFP-LDLR plasmid using Lipofectamine 2000. After 24 hours in growth media (Dulbecco’s Modified Eagle Medium (DMEM) with 10% Fetal bovine serum (FBS)) at 37°C, enzymatic ligation of 11-Br was performed in DPBS containing 10 μM LplA(W37A), 500 μM 11-Br, 1 mM ATP, 5 mM Mg(OAc)2 and 1% (w/v) BSA (Fraction V, EMD) as a blocking agent for 5 minutes at room temperature. Cells were then rinsed three times with DPBS followed by treatment with 50 nM HaloTag-QD60531 in DPBS containing 1% BSA for 5 minutes at room temperature. After another three rinses with DPBS, cells were imaged in the same buffer on a Zeiss Axio Observer.Z1 inverted epifluorescence microscope using a 40X oil-immersion lens. GFP (493/16 excitation, 525/30 emission, 488 dichroic, 300 ms exposure), QD605 (400/120 excitation, 605/30 emission, 488 dichroic, 200 ms exposure), and DIC images were collected and analyzed using Slidebook software (Intelligent Imaging Innovations). Fluorescence images were normalized to the same intensity ranges.
Cloning and analysis of −4 Phe→Val mutant of LAP4.1 (Figure S3)
pCTCON2 plasmid carrying LAP4.1 was isolated from yeast clone using the Zymoprep Yeast Plasmid Miniprep kit. Phe at position −4 was mutated to Val using the QuikChange primer 5′GGAGGGTCGGCTAGCGGA GTG GAACTTGATAAAGTATGGTTTGATGTCG and its reverse complement primer. This construct was subsequently transformed into S. cerevisiae EBY100, grown and induced as described above (see “Model selections”). To compare the yeast cell surface lipoylation of the Phe→Val mutant with the original LAP4.1 clone, clones from Gate A and the clones from Gate B, cells were lipoylated as described above except that 200 nM LplA was used.
Cell surface lipoylation of LAP constructs (Figure S4 and S5)
HEK 293T or HeLa cells were transfected with LAP4.1-, LAP4.3D-, E2p-, LAP2-, or LAP1-CFP-TM13 plasmids using Lipofectamine 2000. After 24 hours in growth media (DMEM with 10% FBS) at 37°C, lipoylation was performed in growth media containing 1 μM LplA, 100 μM (±)-α-lipoic acid, 1 mM ATP, 5 mM Mg(OAc)2 and 1% (w/v) BSA for 10 minutes at room temperature. Cells were then rinsed three times with DPBS followed by incubation with rabbit anti-lipoic acid antibody (1:300 dilution, Calbiochem) in DPBS containing 1–2% BSA for 10 minutes at room temperature. Fluorescence staining was achieved by treatment with either fluorescein-conjugated goat-anti-rabbit antibody (1:100 dilution, Calbiochem) or Alexa Fluor 568-conjugated goat-anti-rabbit antibody (1:100 dilution, Invitrogen) for 10 minutes at room temperature in DPBS with 1–2% BSA. Cells were imaged as described above using CFP (420/20 excitation, 475/40 emission, 450 dichroic, 500 ms exposure), fluorescein (493/120 excitation, 525/30 emission, 488 dichroic, 100 ms exposure) and Alexa Fluor 568 (570/20 excitation, 605/30 emmision, 585 dichroic, 200 ms exposure) filter sets. Slidebook software was used for emission intensity ratio quantitation. Average across-cell fluorescein and CFP intensities were used, after background subtraction.
Measurement of LAP2 kinetics (Figure S7)
Synthetic LAP2 peptide (sequence GFEIDKVWYDLDA) was prepared by the Tufts University Core Facility. To measure the kcat and Km values for lipoylation, 50 nM LplA was combined with 750 μM lipoic acid, 2 mM ATP, and 5 mM magnesium acetate in DPBS. Varying concentrations of LAP2 (5.5, 11, 22, 44, 88, 176 or 352 μM) were used. 60 μL aliquots were removed from the 30 °C reactions at 5 minute intervals, up to 20 minutes, and quenched with 180 mM EDTA (final concentration). HPLC was used to determine the amount of product in each aliquot and kinetic parameters were extracted using the Michaelis-Menten equation as described previously.13;28
Supplementary Material
Acknowledgments
Funding was provided by the NIH (R01 GM072670 and PN2 EY018244), MIT, and the Sloan and Dreyfus Foundations. We thank Karishma Rahman, Dr. Irwin Chen, Yoon-Aa Choi, Prof. Mark Howarth, Dr. Benjamin J. Hackel, and Prof. K. Dane Wittrup for their advice and assistance. We acknowledge the MIT Koch Institute for use of flow cytometry facilities.
Footnotes
Supporting Information Available. Figures S1–7 and the complete author list for Reference 30. This material is available free of charge via the internet at http://pubs.acs.org.
Reference List
- 1.Feng SB, Kasahara C, Rickles RJ, Schreiber SL. Proc Nat Acad Sci U S A. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowland RRR, Schneider P, Fang Y, Wootton S, Yoo D, Benfield DA. Virology. 2003;316:135–145. doi: 10.1016/S0042-6822(03)00482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall JG, Frieden C. Proc Nat Acad Sci U S A. 1989;86:3060–3064. doi: 10.1073/pnas.86.9.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerella R, Chen PY, Evans PA, Raine A, Williams DH. Protein Sci. 2000;9:2142–2150. doi: 10.1110/ps.9.11.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toomik R, Ek P. Biochem J. 1997;322:455–460. doi: 10.1042/bj3220455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummings RT, Salowe SP, Cunningham BR, Wiltsie J, Park YW, Sonatore LM, Wisniewski D, Douglas CM, Hermes JD, Scolnick EM. Proc Nat Acad Sci U S A. 2002;99:6603–6606. doi: 10.1073/pnas.062171599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shamoo Y, Steitz TA. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 8.Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. Nat Struct Biol. 2001;8:998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borghouts C, Kunz C, Groner B. J Pept Sci. 2005;11:713–726. doi: 10.1002/psc.717. [DOI] [PubMed] [Google Scholar]
- 10.Sato AK, Viswanathan M, Kent RB, Wood CR. Curr Opin Biotechnol. 2006;17:638–642. doi: 10.1016/j.copbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Lin MZ, Wang L. Physiology. 2008;23:131–141. doi: 10.1152/physiol.00007.2008. [DOI] [PubMed] [Google Scholar]
- 12.Khan AR, Parrish JC, Fraser ME, Smith WW, Bartlett PA, James MNG. Biochemistry. 1998;37:16839–16845. doi: 10.1021/bi9821364. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell RD, Glass DB, Wong CW, Angelos KL, Walsh DA. Biochemistry. 1995;34:528–534. doi: 10.1021/bi00002a018. [DOI] [PubMed] [Google Scholar]
- 15.Viguera AR, Arrondo JLR, Musacchio A, Saraste M, Serrano L. Biochemistry. 1994;33:10925–10933. doi: 10.1021/bi00202a011. [DOI] [PubMed] [Google Scholar]
- 16.Rudiger S, Schneider-Mergener J, Bukau B. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam KS, Wu JZ, Lou Q. Int J Pept Protein Res. 1995;45:587–592. doi: 10.1111/j.1399-3011.1995.tb01323.x. [DOI] [PubMed] [Google Scholar]
- 18.Reineke U, Volkmer-Engert R, Schneider-Mergener J. Curr Opin Biotechnol. 2001;12:59–64. doi: 10.1016/s0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 19.Marani MM, Ceron MCM, Giudicessi SL, de Oliveira E, Cote S, Erra-Balsells R, Albericio F, Cascone O, Camperi SA. J Comb Chem. 2009;11:146–150. doi: 10.1021/cc800145c. [DOI] [PubMed] [Google Scholar]
- 20.Herman RE, Badders D, Fuller M, Makienko EG, Houston ME, Quay SC, Johnson PH. J Biol Chem. 2007;282:9813–9824. doi: 10.1074/jbc.M610722200. [DOI] [PubMed] [Google Scholar]
- 21.Chen I, Choi YA, Ting AY. J Am Chem Soc. 2007;129:6619–6625. doi: 10.1021/ja071013g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidhu SS, Koide S. Curr Opin Struct Biol. 2007;17:481–487. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Cironi P, Lin AJ, Xu YQ, Hrvatin S, Golan DE, Silver PA, Walsh CT, Yin J. ACS Chem Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 24.Schatz PJ. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 25.Dane KY, Chan LA, Rice JJ, Daugherty PS. J Immunol Meth. 2006;309:120–129. doi: 10.1016/j.jim.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Krauland EM, Peelle BR, Wittrup KD, Belcher AM. Biotechnol Bioeng. 2007;97:1009–1020. doi: 10.1002/bit.21341. [DOI] [PubMed] [Google Scholar]
- 27.Wolkowicz R, Jager GC, Nolan GP. J Biol Chem. 2005;280:15195–15201. doi: 10.1074/jbc.M500254200. [DOI] [PubMed] [Google Scholar]
- 28.Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY. Angew Chem Int Ed Engl. 2008;47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cronan JE, Zhao X, Jiang Y. Adv Microb Physiol. 2005;50:103–146. doi: 10.1016/S0065-2911(05)50003-1. [DOI] [PubMed] [Google Scholar]
- 30.Los GV, et al. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 31.Liu DS, Phipps WS, Howarth M, Puthenveetil S, Ting AY. Irreversible targeting to a peptide in living cells using lipoate ligase and Halotag: application to two-color quantum dot tracking of receptors. 2009 [Google Scholar]
- 32.Baruah H, Uttamapinant C, White KA, Fernández-Suárez M, Puthenveetil S, Thompson S, Ting AY. A fluorophore ligase for site-specific protein labeling in living cells. 2009 doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara K, Toma S, Okamura-Ikeda K, Motokawa Y, Nakagawa A, Taniguchi H. J Biol Chem. 2005;280:33645–33651. doi: 10.1074/jbc.M505010200. [DOI] [PubMed] [Google Scholar]
- 34.Jones DD, Stott KM, Howard MJ, Perham RN. Biochemistry. 2000;39:8448–8459. doi: 10.1021/bi992978i. [DOI] [PubMed] [Google Scholar]
- 35.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara K, Suzuki M, Okumachi Y, Okamura-Ikeda K, Fujiwara T, Takahashi E, Motokawa Y. Eur J Biochem. 1999;260:761–767. doi: 10.1046/j.1432-1327.1999.00204.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim DJ, Kim KH, Lee HH, Lee SJ, Ha JY, Yoon HJ, Suh SW. J Biol Chem. 2005;280:38081–38089. doi: 10.1074/jbc.M507284200. [DOI] [PubMed] [Google Scholar]
- 38.Gai SA, Wittrup KD. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paschke M. Appl Microbiol Biotechnol. 2006;70:2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]
- 40.Martin BR, Giepmans BN, Adams SR, Tsien RY. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 41.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 42.Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. J Biol Chem. 2008;283:14739–14750. doi: 10.1074/jbc.M709116200. [DOI] [PubMed] [Google Scholar]
- 43.Dardel F, Davis AL, Laue ED, Perham RN. J Mol Biol. 1993;229:1037–1048. doi: 10.1006/jmbi.1993.1103. [DOI] [PubMed] [Google Scholar]
- 44.Howard MJ, Chauhan HJ, Domingo GJ, Fuller C, Perham RN. J Mol Biol. 2000;295:1023–1037. doi: 10.1006/jmbi.1999.3391. [DOI] [PubMed] [Google Scholar]
- 45.Ricaud PM, Howard MJ, Roberts EL, Broadhurst RW, Perham RN. J Mol Biol. 1996;264:179–190. doi: 10.1006/jmbi.1996.0632. [DOI] [PubMed] [Google Scholar]
- 46.Reche P, Perham RN. EMBO J. 1999;18:2673–2682. doi: 10.1093/emboj/18.10.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui GF, Nan BY, Hu JC, Wang YP, Jin CW, Xia B. J Biol Chem. 2006;281:20598–20607. doi: 10.1074/jbc.M602660200. [DOI] [PubMed] [Google Scholar]
- 48.Reche PA. Protein Sci. 2000;9:1922–1929. doi: 10.1110/ps.9.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.We note that the surface expression levels of TM fusions to LAP peptides are ~2-fold lower than TM fusions to E2p. However, expression levels of intracellular proteins are similar, whether fused to a LAP sequence or E2p (Figure S6).
- 50.Beckett D, Kovaleva E, Schatz PJ. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antipov E, Cho AE, Wittrup KD, Klibanov AM. Proc Nat Acad Sci U S A. 2008;105:17694–17699. doi: 10.1073/pnas.0809851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colby DW, Kellogg BA, Graff CP, Yeung YA, Swers JS, Wittrup KD. Protein Eng. 2004;388:348–358. doi: 10.1016/S0076-6879(04)88027-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.