Abstract
In transverse orientation, maize (Zea mays) roots are composed of a central stele that is embedded in multiple layers of cortical parenchyma. The stele functions in the transport of water, nutrients, and photosynthates, while the cortical parenchyma fulfills metabolic functions that are not very well characterized. To better understand the molecular functions of these root tissues, protein- and phytohormone-profiling experiments were conducted. Two-dimensional gel electrophoresis combined with electrospray ionization tandem mass spectrometry identified 59 proteins that were preferentially accumulated in the cortical parenchyma and 11 stele-specific proteins. Hormone profiling revealed preferential accumulation of indole acetic acid and its conjugate indole acetic acid-aspartate in the stele and predominant localization of the cytokinin cis-zeatin, its precursor cis-zeatin riboside, and its conjugate cis-zeatin O-glucoside in the cortical parenchyma. A root-specific β-glucosidase that functions in the hydrolysis of cis-zeatin O-glucoside was preferentially accumulated in the cortical parenchyma. Similarly, four enzymes involved in ammonium assimilation that are regulated by cytokinin were preferentially accumulated in the cortical parenchyma. The antagonistic distribution of auxin and cytokinin in the stele and cortical parenchyma, together with the cortical parenchyma-specific accumulation of cytokinin-regulated proteins, suggest a molecular framework that specifies the function of these root tissues that also play a role in the formation of lateral roots from pericycle and endodermis cells.
The maize (Zea mays) root system is composed of primary and seminal roots that are laid down during embryo development and shoot-borne and lateral roots that are initiated after germination (Hochholdinger, 2009). In general, the longitudinal and radial organization of all maize root types is very similar (Hochholdinger et al., 2004b). The longitudinal structure of the maize root can be divided into various zones of development, including the root cap at the terminal end, a subterminal meristematic zone, an elongation zone, and a differentiation zone (Ishikawa and Evans, 1995). Cells formed in the subterminal root meristem move proximally into the elongation zone, where these cells start to elongate and move on to the differentiation zone (Ishikawa and Evans, 1995). Hence, cells of a particular cell type along the longitudinal axis of a root represent a gradient of cell differentiation, with very young undifferentiated cells at the distal end near the root tip and differentiated cells at the proximal end of the root. The differentiation zone of a root can be very easily spotted from outside the root by the presence of epidermal root hairs. In general, the differentiation zone of maize roots can be radially divided into an inner part that comprises the stele and an outer part that is defined by multiple layers of parenchymal tissue and a single epidermal layer that connects the root with the exterior soil environment. The stele of the primary root contains a vascular system with differentiated xylem vessels that function in the transport of water and nutrients and primary phloem elements that function in the transport of photosynthates (Hochholdinger, 2009). The vasculature of the stele surrounds the inner parenchymal pith tissue. The outermost cell layer of the stele is the pericycle. The parenchymal outer part of the root is defined by a single layer of endodermal cells and multiple layers of cortical tissue and is enclosed by a single epidermal layer. The root epidermis functions in the uptake of nutrients, which are then either carried across the cortex into the xylem, where they are transported via the transpiration stream to the shoot, or metabolized in the cortex of the root (Marschner, 2003). The biochemical processes that are taking place in the cortex and thus defining this tissue are only poorly understood. Proteomic technology provides an approach that allows us to dissect and quantify components such as biochemical pathways in a tissue-specific way by combining the resolution of two-dimensional gel electrophoresis (2-DE) with the sensitivity of mass spectrometry and database searching (Hochholdinger et al., 2006). In recent years, maize root proteomes were analyzed in order to better understand development (Hochholdinger et al., 2005), genotypic variation (Hochholdinger et al., 2004a; Wen et al., 2005; Liu et al., 2006; Sauer et al., 2006; Hoecker et al., 2008), environmental interactions (Chang et al., 2000; Zhu et al., 2006, 2007; Li et al., 2007), or subcellular processes (Zhu et al., 2006, 2007) of different root types. Most of these surveys investigated whole roots (Hochholdinger et al., 2004a; Wen et al., 2005; Liu et al., 2006; Li et al., 2007; Hoecker et al., 2008), while a few specifically analyzed the primary root tip (Chang et al., 2000) and the elongation zone (Zhu et al., 2006, 2007). A major drawback of the analysis of whole roots or longitudinal developmental zones of roots is the composite organization of roots, which are made up of distinct radial tissue types, each providing specific gene expression and hence protein accumulation profiles. Tissue- or cell type-specific analyses can overcome this limitation (Schnable et al., 2004). Pericycle-specific transcriptome profiling by combining laser capture microdissection with microarray experiments has been applied to identify genes related to lateral root initiation (Woll et al., 2005) and pericycle specification (Dembinsky et al., 2007). Thus far, cell type-specific proteome analyses are challenging, since proteins cannot be amplified, in contrast to transcripts, prior to their analysis. Hence, surveying the major pericycle proteins of the differentiation zone of maize primary roots was confined to the identification of the 20 most abundant proteins of this cell type (Dembinsky et al., 2007). Mechanical separation of maize tissues (e.g. cortical parenchyma and stele) provides an alternative experimental approach to study the proteome composition of specific maize tissues. This approach can also be applied to directly measure tissue-specific concentrations of phytohormone levels in cortical parenchyma and stele tissue. Thus far, the detection of tissue-specific auxin and cytokinin levels was often limited to indirect evidence. GFP under the control of the synthetic promoter Direct Repeat5 (DR5; Friml, 2003) is used to measure free indole acetic acid (IAA) auxin levels, while GUS under the control of the cytokinin-sensitive promoter ARR5 (Aloni et al., 2004) or TCS (Mueller and Sheen, 2008) is used to visualize free bioactive cytokinin levels. Alternatively, antibodies that recognize trans-zeatin riboside and dihydrozeatin riboside and their derivatives can be employed to measure cytokinin levels (Eberle et al., 1986). Direct measurements of hormones are particularly helpful to discriminate between hormone precursors and conjugates that cannot be detected via marker lines or antibodies.
The goal of this study was to test three hypotheses: first, that there is significant overlap but also differential protein accumulation between cortical parenchyma and stele tissues in the differentiation zone of the maize primary root; second, that these differential proteins are related to the specific functions of these tissues in root development, which might also be reflected in the overall distribution of functional categories or biochemical pathways; and third, that distinct functions of these tissue types are also reflected by the distribution of plant hormones and their intermediates.
RESULTS
Manual Separation of Cortical Parenchyma and Stele Tissues in the Differentiation Zone of Maize Primary Roots
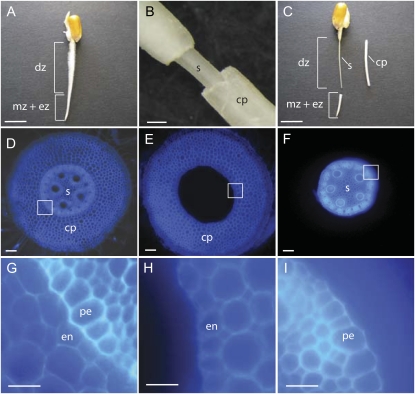
In longitudinal orientation, the maize primary root (Fig. 1A) can be divided into meristematic (mz), elongation (ez), and differentiation (dz) zones. The differentiation zone can be distinguished from the meristematic and elongation zones by the presence of root hairs, which allows spotting this region of the root very easily. In transverse orientation, the differentiation zone of maize roots can be divided into a central stele (Fig. 1D, s), which contains vascular tissues, pith parenchyma, and the pericycle, and the surrounding parenchymous ground tissue (Fig. 1D, cp), which is composed of the epidermis, multiple layers of cortical parenchyma tissue, and the endodermis. Due to the dominance of cortical parenchyma cells in this outer region of the root and the lack of an adequate botanical term, this tissue will subsequently be called cortical parenchyma. Toluidine blue is a dye that polychromatically stains cell walls depending on their chemical composition (O'Brien et al., 1964) and therefore allows differentiating stele and cortex tissues at the boundary of pericycle and endodermis (Fig. 1D). At the junction between pericycle (pe) and endodermis (en) cells (Fig. 1G), it is possible to mechanically separate (Fig. 1, B and C) the outer (Fig. 1E, cp) and inner (Fig. 1F, s) regions of the differentiation zone of the root without damaging endodermis (Fig. 1H, en) or pericycle (Fig. 1I, pe) cells (see “Materials and Methods”). In this study, stele and cortical parenchyma of the differentiation zone of 2.5-d-old primary roots of the inbred line B73 were analyzed. At this developmental stage, primary roots of the maize inbred line B73 had an average total length of 21.9 ± 6.6 mm, and the length of the differentiation zone of these roots was 12.7 ± 5.9 mm (n = 71). At this developmental stage, no lateral roots were initiated, as demonstrated in propidium iodide-stained stele preparations, while in 4-d-old stele preparations, lateral primordia were clearly visible (data not shown). After mechanically separating the inner and outer tissues of the differentiation zone of 2.5-d-old maize primary roots, these functionally diverse tissues were subjected to comparative proteome- and hormone-profiling experiments.
Figure 1.
A to C, A 2.5-d-old seedling root of the inbred line B73 before (A) and after (C) separation of stele and cortical parenchyma in the differentiation zone of the root. A, The differentiation zone (dz) is marked by the presence of root hairs, while the root tip contains the meristematic zone (mz) and the elongation zone (ez). B, Close-up after cutting the cortical parenchyma close to the coleorhiza and removing the cortical parenchyma (cp) from the stele tissue (s). C, Differentiation zone of the same seedling as in A after removing the root tip (mz + ez) and mechanical separation of the cortical parenchyma (cp). Subsequently, these tissues were used for protein and hormone analyses. D to I, Toluidine blue-stained free hand transverse sections of the differentiation zone of 2.5-d-old B73 primary roots before (D and G) and after (E, F, H, and I) manual separation. D, Transverse whole root transverse section before manual separation. E, Transverse section of the cortical parenchyma after separation (compare with cp in B and C). F, Transverse section of the stele after separation (compare with s in B and C). G, Close-up of the region marked by a white box in D, indicating the junction of pericycle (pe) and endodermis (en) cell layers. H, Close-up of the region marked by a white box in E. Note the intact endodermis (en) cell layer after tissue separation. I, Close-up of the region marked by a white box in F. Note the intact pericycle cell layer (pe) after tissue separation. Bars = 1 cm (A and C), 500 μm (B), 100 μm (D–F), 50 μm (G–I). [See online article for color version of this figure.]
Isolation and 2-DE Separation of Cortical Parenchyma and Stele Tissue Proteins
Soluble proteins were isolated in three independent biological replicates from cortical parenchyma and stele tissues of the maize inbred line B73. Isoelectric focusing of the three protein extracts per tissue was performed on linear gradients ranging from pH 4 to 7. Subsequently, these protein extracts were separated according to their molecular masses in a second dimension on 12.5% SDS-PAGE gels. Finally, protein spots were visualized with colloidal Coomassie Brilliant Blue staining. In total, 599 distinct protein spots were reproducibly detected in cortical parenchyma or stele tissue. Only proteins that were detected in all three biological replicates of at least one tissue were further analyzed. In Supplemental Figure S1, representative 2-DE gels of B73 cortical parenchyma and stele proteins are displayed. Intensities of all quantified spots per gel were normalized in silico with PD-Quest software to compensate for non-expression-related variations in spot intensity.
Identification of Differentially Accumulated Proteins in Cortical Parenchyma and Stele
One objective of this survey was to identify proteins that are preferentially accumulated in cortical parenchyma or stele tissue. Overall, 70 of 599 proteins (12%) detected in these two tissue types showed an accumulation difference of ≥2 and were statistically significant after adjusting P values of Student's t test (P < 0.01), controlling the false discovery rate (FDR) at 5% (Tables I and II). Among the 70 proteins differentially accumulated in cortical parenchyma versus stele protein extracts, 59 were exclusively or preferentially expressed in cortical parenchyma, while 11 proteins were predominantly accumulated in the stele (Tables I and II). All 70 protein spots were picked from representative 2-DE gels and digested with trypsin. Peptide analyses and identifications were performed using nano-HPLC-electrospray ionization-tandem mass spectrometry (ESI-MS/MS). Automated MASCOT software (Matrix Science; Perkins et al., 1999) was applied for National Center for Biotechnology Information nonredundant (NCBI.nr) protein database searching of all available higher plant proteins (Viridiplantae). All 70 differentially accumulated proteins were identified via ESI-MS/MS analyses, functionally annotated via the NCBI.nr database (Tables I and II), and classified according to the Munich Information Center for Protein Sequences annotation system (http://mips.gsf.de/) into different functional categories. A list of identified peptides for all proteins is provided in Supplemental Table S1.
Table I.
Proteins preferentially accumulated in cortical parenchyma compared with stele tissue of maize primary roots identified after 2-DE separation and nano-HPLC-ESI-MS/MS analysis of trypsin-digested proteins matched against the NCBI.nr protein database entries
Preferentially accumulated proteins, ≥2-fold change; t test, P < 0.01 in three independent biological replicates.
| Spot No. | Cortex Specificitya | MOWSE Scoreb | Sequence Coveragec | Mr Predicted/Mr Geld | pI Predicted/pI Gele | Function NCBI.nr [Species] Accession No.f | t Test Pg | FDR Ph |
|---|---|---|---|---|---|---|---|---|
| Metabolism | ||||||||
| 1 | Cortex only | 231 | 12 | 41.6/44.1 | 5.3/5.4 | 12-Oxo-phytodienoic acid reductase [Zea mays] AAY26523 | <0.001 | 0.007 |
| 2 | Cortex only | 259 | 16 | 41.6/43.6 | 5.3/5.6 | 12-Oxo-phytodienoic acid reductase [Z. mays] AAY26523 | <0.001 | 0.048 |
| 3 | Cortex only | 697 | 19 | 64.2/65.3 | 6.2/5.5 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | 0.001 | 0.014 |
| 4 | Cortex only | 934 | 23 | 82.0/84.9 | 5.6/5.5 | Hydroxymethylbutenyl 4-diphosphate synthase [Z. mays] AAT70081 | <0.001 | 0.010 |
| 5 | Cortex only | 246 | 3 | 113.1/79.2 | 5.9/6.0 | Glycosyl hydrolase [Oryza sativa] ABG22500 | <0.001 | 0.006 |
| 6 | Cortex only | 319 | 9 | 71.8/70.1 | 7.2/6.2 | Oxidoreductase [Z. mays] CAA12157 | <0.001 | 0.003 |
| 7 | Cortex only | 212 | 6 | 66.5/70.4 | 5.8/6.1 | Asparagine synthetase [Z. mays] P49094 | <0.001 | 0.003 |
| 8 | Cortex only | 140 | 11 | 29.3/25.8 | 6.8/6.3 | Haloacid dehalogenase-like hydrolase [O. sativa] NP_001058303i | 0.003 | 0.023 |
| 9 | Cortex only | 204 | 9 | 44.0/41.7 | 6.0/6.4 | Glutamate dehydrogenase [Z. mays] AAB51595 | 0.003 | 0.022 |
| 10 | Cortex only | 169 | 8 | 43.5/35.2 | 6.1/5.4 | Cysteine synthase 1 [O. sativa] EAZ15034i | <0.001 | 0.002 |
| 11 | Cortex only | 280 | 8 | 102.4/110.5 | 5.7/5.4 | Pyruvate, orthophosphate dikinase [Z. mays] AAA33498 | 0.006 | 0.035 |
| 12 | Cortex only | 334 | 13 | 64.2/66.0 | 6.2/5.6 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | 0.004 | 0.027 |
| 13 | Cortex only | 125 | 5 | 77.5/90.5 | 5.7/5.5 | Acetyl-CoA synthetase [O. sativa] NP_001046995i | <0.001 | 0.004 |
| 14 | Cortex only | 242 | 8 | 51.6/51.2 | 5.5/6.0 | UDP-Glc pyrophosphorylase [O. sativa] NP_001045689i | <0.001 | 0.002 |
| 15 | Cortex only | 542 | 18 | 63.1/57.3 | 8.6/6.2 | ζ-Carotene desaturase [Z. mays] Q9ZTP4 | 0.005 | 0.032 |
| 16 | Cortex only | 278 | 15 | 41.2/44.5 | 5.9/6.0 | Cysteine desulfurase [O. sativa] ABA97759 | <0.001 | 0.002 |
| 17 | Cortex only | 297 | 9 | 60.8/70.3 | 6.0/6.2 | Aspartyl-tRNA synthetase [O. sativa] NP_001047770i | 0.001 | 0.013 |
| 18 | Cortex only | 586 | 23 | 34.5/34.6 | 6.2/6.4 | Aldose reductase [Z. mays] ABF61890 | <0.001 | 0.011 |
| 19 | Cortex only | 386 | 20 | 39.4/33.8 | 7.7/6.4 | Pyridoxine biosynthesis protein [O. sativa] EAZ02455 | <0.001 | 0.006 |
| 20 | Cortex only | 277 | 15 | 41.7/48.3 | 5.9/6.2 | 12-Oxo-phytodienoic acid reductase [Z. mays] AAY26521 | 0.006 | 0.035 |
| 21 | Cortex only | 233 | 12 | 47.5/40.6 | 7.6/6.5 | Aspartate aminotransferase [O. sativa] BAA23815 | <0.001 | 0.006 |
| 22 | Cortex only | 258 | 15 | 40.1/35.0 | 8.4/6.0 | Aspartate/glutamate/uridylate kinase [O. sativa] EAY75934i | <0.001 | 0.002 |
| 23 | 22.9 | 1237 | 34 | 64.2/63.7 | 6.2/5.8 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | <0.001 | 0.002 |
| 24 | 18.2 | 890 | 19 | 64.2/64.5 | 6.2/5.7 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | <0.001 | 0.002 |
| 25 | 14.5 | 181 | 5 | 64.2/60.0 | 6.2/5.6 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | <0.001 | 0.002 |
| 26 | 8.7 | 730 | 32 | 39.2/37.9 | 5.6/5.6 | Glutamine synthetase [Z. mays] BAA03433 | <0.001 | 0.006 |
| 27 | 8.7 | 256 | 14 | 39.2/37.8 | 5.6/5.8 | Glutamine synthetase [Z. mays] BAA03433 | 0.002 | 0.018 |
| 28 | 8.0 | 190 | 7 | 64.2/61.5 | 6.2/5.5 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | <0.001 | 0.006 |
| 29 | 7.1 | 340 | 5 | 113.9/130.0 | 5.9/6.3 | Glycosyl hydrolase [O. sativa] ABG22500 | 0.003 | 0.028 |
| 30 | 6.5 | 135 | 5 | 49.6/42.6 | 6.6/6.4 | Alanine-glyoxylate aminotransferase [O. sativa] EAY89931i | <0.001 | 0.003 |
| 31 | 5.0 | 244 | 9 | 64.2/35.8 | 6.2/5.8 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | 0.005 | 0.032 |
| 32 | 4.1 | 518 | 12 | 64.5/71.3 | 6.5/6.0 | Phosphoglycerate dehydrogenase [O. sativa] BAD09434 | 0.003 | 0.026 |
| 33 | 3.9 | 149 | 7 | 44.5/44.8 | 6.4/6.1 | Glutamate dehydrogenase A [O. sativa] EAY86911i | <0.001 | 0.010 |
| 34 | 3.3 | 581 | 20 | 64.2/59.9 | 6.2/5.7 | β-d-Glucoside glucohydrolase [Z. mays] P49235 | 0.002 | 0.017 |
| 35 | 3.2 | 1104 | 22 | 102.4/108.4 | 5.7/5.4 | Pyruvate, orthophosphate dikinase [Z. mays] AAA33498 | 0.001 | 0.013 |
| 36 | 3.0 | 390 | 8 | 77.5/88.7 | 5.7/5.6 | Acetyl-CoA synthetase [O. sativa] NP_001046995i | 0.004 | 0.029 |
| 37 | 2.8 | 411 | 18 | 42.1/42.5 | 5.6/5.6 | 3-Phosphoglycerate kinase [Z. mays] P12783 | 0.001 | 0.013 |
| 38 | 2.5 | 266 | 11 | 54.8/57.2 | 5.6/5.6 | Aldehyde dehydrogenase RF2D [Z. mays] AAL99611 | 0.006 | 0.036 |
| 39 | 2.5 | 303 | 7 | 76.3/61.5 | 5.7/5.6 | Ketol-acid reductoisomerase [O. sativa] EAY75202i | <0.001 | 0.005 |
| 40 | 2.2 | 132 | 8 | 44.0/43.3 | 6.1/6.1 | Glutamate dehydrogenase [Z. mays] Q43260 | 0.003 | 0.023 |
| 41 | 2.1 | 597 | 22 | 44.0/41.8 | 6.1/6.3 | Glutamate dehydrogenase [Z. mays] Q43260 | 0.003 | 0.023 |
| 42 | 2.1 | 306 | 15 | 32.4/25.8 | 7.0/5.0 | Triosephosphate isomerase [O. sativa] NP_001063777 | 0.002 | 0.017 |
| 43 | 2.1 | 737 | 27 | 52.5/47.8 | 5.8/5.9 | Glutathione synthetase [Z. mays] CAE18179 | 0.009 | 0.047 |
| Signal transduction | ||||||||
| 44 | Cortex only | 127 | 3 | 69.8/85.3 | 5.9/5.7 | Metallophosphatase [O. sativa] NP_001062330i | <0.001 | 0.002 |
| Protein fate | ||||||||
| 45 | Cortex only | 524 | 11 | 105.6/115.8 | 5.5/5.4 | Lon protease homolog 2 [Z. mays] NP_001105895 | 0.002 | 0.018 |
| 46 | Cortex only | 278 | 20 | 32.6/41.4 | 4.6/5.5 | Histone deacetylase 2b [Z. mays] Q9M4U5 | 0.007 | 0.041 |
| Protein synthesis | ||||||||
| 47 | Cortex only | 589 | 29 | 46.5/50.4 | 5.4/5.4 | Translational initiation factor eIF-4A [Z. mays] NP_001105372 | <0.001 | 0.011 |
| Disease/defense/stress response | ||||||||
| 48 | Cortex only | 1308 | 23 | 102.0/100.0 | 6.6/5.6 | ATP-dependent Clp protease [O. sativa] NP_001066442 | 0.001 | 0.012 |
| 49 | 3.5 | 198 | 22 | 25.8/26.1 | 5.4/5.4 | Glutathione S-transferase GST 26 [Z. mays] AAG34834 | 0.008 | 0.041 |
| 50 | 2.3 | 433 | 16 | 50.2/45.5 | 5.2/5.1 | UDP-Glucosyltransferase BX9 [Z. mays] AAL57038 | 0.006 | 0.035 |
| 51 | 2.3 | 237 | 19 | 26.7/26.4 | 6.1/6.3 | Aluminum-induced protein-like protein [Setaria italica] ABM97609 | <0.001 | 0.012 |
| Energy | ||||||||
| 52 | Cortex only | 511 | 17 | 61.9/75.5 | 5.9/5.4 | Vacuolar ATP synthase catalytic subunit A [Z. mays] P49087 | <0.001 | 0.002 |
| 53 | 2.4 | 834 | 25 | 61.9/74.1 | 5.9/5.5 | Vacuolar ATP synthase catalytic subunit A [Z. mays] P49087 | 0.004 | 0.030 |
| Cell fate | ||||||||
| 54 | Cortex only | 291 | 10 | 61.1/46.1 | 6.9/5.4 | Actin-interacting protein [O. sativa] BAC79943 | 0.001 | 0.013 |
| Unknown function | ||||||||
| 55 | Cortex only | 151 | 14 | 22.5/20.8 | 7.8/6.5 | Unknown protein [O. sativa] NP_001042770i | <0.001 | 0.003 |
| 56 | Cortex only | 163 | 14 | 22.5/20.3 | 7.8/6.5 | Unknown protein [O. sativa] NP_001042770i | <0.001 | 0.012 |
| 57 | Cortex only | 171 | 4 | 66.2/117.0 | 4.6/4.7 | Unknown protein [O. sativa] EAY76673i | <0.001 | 0.002 |
| 58 | Cortex only | 180 | 6 | 52.1/54.8 | 5.2/5.6 | Unknown protein [O. sativa] AAM93677i | 0.005 | 0.032 |
| 59 |
2.7 |
215 |
9 |
40.2/37.4 |
5.7/5.9 |
Herbicide safener-binding protein [Z. mays] AAC12715 |
0.009 |
0.048 |
Ratio of accumulation (fold change) of a particular protein between primary root cortex versus stele tissue.
Probability-based MOWSE score is defined as –10 × log(P), where P is the probability that the observed match is a random event. Scores > 43 indicate identity or extensive homology (P < 0.05). Protein scores are derived from ion scores as a nonprobabilistic basis for ranking protein hits.
Percentage of predicted protein sequence covered by matched peptides.
Theoretical molecular mass of predicted protein/experimental molecular mass of protein on the gel.
Theoretical pI of predicted protein/experimental pI of protein on the gel.
Function of sequenced peptides obtained via automated algorithm of the MASCOT software (www.matrixscience.com) from the NCBI.nr database.
P values obtained via Student's t test after comparing protein accumulation of three biological replicates of primary root cortex versus stele tissue (cutoff, P ≤ 0.01).
P values of the linear contrasts were adjusted according to Wald tests (cutoff, P ≤ 0.05).
For protein accessions identified by MASCOT that had no functional annotation, a function was assigned via BLASTP searches of the corresponding protein accession numbers (cutoff, E ≤ 1e−10).
Table II.
Proteins preferentially accumulated in stele tissue of maize primary roots compared with cortex identified after 2-DE separation and nano-HPLC-ESI-MS/MS analysis of trypsin-digested proteins matched against the NCBI.nr protein database entries
Preferentially accumulated proteins, >2-fold change; t test, P < 0.01 in three independent biological replicates.
| Spot No. | Stele Specificitya | MOWSE Scoreb | Sequence Coveragec | Mr Predicted/Mr Geld | pI Predicted/pI Gele | Function NCBI.nr [Species] Accession No.f | t Test Pg | FDR Ph |
|---|---|---|---|---|---|---|---|---|
| Metabolism | ||||||||
| 60 | 3.2 | 1705 | 33 | 92.9/93.8 | 6.0/6.6 | Sucrose synthase 1 [Z. mays] NP_001105323 | <0.001 | 0.007 |
| 61 | 2.5 | 828 | 17 | 83.7/86.0 | 5.9/6.2 | Methionine synthase protein [Sorghum bicolor] AAL73979 | <0.001 | 0.003 |
| 62 | 2.3 | 675 | 22 | 52.9/56.3 | 6.1/5.8 | UDP-Glc dehydrogenase [Colocasia esculenta] AAO62313 | 0.004 | 0.029 |
| Disease/defense/stress response | ||||||||
| 63 | Stele only | 636 | 19 | 57.2/60.8 | 5.6/5.8 | TCP-1/cpn60 chaperonin family protein [O. sativa] NP_001050672i | <0.001 | 0.010 |
| 64 | Stele only | 881 | 20 | 59.0/67.9 | 5.4/5.7 | T-complex protein 1 subunit ε (TCP-1/cpn60 chaperonin family) [Avena sativa] P54411 | <0.001 | 0.002 |
| 65 | 3.4 | 183 | 14 | 22.4/29.4 | 5.5/5.6 | Remorin 1 [O. sativa] NP_001053409i | 0.004 | 0.030 |
| 66 | 2.4 | 111 | 8 | 35.9/41.7 | 5.8/6.6 | Peroxidase [O. sativa] NP_001058379i | <0.001 | 0.006 |
| 67 | 2.1 | 379 | 10 | 62.0/76.3 | 5.3/5.4 | Peptidyl-prolyl isomerase [O. sativa] Q43207 | <0.001 | 0.039 |
| Energy | ||||||||
| 68 | Stele only | 162 | 9 | 19.8/17.9 | 5.3/5.1 | ATP synthase D chain [Solanum demissum] AAT40531 | 0.004 | 0.031 |
| Cell fate | ||||||||
| 69 | 2.4 | 246 | 19 | 41.7/44.8 | 5.2/5.1 | Actin [Saccharum officinarum] AAU93346 | <0.001 | 0.049 |
| 70 |
4.3 |
94 |
8 |
41.6/14.5 |
5.2/5.7 |
Actin3 [Glycine max] P02580 |
0.001 |
0.013 |
Ratio of accumulation (fold change) of a particular protein between primary root stele versus cortex tissue.
Probability-based MOWSE score is defined as –10 × log(P), where P is the probability that the observed match is a random event. Scores > 43 indicate identity or extensive homology (P < 0.05). Protein scores are derived from ion scores as a nonprobabilistic basis for ranking protein hits.
Percentage of predicted protein sequence covered by matched peptides.
Theoretical molecular mass of predicted protein/experimental molecular mass of protein on the gel.
Theoretical pI of predicted protein/experimental pI of protein on the gel.
Function of sequenced peptides obtained via automated algorithm of the MASCOT software (www.matrixscience.com) from the NCBI.nr database.
P values obtained via Student's t test after comparing protein accumulation of three biological replicates of primary root stele versus cortex tissue (cutoff, P ≤ 0.01).
P values of the linear contrasts were adjusted according to Wald tests (cutoff, P ≤ 0.05).
For protein accessions identified by MASCOT that had no functional annotation, a function was assigned via BLASTP searches of the corresponding protein accession numbers (cutoff, E ≤ 1e−10).
Identification of Protein Accessions Represented by Multiple Protein Spots
The 70 differentially accumulated protein spots that were identified in cortical parenchyma and stele tissues represented 56 different GenBank accessions. In total, six of the 56 different accessions were represented by more than one protein spot in the cortical parenchyma tissue. Among those, eight different cortical parenchyma-specific isoforms (proteins 3, 12, 23, 24, 25, 28, 31, and 34) of a β-glucosidase (GenBank accession no. P49235) were identified. Among these proteins, the isoform represented by protein 23 was the most abundant protein detected in this study. These different isoforms could represent different modifications of the same gene product or different members of gene families.
Preferential Accumulation of Proteins and Transcripts of a β-Glucosidase in Cortical Parenchyma Cells
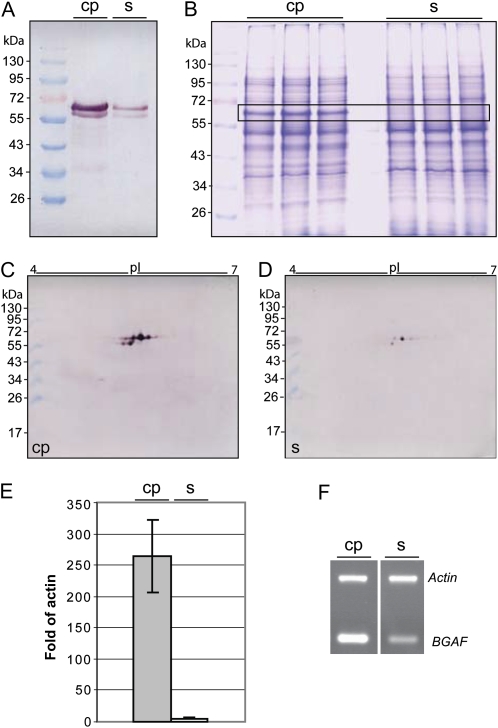
The accumulation of the β-glucosidase that displayed preferential accumulation of eight different isoforms in the cortical parenchyma was analyzed in more detail. First, western immunoblots with a specific antibody against this enzyme confirmed preferential accumulation of this protein in cortical parenchyma versus stele tissue (Fig. 2A). Interestingly, the two major bands in this western immunoblot, which are located between the size markers of 55 and 72 kD, correspond in size to seven of the differential proteins. While the upper and more abundant of these two bands comprises most likely proteins 3, 12, 23, and 24, which range in size between 63.7 and 66 kD (Table I), the lower band corresponds in size to isoforms 25, 28, and 34, which range in size between 59.9 and 61.5 kD (Table I). Isoform 31, which is represented by a 35.8-kD isoform in Supplemental Figure S1 and might be a specific degradation product or another related protein, is most likely displayed on the western immunoblot by a band slightly above the 34-kD marker. The maize β-glucosidase gene encodes a 566-amino acid protein that contains a 54-amino acid transit peptide for plastid targeting (Cicek and Esen, 1999). The precursor protein has a mass of 64.2 kD, while the mass of the mature monomer is 58.4 kD, which suggests that seven of the differential β-glucosidase proteins represent premature and mature proteins. The band that most likely corresponds to the upper band in Figure 2A can already be detected as cortex specific in Coomassie Brilliant Blue-stained gels of total protein extracts (Fig. 2B). 2-DE western immunoblots (Fig. 2, C and D) confirmed the results obtained by the mass spectrometric identification of cortical parenchyma-specific proteins (Table I) and the one-dimensional western immunoblots (Fig. 2A). All protein spots detected by a β-glucosidase-specific antibody were preferentially accumulated in cortical parenchyma tissue. The most abundant protein spots correspond to the two major bands in Figure 2A, while the 35.8-kD marker is represented by a very faint spot slightly above the 34-kD marker. Accumulation rates of mRNA and proteins depend on the synthesis and degradation rates of these molecules. Therefore, protein accumulation differences do not necessarily correlate with differences of the corresponding transcripts. To investigate if the accumulation differences of the different β-glucosidase isoforms are already manifested on the RNA level, quantitative real-time PCR (qRT-PCR) was performed (Fig. 2E). Transcript levels of β-glucosidase confirmed highly preferential expression of the β-glucosidase gene in cortical parenchyma cells. β-Glucosidases can specifically interact with β-glucosidase aggregation factor proteins (BGAF; GenBank accession no. NM_001111494) to form high-mass heterocomplexes (Blanchard et al., 2001). Binding of BGAF to β-glucosidase does not affect enzyme activity and most likely plays a protective role shielding the enzymes from proteases. In line with the preferential accumulation of β-glucosidase in cortical parenchyma, BGAF is also preferentially expressed in that tissue compared with the stele (Fig. 2F).
Figure 2.
Cortical parenchyma-specific expression of a β-glucosidase gene is confirmed at the protein (A–D) and RNA (E) levels in 2.5-d-old primary roots. A, Western blot of total cortical parenchyma (cp) and stele (s) protein extracts separated on a 10% SDS-PAGE gel and probed with an anti-β-glucosidase antibody. B, Three biological replicate protein extracts of cortical parenchyma and stele separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue. Bands highlighted in the box correspond in size to the β-glucosidase bands in A. C and D, Western blot of 2-DE-separated cortical parenchyma (C) and stele (D) protein extracts probed with the same anti-β-glucosidase antibody as in A. E, Cortical parenchyma-specific expression of the β-glucosidase gene as confirmed via qRT-PCR. F, Semiquantitative RT-PCR demonstrates preferential expression of the BGAF (GenBank accession no. NM_001111494) in cortical parenchyma versus stele tissue. [See online article for color version of this figure.]
The Cortical Parenchyma-Specific β-Glucosidase Is Preferentially Expressed in Roots and Seedling Tissue at the Shoot/Root Junction
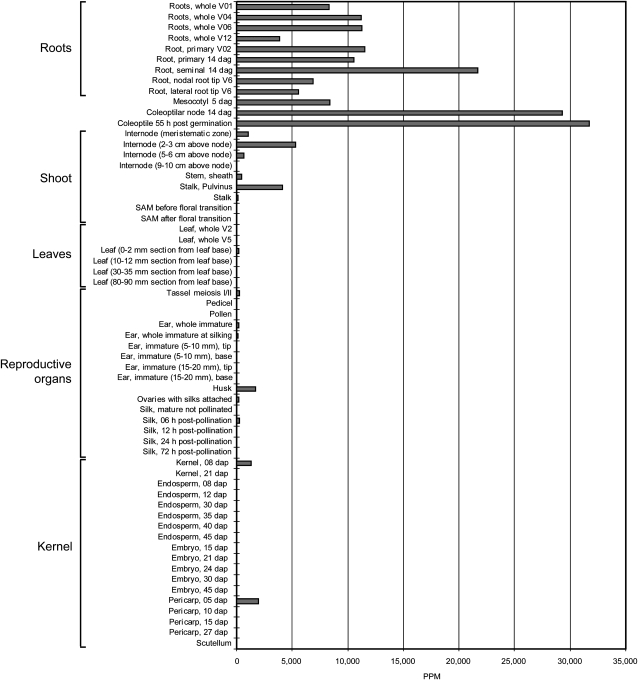
After demonstrating that β-glucosidase proteins and transcripts are preferentially accumulated in the parenchymal cortex of the differentiation zone of roots, we assayed the expression of this gene in 61 different maize tissues (Fig. 3). Massively Parallel Signature Sequencing (MPSS) is an open-ended platform that analyzes gene expression levels in a tissue by counting the number of individual 17-bp mRNA sequences in populations of 2 × 105 to 2 × 106 cDNAs. These 17-bp signature sequences often correspond to unique cDNAs, thus allowing for the quantification of the abundance of a particular cDNA in a sample representing a particular organ and developmental stage in a defined genetic background (Christensen et al., 2003). Expression of the β-glucosidase gene was observed in all analyzed root types at remarkable levels (Fig. 3) that exceeded in some root types 20,000 ppm. Other seedling organs at the root/shoot junction, including mesocotyl, coleoptilar node, and coleoptile, also displayed very high expression levels of this gene, indicating that in the coleoptile, more than 3% of all transcripts (>30,000 ppm) were synthesized by this β-glucosidase gene.
Figure 3.
Relative expression levels (in ppm) of a β-glucosidase gene in 61 different tissues and developmental stages from wild-type B73 inbred using the Solexa-MPSS system. dag, days after germination; dap, days after pollination.
Cortical Parenchyma-Specific Accumulation of Enzymes That Participate in Ammonium Assimilation
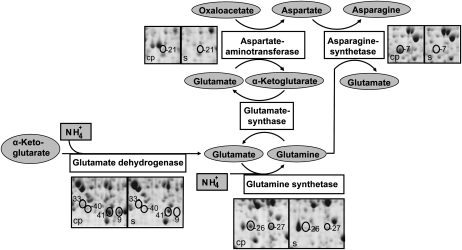
The distinct functions of cortical parenchyma and stele tissues are not only reflected by the differential accumulation of individual proteins but also by multiple enzymes that belong to specific biochemical pathways. In this study, four cortical parenchyma-specific enzymes (Asp aminotransferase, protein 21; Asn synthetase, protein 7; Glu dehydrogenase, proteins 9, 33, 40, and 41; and Gln synthetase, proteins 26 and 27), which are represented by eight protein spots (Fig. 4), are involved in ammonium assimilation into the nitrogen-transport amino acids Glu, Gln, Asp, and Asn (for review, see Lam et al., 1996).
Figure 4.
Cortical parenchyma-specific expression of several enzymes of ammonium assimilation. The numbering of the protein spots is according to Tables I and II.
Tissue-Specific Differences in Hormone Content of Cortical Parenchyma and Stele
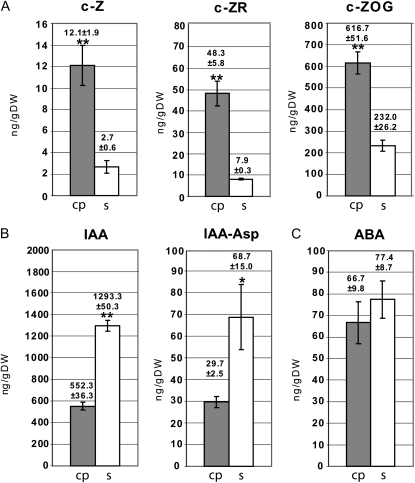
The specific action of phytohormones in growth, development, and differentiation of roots requires their precise tissue-specific distribution. In this study, the plant hormones auxin (IAA), cytokinin, and abscisic acid and selected intermediates and conjugates have been quantified in a tissue-specific fashion in 2.5-d-old cortical parenchyma and the stele of the differentiation zone of maize primary roots. These metabolites have been analyzed via HPLC-ESI-MS/MS using deuterated internal standards. The results of this survey are summarized in Figure 5 and are expressed in nanograms per gram dry weight. Various cytokinins are synthesized from different precursors. The three naturally occurring cytokinins trans-zeatin, cis-zeatin (c-Z), and dihydrozeatin and their precursors are widely found in nature (Sakakibara 2006). c-Z and its related metabolites are the predominant cytokinins in maize (Veach et al., 2003). While the concentrations of trans-zeatin and dihydrozeatin and their precursors and conjugates were below the limit of quantification, c-Z (fold change = 4.5), its precursor c-Z riboside (fold change = 6.1), and the conjugated O-glucoside cis-zeatin O-glucoside (c-ZOG; fold change = 2.7) were preferentially accumulated in cortical parenchyma versus stele tissue (Fig. 5A). The precursor c-Z riboside displayed a higher concentration than c-Z. Notably, most cytokinin detected in the differentiation zone of 2.5-d-old maize primary roots was present in the conjugated form of c-ZOG, which can be reversibly transferred into c-Z by β-glucosidase. The auxin IAA displayed a 2.3 times higher accumulation in stele versus cortical parenchyma tissue (Fig. 5B). Similarly, inactive IAA conjugated with the amino acid Asp was accumulated 2.3 times higher in stele versus cortical parenchyma tissues (Fig. 5B). IAA conjugated with other amino acids was below the limit of quantification (Ala, Glu) or detection (Leu). The amount of free IAA in the stele was 18.8 times higher than its most abundant inactive IAA form conjugated with an amino acid (IAA-Asp). For abscisic acid, no significant concentration differences were observed between cortical parenchyma and stele (Fig. 5C).
Figure 5.
Cortical parenchyma (cp)- and stele (s)-specific profiling of phytohormones and phytohormone precursors revealed cortical parenchyma-specific accumulation of cytokinin and stele-specific accumulation of auxin and their precursors. A, Cytokinin (c-Z), cytokinin (c-ZR), and a cytokinin conjugate (c-ZOG). B, IAA and the IAA conjugate IAA-Asp. C, Abscisic acid (ABA). DW, Dry weight. **P ≤ 0.01; *P ≤ 0.05.
DISCUSSION
In recent years, high-resolution cell type-specific transcriptome analyses have provided novel insights into functional domains of the Arabidopsis (Arabidopsis thaliana) primary roots during development (Birnbaum et al., 2003; Nawy et al., 2005; Brady et al., 2007). These analyses combined the isolation of specific GFP-tagged root cell types via protoplasting and fluorescence-activated cell sorting with downstream microarray analyses. In maize, due to the lack of GFP marker lines, cell type-specific analyses have been limited thus far to the analysis of pericycle cells that have been isolated via laser capture microdissection (Schnable et al., 2004) prior to downstream gene expression analyses (Woll et al., 2005; Dembinsky et al., 2007). In order to obtain sufficient pericycle transcripts for gene expression analyses from laser capture microdissection-captured cells, it was necessary to amplify RNA linearly prior to gene expression analyses by RT. In contrast to RNA, proteins cannot be amplified. Therefore, most proteome analyses have been confined to whole roots (Hochholdinger et al., 2004a, 2005; Wen et al., 2005; Liu et al., 2006; Hoecker et al., 2008) or longitudinal segments of roots (Chang et al., 2000; Zhu et al., 2006, 2007). In this study, therefore, we developed an approach that allowed separating and subsequently analyzing the functionally diverse internal stele tissue and the surrounding cortical parenchyma tissues (endodermis, cortex, and epidermis) in the differentiation zone of the primary root without damaging pericycle or endodermis cells.
Phytohormones are important regulators of root form and function. In this study, it was demonstrated that auxin (IAA) and cytokinin (c-Z) have antagonistic accumulation levels in cortical parenchyma and stele tissues of the differentiation zone, with preferential accumulation of auxin in the stele and c-Z in the cortical parenchyma. Asymmetric auxin distribution in stele and cortical parenchyma is in line with the generally accepted model of polar auxin transport that suggests that auxin is transported toward the root tip through the stele, as indirectly demonstrated by the activity of the auxin response reporter DR5::GFP or DR5::GUS in Arabidopsis (Blilou et al., 2005). More recently, fluorescence-activated cell sorting of GFP-marked cell types in combination with highly sensitive mass spectrometry allowed the direct measurement of IAA distribution and biosynthesis at the cellular level in Arabidopsis primary roots and suggested a local IAA biosynthesis maximum in the root apex (Petersson et al., 2009). In contrast, the distribution of free cytokinin has not been resolved at the tissue level in the differentiation zone of roots. This might be due to the limited sensitivity of cytokinin antibodies (Eberle et al., 1986) and marker lines (Aloni et al., 2004) that cannot distinguish between different cytokinin types, precursors, and conjugated forms. Only recently has it been demonstrated that c-Z is the major active cytokinin isomer in maize roots (Veach et al., 2003). Our study extends these initial observations by demonstrating that c-Z, its precursor cis-zeatin riboside, and the conjugated c-ZOG display a significantly higher concentration in the cortical parenchyma than in the stele. This tissue-specific distribution of phytohormones is laid down very early in root development (Dello Ioio et al., 2008) and implies an important role in determining the specificity and functions of these tissues by regulating downstream gene expression cascades. It should be noted that the plant hormones auxin and cytokinin, which display tissue-specific accumulation patterns in cortical parenchyma and stele, act antagonistically toward lateral root development in the differentiation zone of the root, which takes place in pericycle and endodermis cells at the interface of cortical parenchyma and stele (Esau, 1965). While auxin mediates cell cycle activation, which results in lateral root initiation (Himanen et al., 2002), cytokinin counteracts the promotive action of auxin on lateral root formation (Werner et al., 2003) by directly acting on lateral root founder cells and inhibiting lateral root initiation (Laplaze et al., 2007). Moreover, in Arabidopsis, auxin is a negative regulator of cytokinin levels (Nordström et al., 2004), while on the other hand, cytokinin modulates organogenesis by reducing polar auxin transport by acting on auxin efflux (Pernisová et al., 2008) and influx (Ruzicka et al., 2009) carriers.
Among the cortical parenchyma-specific metabolic proteins, several cytokinin-related enzymes involved in ammonium assimilation and the conversion of storage and active forms of cytokinin and benzoxazinoids were detected, which is in line with the increased cytokinin levels in this tissue. Inorganic nitrogen that is typically taken up from the soil in the form of nitrate or ammonium (Takei et al., 2002; Tabuchi et al., 2007) is assimilated into the nitrogen-transport amino acids Glu, Gln, Asp, and Asn (Sakakibara et al., 2006). These compounds are used to build up nitrogen reserves for subsequent use in growth and development. In this study, four enzymes involved in the assimilation of ammonium, Asp aminotransferase (protein 21), Asn synthetase (protein 7), Glu dehydrogenase (proteins 9, 33, 40, and 41), and Gln synthetase (proteins 26 and 27), were represented by eight protein isoforms that were preferentially accumulated in the cortical parenchyma of the primary root differentiation zone of maize (Fig. 4). Remarkably, all enzymes of ammonium assimilation are induced by cytokinin in Arabidopsis (for summary, see Sakakibara et al., 2006). This suggests that the maize cortical parenchyma is a primary tissue of ammonium assimilation in young maize seedlings before they are in contact with exogenous supplies of nitrogen provided by the soil. This is in line with the observation that in most plants, nitrate assimilation during the early stages of development takes place preferentially in roots, while later in development nitrate is typically translocated to the aerial parts for assimilation (Heldt, 2003). Nitrogen assimilated in these nitrogen-transport amino acids is later donated to the biosynthesis of other amino acids, nucleic acids, and other nitrogen-containing compounds required during root development. An Asp kinase (protein 22) that was exclusively accumulated in the cortical parenchyma provides an example for the entry point in such a pathway, catalyzing the first reaction that phosphorylates Asp and finally leading to the synthesis of the amino acids Thr, Met, Lys, and Ile (Buchanan et al., 2000). Moreover, an Asn synthetase protein (spot 7) is exclusively accumulated in the cortex. Asn acts as a nitrogen storage compound synthesized under high concentrations of ammonium and thus acts in ammonium detoxification (Givan, 1979; Sieciechowicz et al., 1988). Higher levels of ammonium cannot only be due to uptake from the environment but also result from the catabolism of soluble organic nitrogen in maize roots (Feng et al., 1998). This emphasizes a role of Asn synthetase in ammonium assimilation in the cortex.
Other enzymes specific to the cortical parenchyma related to Asp synthesis or action are Asp aminotransferase (protein 21) and aspartyl tRNA synthetase (protein 17). Other enzymes that are involved in the biosynthesis of amino acids that were preferentially accumulated in the cortical parenchyma are a Cys desulfurase (protein 16), which catalyzes the conversion of Cys to Ala, and a Ala glyoxylate aminotransferase (protein 30), which converts glyoxylate to Gly.
Moreover, multiple isoforms of a β-glucosidase (proteins 3, 12, 23, 24, 25, 28, 31, and 34) displayed preferential accumulation in the cortical parenchyma of the differentiation zone of the primary root. These proteomics results were confirmed by western immunoblots and qRT-PCR experiments demonstrating that this protein is accumulated in the differentiation zone in addition to its previously detected expression in meristematically active maize root tips (Brzobohatý et al., 1993). Moreover, MPSS analyses revealed that this gene is preferentially expressed at very high levels in roots, suggesting an important function in this organ. The enzyme β-glucosidase functions in the release of free bioactive cytokinin (Brzobohatý et al., 1993), benzoxazinoids (Cuevas et al., 1992), and monolignols (Escamilla-Treviño et al., 2006) from their glucosylated inactive storage forms and hence correlates with the predominant accumulation of c-ZOG, which is one of the substrates of β-glucosidase, and free c-Z, which is released from c-ZOG by this β-glucosidase. That this β-glucosidase is also involved in the release of free benzoxazinoids from their storage form is supported by the previous observation that the benzoxazinoid 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one was released in the cortical parenchyma during the penetration of emerging lateral roots (Park et al., 2004). Although the release of DIMBOA (benzoxazinoid 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one) from DIMBOA-glucoside might be dominant in the cortical parenchyma during lateral root emergence, it is remarkable that in our experiments (i.e. before the initiation of lateral roots), the enzyme UDP-glucosyltransferase BX9 (protein 50; von Rad et al., 2001), which catalyzes the converse reaction, is also preferentially accumulated in the cortex. Hence, this β-glucosidase might not only function in the release of free cytokinin in the root tip to stimulate cell division activity (Brzobohatý et al., 1993) but also in the release of cytokinin and benzoxazinoids in the cortical parenchyma of the differentiation zone. The notion that the interplay of β-glucosidase and c-Z levels might play an important role in the organization of root stock architecture by influencing lateral root formation and primary root length is supported by transgenic maize plants overexpressing a zeatin O-glucosylation gene (ZOG1; Pineda Rodo et al., 2008). O-Glucosyltransferases convert bioactive cytokinin into their inactive O-glucoside storage products and hence catalyze the converse reaction of β-glucosidase, thus controlling the level of free cytokinin. Inactivation of cytokinin by ZOG1 induced primary root growth and increased the number of lateral roots (Pineda Rodo et al., 2008). Consistent with the preferential accumulation of this β-glucosidase, a BGAF is also preferentially accumulated in the cortical parenchyma. BGAF plays a protective role for β-glucosidase in binding of this enzyme without affecting its activity or kinetic parameters (Blanchard et al., 2001).
While most differential proteins detected in this study were related to the development and function of the cortical parenchyma, 11 proteins were preferentially accumulated in the stele. Among those proteins was a Suc synthase 1 protein (protein 60). Suc synthase catalyzes the conversion of Suc and UDP to Fru and UDP-Glc, which is a precursor for the synthesis of UDP-Gal, starch, cellulose, and callose and thus plays an important role in energy metabolism (Chourey et al., 1998). It has been demonstrated that this enzyme is localized in the outermost cell layers of the maize root cap (Kladnik et al., 1994) and is most abundant in the stele, in particular in association with vascular tissues of the elongation zone (Koch et al., 1992). In this study, we demonstrated that Suc synthase 1 is not only accumulated in the stele of the elongation zone but also in the stele of the differentiation zone in the maize primary root. In line with the preferential accumulation of a Suc synthase, a UDP-Glc dehydrogenase that metabolizes one of the products of Suc synthase, UDP-Glc, is preferentially accumulated in the stele (protein 62). UDP-Glc dehydrogenase plays a role in cell wall pentose biosynthesis (Kärkönen et al., 2005). A different chemical composition of the stele and cortical parenchyma cell walls is also suggested by the distinct polychromatic staining of stele and cortical parenchyma cell walls by toluidine blue, which is attributed to differences in the chemical composition of these cell walls (O'Brien et al., 1964). Remarkably, supply of sugar as provided by Suc synthase (Dennis and Blakelely, 2000) down-regulates the expression of Asn synthase (Chevalier et al., 1996). Hence, the preferential expression of Suc synthase in the stele might, on the other hand, also explain the low expression of Asn synthase in this tissue (Chevalier et al., 1996), which was previously discussed in the context of cortex specific protein accumulation.
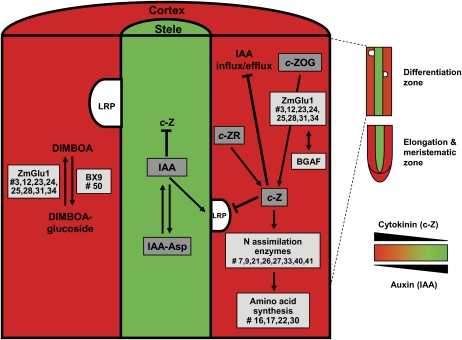
In summary, the antagonistic distribution of auxin and cytokinin in the stele and cortical parenchyma, and the tissue-specific accumulation of cytokinin-related and other proteins, suggest a molecular framework that specifies the function of these tissues. A model summarizing these tissue-specific functions is provided in Figure 6.
Figure 6.
Asymmetric distribution of cytokinin (red) and auxin (green) in the cortical parenchyma and stele in the differentiation zone of the young maize primary root and related cortical parenchyma-specific protein accumulation patterns. Proteins preferentially accumulated in the cortical parenchyma are highlighted in light gray boxes. Numbers (#) correspond to proteins in Tables I and II. Differentially accumulated phytohormones detected in this study, including cytokinin (c-Z), its precursor (c-ZR), its glucoside conjugate (c-ZOG), auxin (IAA), and its Asp conjugate (IAA-Asp), are highlighted in dark gray boxes. BX9, UDP-glucosyltransferase; c-ZR, cis-zeatin riboside; DIMBOA, 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one; LRP, lateral root primorida; ZmGlu-1, β-glucosidase. [See online article for color version of this figure.]
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All experiments were performed with primary roots of the maize (Zea mays) inbred line B73. Cortical parenchyma and stele tissues of the differentiation zone of maize primary roots were sampled from 2.5-d-old seedlings that were germinated between two vertically oriented layers of germination paper (Anchor Paper) in twice distilled water at 28°C in the dark as described previously (Woll et al., 2005).
Separation and Microscopic Analysis of Cortical Parenchyma and Stele Tissue
The first step of separating cortical parenchyma and stele tissues in the differentiation zone of 2.5-d-old primary roots (Fig. 1A) was to detach the first 1 cm of the primary root tip, which comprises the meristematic and differentiation zones, with a razor blade (Fig. 1C). Second, the outermost cell layers (cortical parenchyma) of the root were cut with a razor blade close to the coleorhiza without touching the stele (Fig. 1B). Third, the cortical parenchyma tissue was pulled off the stele by gently bending the root and finally pulling the cortical parenchyma off the stele (Fig. 1C). Finally, the stele that was still attached to the remainder of the seedling was cut close to the coleorhiza. Free hand sections of 2.5-d-old primary roots embedded in 3% agarose, before and after separating cortical parenchyma and stele tissues, were stained for 8 min in 0.001% (w/v) toluidine blue O according to a modified protocol of O'Brien et al. (1964). Specimens were analyzed with a Zeiss-Axioskop HBO 100W/2 microscope and documented with a Canon PowerShot G2 camera. The filter set used for visualizing the toluidine blue O staining contained an excitation filter (BP 450–490), a chromatic beam splitter (FT510), and a barrier filter (LP520).
Protein Isolation
For proteomics experiments, cortical parenchyma and stele tissues were collected from the differentiation zone of approximately 40 to 60 primary roots per biological replicate and immediately frozen in liquid nitrogen. For each tissue type, three independent biological replicates were prepared. Total soluble proteins were extracted from cortical parenchyma or stele tissue via TCA precipitation (10% TCA in acetone containing 0.07% β-mercaptoethanol) at −20°C (Damerval et al., 1986) in 2-mL microfuge tubes overnight. The precipitated proteins were washed five times by incubation in acetone containing 0.07% β-mercaptoethanol for 1 h at −20°C and centrifugation at 10,000 rpm for 15 min at 4°C. Vacuum-dried protein pellets were dissolved in 800 μL of a solution containing 7 m urea, 2 m thiourea, 2% (w/v) CHAPS, 1.25% (v/v) Bio-Lytes 3/10 (Bio-Rad), 50 mm dithiothreitol, and one tablet of protease inhibitor complete (Roche) for 10 mL of extraction solution. For solubilization, the samples were ultrasonicated for 10 min and subsequently treated with 150 units of benzonase endonuclease (Sigma). Sonication was repeated for an additional 10 min followed by incubation for 15 min at room temperature. After centrifugation at 14,000 rpm for 40 min, supernatant containing soluble proteins was taken and the quality and quantity of the soluble protein extracts in the supernatant were determined on preparative SDS-PAGE gels and with the Bradford assay.
2-DE Separation of Cortical Parenchyma and Stele Proteins
Isoelectric focusing of protein extracts was performed with approximately 600 μg of total soluble protein extract on 18-cm immobilized, linear pH 4 to 7 gradients (Imobiline drystrips; Amersham Biosciences) using an Ettan IPGPhor isoelectric focusing unit (Amersham Biosciences). The immobilized pH gradients were rehydrated by applying the protein extracts at 50 V overnight. The following settings of isoelectric focusing was used: a 0- to 300-V gradient for 1 min, 300 V for 2 h, a 300- to 1,000-V gradient for 20 min, 1,000 V for 30 min, a 1,000- to 4,000-V gradient for 1 h, 4,000 V for 30 min, a 4,000- to 8,000-V gradient for 1 h, 8,000 V for 5 h, and an 8,000- to 100-V gradient. Isoelectric focusing was stopped when a total of about 90,000 Vh was reached. The immobilized pH gradients were equilibrated for 15 min in a buffer containing 6 m urea, 30% glycerin, 2% SDS, 0.05 m Tris, pH 8.8, and 1% dithiothreitol, followed by incubation for 15 min in a buffer containing 6 m urea, 30% glycerin, 2% SDS, 0.05 m Tris, pH 8.8, and 4% iodoacetamide. Proteins were subsequently resolved on the basis of their masses on 12.5% SDS-PAGE gels using the Ettan DALTsix electrophoresis system (Amersham Biosciences). In total, six samples representing three biological replicates of cortical parenchyma and stele tissue of maize primary roots were subjected to isoelectric focusing and subsequent SDS-PAGE separation in parallel.
Visualization and Identification of Differential Cortical Parenchyma and Stele Proteins
Proteins on 2-DE SDS-PAGE gels were stained with a colloidal Coomassie Brilliant Blue stain (Neuhoff et al., 1988) for 2.5 d on an orbital shaker as described previously (Sauer et al., 2006). Gel images were obtained using the ImageScanner (Amersham) high-resolution scanner. PD-Quest software version 7.1 (Bio-Rad) was used for analysis of the 2-DE gels. The intensities of spots were normalized to the total intensity of all matched spots on each gel by PD-Quest. These relative spot intensities were subsequently used for statistical analyses. After normalization, Student's t test (independent samples, pooled variance, P ≤ 0.01) was used to identify differentially expressed spots between cortical parenchyma and stele tissues (≥2-fold change). The resulting P values were subsequently adjusted controlling the FDR (Benjamini and Hochberg, 1995) at 5% to account for multiple testing. Tables I and II represent results for proteins with significant differential expression after FDR adjustment. These analyses were generated with SAS version 9.1.3 Service Pack 4.
Nano-HPLC-ESI-MS/MS and Analysis of Spectrometric Data
Proteins that were identified to be statistically differentially accumulated between cortical parenchyma and stele were excised from representative gels and digested in-gel using trypsin (sequencing grade; Promega). Peptide analyses were performed with a Dionex LC Packings HPLC System containing the components Famos (autosampler), Switchos (loading pump and switching valves), and Ultimate (separation pump and UV light detector). A high-performance QStar Pulsar i (Applied Biosystems, Applera) quadrupole time-of-flight mass spectrometer equipped with a nano-ESI source (Column Adapter [ADPC-PRO] and distal coated SilicaTips [FS360-20-10-D-20]; both from New Objective) was used to record the ESI-MS/MS spectra. The mobile phases and gradients were used as described previously (Sauer et al., 2006). Protein identification was performed by correlating the mass spectra with the NCBI.nr protein sequence database Viridiplantae (higher plant) as of September 24, 2008, using the MOWSE algorithm (Perkins et al., 1999) as implemented in the MS search engine MASCOT (Matrix Science). Results from 2-DE analyses and MS experiments and all search results were stored in a Laboratory Information Management System database (Proteinscape 1.3.0; Bruker Daltonics).
Western-Blot Immunodetection
For immunodetection of specific proteins, total protein extracts were separated on homogenous 10% acrylamide SDS-PAGE gels according to Laemmli (1970). Per lane, 20 μg of protein extract was loaded on one-dimensional electrophoresis gels. Alternatively, 40 μg of total protein extracts was separated on 2-DE gels. After gel electrophoresis, proteins were transferred to a nitrocellulose membrane (0.45 mm pore size; Protran BA 85; Whatman) for 60 min at 300 mA with the PerfectBlue (Peqlab Biotechnology) semidry electroblotting device. Immunodetection was performed by incubating the nitrocellulose membranes in Tris-buffered saline (TBS; pH 7.6) blocking solution containing 0.05% (v/v) Tween 20 (TBST) and 5% milk powder for 2 h. The membrane was incubated with anti-β-glucosidase primary antibody (diluted 1:10,000 in TBS buffer containing 1.5% milk powder) for 1 h. After three rounds of washing in TBST, the membranes were incubated with an alkaline phosphatase-conjugated anti-rabbit IgG secondary antibody (1:10,000; Roche) for 1 h. Detection with a substrate solution containing nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate was performed according to the manufacturer's instructions (Roche).
Gene Expression Analysis via Reverse Transcription-PCR
Total RNA was isolated with peqGOLD TriFast (Peqlab Biotechnology) reagent. Extracted RNA was treated with DNase I (Ambion) and followed by inactivation with slurry (Ambion). cDNA for real-time PCR experiments was synthesized from 4 μg of RNA with peqGOLD Moloney murine leukemia virus reverse transcriptase and protocols following the manufacturer's instructions. qRT-PCR was performed with a Bio-Rad iCycler and QuantiTect SYBR Green PCR master mix (Qiagen). Relative expression of β-glucosidase (GenBank accession no. P49235; forward primer, 5′-CCTCATGATGTGGGTGCAG-3′; reverse primer, 5′-GCATGACAAGGCCAGACTG-3′) was compared in cortical parenchyma and stele tissues with the expression levels of a constitutively expressed actin gene (GenBank accession no. J01238; forward primer, 5′-AGGCCACGTACAACTCCATC-3′; reverse primer, 5′-CCACCGATCCAGACACTGTA-3′). Relative expression of the β-glucosidase gene compared with the actin gene was calculated as mean normalized expression compared with actin, using threshold cycle values and averaged primer efficiencies calculated from three biological replications as described previously (Hoecker et al., 2008). Semiquantitative reverse transcription (RT)-PCR was performed to determine the relative expression of BGAF (GenBank accession no. NM_001111494; forward primer, 5′-AGCTGCTCTGCTAGCTCCAC-3′; reverse primer, 5′-GATCGATTCAAGTGCCAATC-3′) expression in cortical parenchyma and stele with reference to a constitutively expressed actin gene (GenBank accession no. J01238; forward primer, 5′-TTGGGTCAGAAAGGTTCAGG-3′; reverse primer, 5′-GCACTTCATGTGGACAATGC-3′) that was employed as an internal control. The semiquantitative RT-PCR was performed as described previously (Dembinsky et al., 2007).
Hormone Profiling of Cortical Parenchyma Versus Stele Tissues
For the hormone-profiling experiments, cortical parenchyma and stele tissues from the differentiation zone of 2.5-d-old primary roots was collected. Cortical parenchyma tissue of approximately 100 primary roots and stele tissue of approximately 300 primary roots were harvested per biological replicate and freeze dried. For each tissue type, three independent biological replicates were prepared. Plant hormone analyses were performed at the Plant Biotechnology Institute of the National Research Council of Canada by HPLC-ESI-MS/MS using deuterated internal standards, as described previously (Chiwocha et al., 2003, 2005). Results are expressed in nanograms per gram dry weight. Hormones or intermediates were considered differentially accumulated between cortical parenchyma and stele if a significant difference was confirmed in Student's t test. The significance level is indicated in Figure 5 by one (P < 0.05) or two (P < 0.01) asterisks.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Representative proteomic 2-DE maps of soluble proteins extracted from 2.5-d-old cortical parenchyma (A) and stele (B) tissues from the differentiation zone of maize primary roots.
Supplemental Table S1. Peptide fragments identified via ESI-MS/MS and summarized in Tables I and II.
Supplementary Material
Acknowledgments
We appreciate the assistance of Dr. Iriana Zaharia at the Plant Biotechnology Institute (National Research Council, Saskatoon, Saskatchewan, Canada) in performing the hormone-profiling assays. We also thank Dr. Asim Esen (Virginia Tech Plant Molecular Biology and Biochemistry Laboratory, Blacksburg, Virginia) for the gift of the anti-β-glucosidase antibody.
This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 446 (“Mechanisms of Cell Behavior in Eukaryotes”), by the Deutscher Akademischer Austauschdienst (to M.S.), and by the Ministerium für Wissenschaft und Kunst, Landesregierung Baden-Württemberg (to Proteome Centrum Tuebingen).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Frank Hochholdinger (frank.hochholdinger@zmbp.uni-tuebingen.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aloni R, Langhans M, Aloni E, Ullrich CI (2004) Role of cytokinin in the regulation of root gravitropism. Planta 220 177–182 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57 289–300 [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blanchard DJ, Cicek M, Chen J, Esen A (2001) Identification of beta-glucosidase aggregating factor (BGAF) and mapping of BGAF binding regions on maize beta-glucosidase. J Biol Chem 276 11895–11901 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K (1993) Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262 1051–1054 [DOI] [PubMed] [Google Scholar]
- Buchanan B, Gruissem W, Jones RL, editors (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD
- Chang WWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JK (2000) Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol 122 295–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Just D, Raymond P (1996) Metabolic regulation of asparagine synthatase gene expression in maize (Zea mays L.) root tip. Plant J 9 1–11 [DOI] [PubMed] [Google Scholar]
- Chiwocha SDS, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross ARS, Kermode AR (2003) A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J 35 405–417 [DOI] [PubMed] [Google Scholar]
- Chiwocha SDS, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross ARS, Kermode AR (2005) The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J 42 35–48 [DOI] [PubMed] [Google Scholar]
- Chourey PS, Taliercio EW, Carlson SJ, Ruan YL (1998) Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet 259 88–96 [DOI] [PubMed] [Google Scholar]
- Christensen TM, Veijlupkova Z, Sharma YK, Arthur KM, Spatafora JW, Albright CA, Meeley RB, Duvick JP, Quantrano RS, Fowler JE (2003) Conserved subgroups and developmental regulation in the monocot rop gene family. Plant Physiol 133 1791–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek M, Esen A (1999) Expression of soluble and catalytically active plant (monocot) β-glucosidases in E. coli. Biotechnol Bioeng 63 392–400 [DOI] [PubMed] [Google Scholar]
- Cuevas L, Niemeyer HM, Jonsson LMV (1992) Partial purification and characterization of a hydroxamic acid glucoside β-image-glucosidase from maize. Phytochemistry 31 2609–2612 [Google Scholar]
- Damerval C, Vienne D, Zivy M, Thiellement H (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7 52–54 [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B, et al (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Blakelely SD (2000) Carbohydrate metabolism. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 630–675
- Eberle J, Arnscheidt A, Klix D, Weiler EW (1986) Monoclonal antibodies to plant growth regulators. III. Zeatinriboside and dihydrozeatinriboside. Plant Physiol 81 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K (1965) Plant Anatomy, Ed 2. John Wiley and Sons, New York
- Escamilla-Treviño LL, Chen W, Card ML, Shih MC, Cheng CL, Poulton JE (2006) Arabidopsis thaliana beta-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry 67 1651–1660 [DOI] [PubMed] [Google Scholar]
- Feng J, Volk RJ, Jackson WA (1998) Source and magnitude of ammonium generation in maize roots. Plant Physiol 118 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J (2003) Auxin transport: shaping the plant. Curr Opin Plant Biol 6 7–12 [DOI] [PubMed] [Google Scholar]
- Givan CV (1979) Metabolic detoxification of ammonia in tissue of higher plants. Phytochemistry 18 375–382 [Google Scholar]
- Heldt HW (2003) Pflanzenbiochemie, Ed 3. Spektrum Akademischer Verlag, Heidelberg
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F (2009) The maize root system: morphology, anatomy and genetics. In JL Bennetzen, SC Hake, eds, The Maize Handbook: Its Biology. Springer, Berlin, pp 145–160
- Hochholdinger F, Guo L, Schnable PS (2004. a) Lateral roots affect the proteome of the primary root of maize (Zea mays L.). Plant Mol Biol 56 397–412 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K (2004. b) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Sauer M, Dembinsky D, Hoecker N, Muthreich N, Saleem M, Liu Y (2006) Proteomic dissection of plant development. Proteomics 6 4076–4083 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Guo L, Schnable PS (2005) The accumulation of abundant soluble proteins changes early in the development of the primary roots of maize (Zea mays L.). Proteomics 5 4885–4893 [DOI] [PubMed] [Google Scholar]
- Hoecker N, Lamkemeyer T, Sarholz B, Paschold A, Fladerer C, Madlung J, Wurster K, Stahl M, Piepho H-P, Nordheim A, et al (2008) Analysis of non-additive protein accumulation in young primary roots of a maize (Zea mays L.) F1-hybrid compared to its parental inbred lines. Proteomics 8 3882–3894 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML (1995) Specialized zones of development in roots. Plant Physiol 109 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkönen A, Murigneux A, Martinant JP, Pepey E, Tatout C, Dudley BJ, Fry SC (2005) UDP-glucose dehydrogenases of maize: a role in cell wall pentose biosynthesis. Biochem J 391 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladnik A, Vilhar B, Chourey PS, Dermastia M (1994) Sucrose synthase isozyme SUS1 in the maize root cap is preferentially localized in the endopolyploid outer cells. Can J Bot 82 96–103 [Google Scholar]
- Koch KE, Nolte KD, Duke ER, McCarty DR, Avigne WR (1992) Sugar levels modulate differential expression of maize sucrose synthase genes. Plant Cell 4 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47 569–593 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xu C, Zhang K, Yang A, Zhang J (2007) Proteomic analysis of root growth and metabolic changes under phosphorus deficit in maize (Zea mays L.) plants. Proteomics 7 1501–1512 [DOI] [PubMed] [Google Scholar]
- Liu Y, Lamkemeyer T, Jakob A, Mi G, Zhang F, Nordheim A, Hochholdinger F (2006) Comparative proteome analyses of maize (Zea mays L.) primary roots prior to lateral root initiation reveal differential protein expression in the lateral root initiation mutant rum1. Proteomics 6 4300–4308 [DOI] [PubMed] [Google Scholar]
- Marschner H (2003) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Mueller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V, Arnold N, Taube D, Erhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie blue. Electrophoresis 9 255–262 [DOI] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, Dolezal K, Sandberg G (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TP, Feder N, McCully ME (1964) Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma 59 368–373 [Google Scholar]
- Park WJ, Hochholdinger F, Gierl A (2004) Release of the benzoxazinoids defense molecules during lateral- and crown root emergence in Zea mays. J Plant Physiol 161 981–985 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pernisová M, Klíma P, Horák J, Válková M, Malbeck J, Soucek P, Reichman P, Hoyerová K, Dubová J, Friml J, et al (2008) Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci USA 106 3609–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda Rodo A, Brugière N, Vankova R, Malbeck J, Olson JM, Haines SC, Martin RC, Habben JE, Mok DW, Mok MC (2008) Over-expression of a zeatin O-glucosylation gene in maize leads to growth retardation and tasselseed formation. J Exp Bot 59 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57 431–449 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11 440–448 [DOI] [PubMed] [Google Scholar]
- Sauer M, Jakob A, Nordheim A, Hochholdinger F (2006) Proteomic analysis of shoot-borne root initiation in maize (Zea mays L.). Proteomics 6 2530–2541 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7 50–56 [DOI] [PubMed] [Google Scholar]
- Sieciechowicz KA, Joy KW, Ireland RJ (1988) The metabolism of asparagine in plants. Phytochemistry 27 663–671 [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T (2007) Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J Exp Bot 58 2319–2327 [DOI] [PubMed] [Google Scholar]
- Takei K, Takahashi T, Sugiyama T, Yamaya T, Sakakibara H (2002) Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot 53 971–977 [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DW, Malbeck J, Vankova R, Mok MC (2003) O-Glucosylation of cis-zeatin in maize: characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol 131 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rad U, Hüttl R, Lottspeich F, Gierl A, Frey M (2001) Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J 28 633–642 [DOI] [PubMed] [Google Scholar]
- Wen TJ, Hochholdinger F, Sauer M, Bruce W, Schnable PS (2005) The roothairless1 gene of maize (Zea mays) encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol 138 1637–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll K, Borsuk L, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization and pericycle specific transcriptome analyses of the novel maize (Zea mays L.) lateral and seminal root initiation mutant rum1. Plant Physiol 139 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Alvarez S, Marsh EL, Lenoble ME, Cho IJ, Sivaguru M, Chen S, Nguyen HT, Wu Y, Schachtman DP, et al (2007) Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol 145 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen S, Alvarez S, Asirvatham VS, Schachtman DP, Wu Y, Sharp RE (2006) Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiol 140 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.