Abstract
Nitrogen fixation in legumes requires the development of root organs called nodules and their infection by symbiotic rhizobia. Over the last decade, Medicago truncatula has emerged as a major model plant for the analysis of plant-microbe symbioses and for addressing questions pertaining to legume biology. While the initiation of symbiosis and the development of nitrogen-fixing root nodules depend on the activation of a protein phosphorylation-mediated signal transduction cascade in response to symbiotic signals produced by the rhizobia, few sites of in vivo phosphorylation have previously been identified in M. truncatula. We have characterized sites of phosphorylation on proteins from M. truncatula roots, from both whole cell lysates and membrane-enriched fractions, using immobilized metal affinity chromatography and tandem mass spectrometry. Here, we report 3,457 unique phosphopeptides spanning 3,404 nonredundant sites of in vivo phosphorylation on 829 proteins in M. truncatula Jemalong A17 roots, identified using the complementary tandem mass spectrometry fragmentation methods electron transfer dissociation and collision-activated dissociation. With this being, to our knowledge, the first large-scale plant phosphoproteomic study to utilize electron transfer dissociation, analysis of the identified phosphorylation sites revealed phosphorylation motifs not previously observed in plants. Furthermore, several of the phosphorylation motifs, including LxKxxs and RxxSxxxs, have yet to be reported as kinase specificities for in vivo substrates in any species, to our knowledge. Multiple sites of phosphorylation were identified on several key proteins involved in initiating rhizobial symbiosis, including SICKLE, NUCLEOPORIN133, and INTERACTING PROTEIN OF DMI3. Finally, we used these data to create an open-access online database for M. truncatula phosphoproteomic data.
Medicago truncatula has become a model for studying the biology of leguminous plants such as soybean (Glycine max), alfalfa (Medicago sativa), and clover (Trifolium spp.; Singh et al., 2007). Most members of this vast family have the ability to fix atmospheric nitrogen by virtue of an endosymbiotic association with rhizobial bacteria, through which legumes undergo nodulation, the process of forming root nodules (Jones et al., 2007). Legumes are central to modern agriculture and civilization because of their ability to grow in nitrogen-depleted soils and replenish nitrogen through crop rotation. Consequently, there is great interest in understanding the molecular events that allow legumes to recognize their symbionts, develop root nodules, and fix nitrogen. Nod factors are lipochitooligosaccharidic signals secreted by the rhizobia and are required, in most legumes, for intracellular infection and nodule development. In recent decades, an elegant combination of genetics, biochemistry, and cell biology has shown that Nod factors activate intricate signaling events within cells of legume roots, including protein phosphorylation cascades and intracellular ion fluxes (Oldroyd and Downie, 2008).
Protein phosphorylation is a central mechanism of signal transfer in cells (Laugesen et al., 2006; Peck, 2006; Huber, 2007). Several characterized protein kinases are required for symbiosis signal transduction in M. truncatula roots (Lévy et al., 2004; Yoshida and Parniske, 2005; Smit et al., 2007). A recent antibody-based study of cultured M. truncatula cells observed protein phosphorylation changes at the proteomic level in response to fungal infection (Trapphoff et al., 2009); however, the target residues of the phosphorylation events were not determined. A variety of studies have determined in vitro phosphorylation sites on legume proteins and demonstrated the biological importance of the target residues by mutagenesis (Yoshida and Parniske, 2005; Arrighi et al., 2006; Lima et al., 2006; Miyahara et al., 2008; Yano et al., 2008). To our knowledge, only six sites of in vivo protein phosphorylation have been detected for M. truncatula (Laugesen et al., 2006; Lima et al., 2006; Wienkoop et al., 2008), demonstrating the need for the identification of endogenous protein phosphorylation sites in legume model organisms on a proteome-wide scale.
While considerable advancements have been made in the global analysis of protein phosphorylation (Nita-Lazar et al., 2008; Macek et al., 2009; Piggee, 2009; Thingholm et al., 2009), phosphoproteomics in plants has lagged years behind that of the mammalian systems (Kersten et al., 2006, 2009; Peck, 2006), which have more fully sequenced genomes and better annotated protein predictions. Arabidopsis (Arabidopsis thaliana), the first plant genome sequenced (Arabidopsis Genome Initiative, 2000), is now predicted to have over 1,000 protein kinases (Finn et al., 2008), approximately twice as many as in human (Manning et al., 2002). Because many of the kinases in the commonly studied mammalian systems are not conserved in the plant kingdom, there is significant need for large-scale phosphoproteomic technologies to discern the intricacies of phosphorylation-mediated cell signaling in plants. With the high mass accuracy afforded by the linear ion trap-orbitrap hybrid mass spectrometer (Makarov et al., 2006; Yates et al., 2006), recent studies in Arabidopsis have reported 2,597 phosphopeptides from suspension cell culture (Sugiyama et al., 2008) and 3,029 phosphopeptides from seedlings (Reiland et al., 2009).
All previous large-scale plant phosphoproteomic studies have relied solely on collision-activated dissociation (CAD) during tandem mass spectrometry (MS/MS) and have not taken advantage of the more recently developed methods (Kersten et al., 2009) electron capture dissociation (Kelleher et al., 1999) or electron transfer dissociation (ETD; Coon et al., 2004; Syka et al., 2004). Mapping sites of posttranslational modifications, such as phosphorylation, is often more straightforward using electron-based fragmentation methods, as they frequently produce a full spectrum of sequence-informative ions without causing neutral loss of the modifying functional groups (Meng et al., 2005; Chi et al., 2007; Khidekel et al., 2007; Molina et al., 2007; Wiesner et al., 2008; Chalkley et al., 2009; Swaney et al., 2009). With an ETD-enabled hybrid orbitrap mass spectrometer (McAlister et al., 2007, 2008), we previously compared the performance of CAD and ETD tandem MS for large-scale identification of phosphopeptides (Swaney et al., 2009). ETD identified a greater percentage of unique phosphopeptides and more frequently localized phosphorylation sites. Still, the low overlap of identified phosphopeptides indicates that the two methods are highly complementary. With this in mind, we recently developed a decision tree-driven tandem MS algorithm to select the optimal fragmentation method for each precursor (Swaney et al., 2008).
Here, we utilize this technology to map sites of in vivo protein phosphorylation in roots of M. truncatula Jemalong A17 plants. Phosphoproteins, from both whole-cell lysate and membrane-enriched fractions, were analyzed after digestion with a variety of different enzymes individually. Utilizing the complementary fragmentation methods of ETD and CAD, we report 3,404 nonredundant phosphorylation sites at an estimated false discovery rate (FDR) of 1%. Analysis of these data revealed several phosphorylation motifs not previously observed in plants. The phosphorylation sites identified provide insight into the potential regulation of key proteins involved in rhizobial symbiosis, potential consensus sequences by which kinases recognize their substrates, and critical phosphorylation events that are conserved between plant species.
RESULTS
Phosphopeptide Identifications
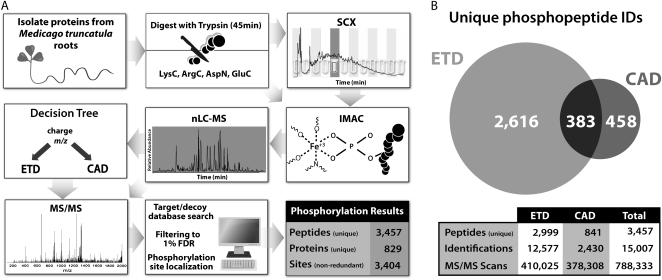
Proteins from M. truncatula Jemalong A17 roots were isolated and analyzed as depicted in Figure 1A, resulting in the identification of 3,457 unique phosphopeptides spanning 829 proteins with 3,404 nonredundant sites of phosphorylation. Proteins, either from whole-cell lysate or membrane-enriched fractions, were isolated from the roots of plants grown in aeroponic conditions and digested using a variety of enzymes individually. Peptides were generated from limited tryptic digestion for 45 min or digestion with ArgC, AspN, GluC, or LysC as indicated. The trypsin-digested peptides were separated by strong cation-exchange chromatography (SCX). For each experiment, the peptides were subjected to immobilized metal affinity chromatography (IMAC) for phosphopeptide enrichment (Ficarro et al., 2002; Ndassa et al., 2006), followed by nanoflow reverse-phase liquid chromatography (LC)-MS/MS using an ETD-enabled hybrid linear ion trap-orbitrap mass spectrometer (McAlister et al., 2007, 2008). After obtaining MS1 survey scans in the orbitrap, data-dependent MS/MS scans utilizing either ETD or CAD, in a decision tree-driven fashion (Swaney et al., 2008) for most analyses, were performed in the ion trap (several analyses of LysC-generated peptides used ETD only). The tandem MS spectra were searched using the Open Mass Spectrometry Search Algorithm (OMSSA; Geer et al., 2004) against a concatenated target-decoy database consisting of M. truncatula protein sequences. The results were filtered to a 1% FDR at the unique peptide level (Elias and Gygi, 2007), with 93% of all identified peptides containing one or more phosphoryl modifications. The phosphorylation sites were validated using custom software (Swaney et al., 2009), the phosphopeptides were grouped to parsimonious protein groups, and the resulting protein list was filtered to 1% FDR. Confident phosphopeptide identifications were produced from 15,007 MS/MS spectra generated by both ETD and CAD (Fig. 1B).
Figure 1.
M. truncatula phosphoproteomic analysis work flow and summary of identified phosphopeptides. A, Root proteins isolated from plants grown under aeroponic conditions were subjected to limited digestion (45 min) with trypsin, and the resulting peptides were separated by SCX. Additional aliquots of protein were digested with ArgC, AspN, GluC, or LysC (and not subjected to SCX). The samples were enriched for phosphopeptides via IMAC and subjected to reverse-phase nano-LC-MS/MS using both ETD and CAD on an ETD-enabled orbitrap mass spectrometer. OMSSA was used to search the data against a concatenated target-decoy M. truncatula database, and the unique peptide identifications were filtered to 1% FDR, resulting in the indicated identifications. B, The Venn diagram indicates the distribution of unique phosphopeptides sequenced by ETD and CAD, with the overlap shown.
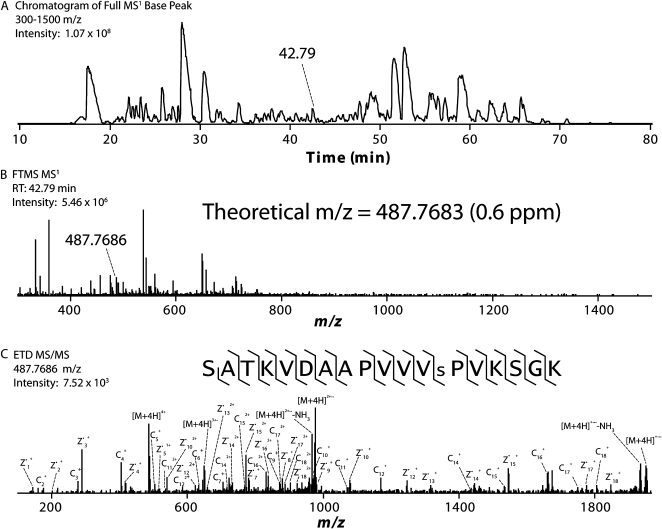
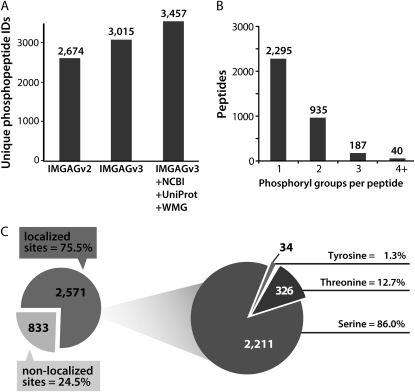
A representative chromatogram, with corresponding spectra from one MS1 scan and one MS2 scan, is displayed in Figure 2. The complexity of the IMAC-eluted peptide mixture is evident from the MS1 chromatogram indicated (Fig. 2A). An example peptide having an observed mass-to-charge ratio (m/z) value of 487.7686 is indicated in the MS1 spectrum (Fig. 2B), which is within 0.6 ppm of the theoretical m/z value for the peptide that was ultimately identified from the ETD-tandem MS scan. The corresponding MS2 spectrum (Fig. 2C) contains 31 out of 32 theoretical fragment ions, enabling localization of the phosphoryl group to the indicated Ser. The number of unique phosphopeptides identified from all the MS/MS data collected was maximized by searching against a protein database containing M. truncatula sequences from a variety of sources (Fig. 3A). Including other legume species besides M. truncatula did not significantly increase the number of confident identifications (data not shown). Of the 3,457 unique phosphopeptides identified to 1% FDR, the majority (66.4%) contained a single phosphoryl group (Fig. 3B). Some multiply phosphorylated peptides, however, were also identified: 27.0% contained two, 5.4% contained three, and 1.1% contained four or more phosphoryl groups. Consistent with previous reports of increased localization efficiency with ETD, 87.4% of the phosphopeptides identified following ETD had all their phosphoryl groups localized, compared with 79.1% for CAD and 86.1% overall (Swaney et al., 2009). The phosphoryl groups on all the identified peptides span 3,404 nonredundant sites in the M. truncatula proteome, with the localization of 2,572 (75.5%) of these sites confirmed (Fig. 3C). Of all the residues observed as nonredundant sites of localized in vivo phosphorylation, 86.0% were Ser, 12.7% were Thr, and 1.3% were Tyr.
Figure 2.
Representative nano-LC-MS/MS data utilized for identifying M. truncatula phosphopeptides. A, The chromatogram shown represents MS1 base-peak signal intensities throughout a decision tree-driven nano-LC-MS/MS analysis of SCX/IMAC-enriched phosphopeptides (partial tryptic digest of proteins from whole cell lysate). B, The MS1 spectrum is from a high-resolution Fourier transform MS (FTMS) scan of peptides eluting at approximately 43 min. The peptide of m/z value 487.7686 (charge state +4) was selected for fragmentation by ETD. C, The MS2 spectrum was produced from a subsequent ion-trap MS/MS scan. Database searching and FDR analysis identified the peptide sequence shown, with a theoretical mass that is within 0.6 ppm of the precursor selected for MS/MS. Using in-house-written software, the site of phosphorylation was localized to the Ser indicated by the lowercase s. Manual validation of fragment ions shown is in agreement with the automated analysis (for Microsoft Excel spreadsheet, see Supplemental Table S1), matching 31 out of 32 possible fragment ions observed.
Figure 3.
Phosphoryl modifications at the peptide and proteome levels. A, In-house software was used to create a nonredundant protein database using M. truncatula protein sequences from a number of different sources: the International Medicago Genome Annotation Group (IMGAG; versions 2.0 and 3.0), the National Center for Biotechnology Information (NCBI), the Universal Protein Resource (UniProt), and the Wisconsin Medicago Group (WMG). The effect of including sequences from the various sources on the unique phosphopeptides identified is depicted. Including other legume species besides M. truncatula did not significantly increase the number of confident identifications (data not shown). B, The distribution of the number of phosphoryl groups per peptide is shown for all 3,457 unique phosphopeptides. C, The localization efficiency for the 3,404 nonredundant phosphorylation sites identified in the M. truncatula proteome using in-house-written software is indicated. The distribution of phosphoryl groups on Ser, Thr, and Tyr was assessed for all nonredundant localized phosphorylation sites.
Species-Specific Phosphorylation Site Motif Analysis
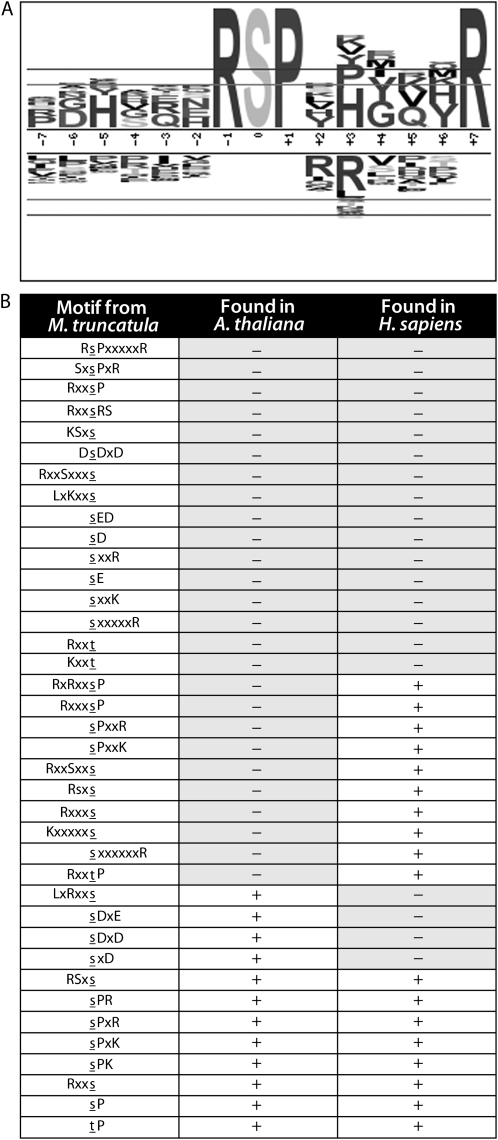
To determine potential consensus sequences for M. truncatula kinases, motif analysis of the identified phosphopeptides was performed using a prerelease of version 2.0 of motif-x (Schwartz and Gygi, 2005). The analysis was run at a high significance threshold (P < 0.0005 following Bonferroni correction) and restricted to the phosphorylation sites for which localization was confirmed, resulting in the identification of 38 phosphorylation motifs containing at least one fixed position aside from the central phosphorylated residue. A probability log-based logo (pLOGo), representing the statistical significance of one identified motif, is shown in Figure 4A (for pLOGos for all M. truncatula phosphorylation motifs, see Supplemental Fig. S1). All 38 of the phosphorylation motifs identified in M. truncatula are presented in Figure 4B. Since an occurrence threshold of 20 was used in the analysis, each motif is represented by a minimum of 20 unique localized phosphorylation sites in the large-scale M. truncatula phosphopeptide data set.
Figure 4.
M. truncatula phosphorylation motif analysis. A, A pLOGo, representing the statistical significance of one motif identified by performing phosphorylation motif analysis using version 2.0 of motif-x (for pLOGos for all motifs identified, see Supplemental Fig. S1). The taller a residue is, the more statistically significant it is. Residues over the midline are overrepresented, while those below the midline are underrepresented. The residues closest to the midline are the most significant. The inner horizontal line represents the 0.01 statistical significance threshold (following Bonferroni correction), and the outer line represents the significance threshold used to fix positions within the motif. B, All phosphorylation motifs identified in M. truncatula and their overlap with those present in data sets from previous studies are shown (Heazlewood et al., 2008; Swaney et al., 2009). The localized phosphorylated residue is indicated in lowercase underlined letters (s or t), and x indicates any amino acid.
To investigate the specificity of the identified motifs to legumes, parallel analysis was performed for a nonlegume plant species with data obtained from the PhosPhAt 2.2 database of published Arabidopsis phosphopeptides (Heazlewood et al., 2008). Of the 38 phosphorylation motifs in the M. truncatula data set described herein, 26 were not present in the Arabidopsis data. In addition to the biological differences between M. truncatula and Arabidopsis, some of the discrepancy in the motifs observed could be because the Arabidopsis phosphopeptides are from studies that did not use ETD. For instance, in our previous study using both ETD and CAD for MS/MS phosphoproteomic analysis of human cells, we found that ETD identified novel phosphorylation motifs that were basophilic in nature and not detected by CAD (Swaney et al., 2009), consistent with the compatibility of ETD with peptides of high charge density (Swaney et al., 2007). With that in mind, we also compared the overlap in motifs with those present in our previously published human phosphopeptide data set generated using both ETD and CAD (Swaney et al., 2009). Interestingly, all of the M. truncatula motifs that were not found in Arabidopsis but were found in human contained a basic residue, consistent with the notion that these motifs are missing from the Arabidopsis data sets because ETD was not used in these previous plant studies (and not necessarily due to biological differences between M. truncatula and Arabidopsis). Twenty of the M. truncatula motifs were not present in the human data set, and 16 were completely unique to M. truncatula.
Phosphoprotein Assessment
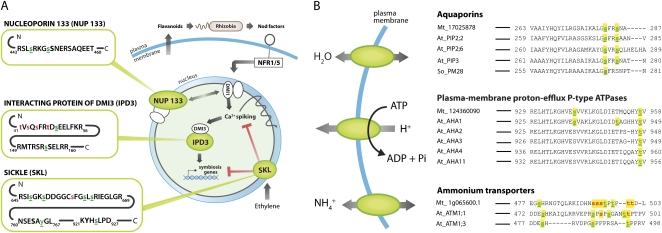
Phosphopeptide identifications were sorted into 829 parsimonious protein groups (Supplemental Table S1) or sets of proteins identified by the exact same collection of phosphopeptides. Interestingly, multiple phosphorylation sites were identified on several proteins with key roles in legume/rhizobia symbiosis. Figure 5A displays the sites of phosphorylation on the symbiotic proteins SICKLE (SKL), INTERACTING PROTEIN OF DMI3 (IPD3), and NUCLEOPORIN133 (NUP133). The role of the identified phosphoproteins in nodulation signal transduction is depicted. Phosphorylation sites are indicated by a lowercase s, t, or y, with localized sites of phosphorylation shown in underlined green lettering and nonlocalized sites from ambiguous positional isomers indicated by red lettering without underlining. A phosphorylation site is considered localized if a potential positional isomer with a phosphoryl group in the corresponding position has the greatest number of theoretical fragments that match to ions observed in a given tandem MS spectrum when compared with all other possible positional isomers. Several of the M. truncatula protein phosphorylation motifs (Fig. 5) are present on the nodulation signaling proteins highlighted in Figure 5A. IPD3 contains localized phosphorylation sites fitting the motifs RSxs, Rxxs, sE, and sxxxxxR; NUP133 contains LxRxxs, RSxs, RxxSxxxs, and Rxxs; and RSxs, Rxxs, sD, RxxSxxs, sxD, sxxxxxxR, and sxxR are present on SKL.
Figure 5.
Phosphorylation sites on key proteins for symbiosis in legumes and conserved functions in plants. Localized (green, underlined) and nonlocalized (red) phosphorylation sites are shown in lowercase letters on proteins whose cellular functions are depicted. A, Phosphorylation sites are indicated for the nodulation signaling proteins NUP133, IPD3, and SKL. B, A plasma-membrane proton-efflux P-type ATPase (accession no. gi|124360090), an aquaporin (accession no. gi|17025878), and an Rh-like ammonium transporter (accession no. IMGA|Medtr1g065600.1), which were identified as membrane phosphoproteins in M. truncatula (Mt_), were aligned with homologous phosphoproteins reported in the PhosPhAt 2.2 database of published Arabidopsis (At_) phosphopeptides (Heazlewood et al., 2008) and a previous spinach (So_) study (Johansson et al., 1998). A phosphorylation site detected by non-MS methods is shown in blue.
Membrane phosphoproteins identified included two aquaporins (gi|17025878 and gi|14538013), members of the plasma membrane proton-efflux P-type ATPase family (gi|124360090 and IMGA|Medtr8g129390.1), and an Rh-like protein/ammonium transporter (IMGA|Medtr1g065600.1), all of which play central roles in the biology of plants (legumes and nonlegumes; Ludewig et al., 2007; Uehlein et al., 2007; Duby and Boutry, 2009) as well as species in other kingdoms. Figure 5B illustrates the identified sites of phosphorylation on one of each of these classes of membrane proteins and shows their alignment with sequences from homologous phosphoproteins in other plant species. The P-type ATPase indicated (gi|124360090) was phosphorylated on its penultimate Thr of the C terminus, phosphorylation of which is required for activation of enzymatic activity in Arabidopsis orthologs (Robertson et al., 2004). For the M. truncatula aquaporin shown (gi|17025878), phosphorylation of one identified Ser near the C terminus (Ser-285) controls water transport activity in spinach (Spinacia oleracea; Johansson et al., 1998) and targeting to the plasma membrane in Arabidopsis (Prak et al., 2008). Another phosphoryl group was localized to Ser-282, a conserved site previously reported in Arabidopsis (Prak et al., 2008). The Rh-like ammonium transporter (IMGA|Medtr1g065600.1) was multiply phosphorylated on its cytosolic C-terminal region, a domain that regulates ammonium transport in response to phosphorylation in Arabidopsis (Loque et al., 2007).
All unique phosphopeptides and phosphoproteins, identified at 1% FDR, are listed in Supplemental Table S1 With these data, we have created an online database for depositing and viewing M. truncatula phosphoproteomic data (http://www.phospho.medicago.wisc.edu). Search functions allow users to find specific proteins of interest or all proteins within certain phosphorylation motifs. The online database utilizes the same scheme for localized (green, underlined) and nonlocalized (red, no underlining) sites of phosphorylation demonstrated in Figure 5. In addition, a summary of the pertinent MS data leading to the identification of each specific phosphorylation site can be directly accessed by selecting the residue of interest.
DISCUSSION
Recent advancements in proteomic technologies provide the ability to discern sites of in vivo protein phosphorylation events in plant tissue in a high-throughput manner. By utilizing decision tree-driven tandem MS (Swaney et al., 2008), employing the complementary dissociation methods ETD and CAD, we have identified 3,457 unique phosphopeptides (at 1% FDR) in M. truncatula Jemalong A17 roots (Fig. 1). The phosphopeptides identified map to 3,404 nonredundant sites of in vivo phosphorylation on 829 parsimonious protein groups at 1% FDR. Localization was confirmed for 75.5% of the phosphorylation sites identified using custom software (Fig. 3C), in agreement with manual validation (Fig. 2). The distribution of Ser, Thr, and Tyr phosphorylation observed for localized phosphorylation sites (Fig. 3C) is generally consistent with a previous large-scale analysis of Arabidopsis (Sugiyama et al., 2008). Of note, however, our data indicate the relative abundance of Tyr phosphorylation as 1.3% in Medicago compared with 4.3% in the Arabidopsis study (Sugiyama et al., 2008). Regardless of whether this discrepancy is from differences in biology or methodology, both studies support the notion that Tyr phosphorylation in plants is more abundant than once thought (de la Fuente van Bentem and Hirt, 2009).
Three of the phosphoproteins identified have roles in nodulation (Fig. 5A). Two localized phosphorylation sites and four nonlocalized (potential) phosphorylation sites were identified on IPD3, a protein that interacts with the calcium/calmodulin-dependent protein kinase (CCaMK) DMI3 (for DOESN'T MAKE INFECTIONS3) of M. truncatula. The Lotus japonicus ortholog of IPD3, named CYCLOPS, is required for intracellular infection and phosphorylated by the L. japonicus CCaMK (Messinese et al., 2007; Yano et al., 2008); however, the exact phosphorylation sites of CYCLOPS by CCaMK are still unknown. The nonlocalized (potential) phosphorylation sites at Ser-43 and the localized phosphorylation site at Ser-50 on IPD3 are conserved in CYCLOPS. While the exact residues on CYCLOPS that are targeted by CCaMK are unknown, Ser-43 and Ser-50 reside in the domain of CYCLOPS that is phosphorylated by CCaMK. Thus, these sites are strong candidate targets of DMI3/CCaMK (Yano et al., 2008). The phosphoryl group localized to Ser-155 of IPD3 may represent a phosphorylation event by another, yet-to-be characterized protein kinase, as the corresponding position of this residue on CYCLOPS is outside the domain targeted by CCaMK. Two localized sites of phosphorylation were identified on NUP133, a protein required for nodule development and perinuclear calcium spiking in L. japonicus (Kanamori et al., 2006). Six localized phosphorylation sites and one nonlocalized (potential) site of phosphorylation were identified on the protein SKL, an ortholog of Arabidopsis EIN2, which is required for ethylene signaling and is a negative regulator of nodule formation (Prayitno et al., 2006; Penmetsa et al., 2008). Phosphorylation of NUP133 and SKL has not been reported in legumes.
Motif analysis revealed 26 phosphorylation motifs not extracted from sequences contained in the PhosPhAt 2.2 database of published Arabidopsis phosphopeptides under identical motif-x parameters (Fig. 4; Heazlewood et al., 2008). Since 10 of these motifs were extracted from the human data set of our previous study using both ETD and CAD (Swaney et al., 2009), the absence of enrichment of these motifs in the Arabidopsis data set is likely due to differences in the phosphoproteomic methodology rather than differences in biology between the two plants. However, 16 of the extracted motifs were completely unique to the M. truncatula data set, suggesting that they may potentially represent phosphorylation events between evolutionarily distinct kinase-substrate pairs. Aside from the relatively nonspecific acidophilic and basophilic motifs extracted (e.g. sD, sE, sxxK, sxxR), analysis of the motifs unique to M. truncatula revealed several interesting motif classes, including two motifs bearing “RS” signatures (RsPxxxxxR and RxxsRS). While inspection of the M. truncatula proteins with phosphorylation sites fitting these motifs indicated a wide variety of proteins involved in splicing and RNA binding, consistent with the characterized role of phosphorylated RS domain-containing proteins in both metazoans and plants (de la Fuente van Bentem et al., 2006; Nikolakaki et al., 2008), the motifs extracted here may provide insight into the yet undetermined specificity of the kinase(s) responsible for phosphorylating these factors. Furthermore, a large number of DNA-binding proteins were also detected among the RS motif-containing proteins, raising the possibility that these motifs are regulators of nucleotide binding more generally. Another class of motifs that were uniquely enriched in the M. truncatula data set contained hydrophobic residues at the –5 position and basic residues at the –3 position, including LxKxxs, (L)xRxxt, (LM)xKxxt, and (L)xKSxs (where residues in parentheses are readily observable from the pLOGos in Supplemental Fig. S1). Although the general motif core, LxRxxs, has been associated with several protein kinases, including PKD, MAPKAPK2, and Chk2 (Stokoe et al., 1993; Nishikawa et al., 1997; O'Neill et al., 2002), to our knowledge the importance of Lys as a conservative amino acid substitution at the –3 position has yet to be reported in in vivo substrates. Thus, the motif, ψx[KR]xx[st] (where ψ denotes any hydrophobic residue) likely represents the specificity of a kinase (or kinases) with significant activity in M. truncatula root cells. Finally, in addition to not being present in the list of motifs extracted from the PhosPhAt database and our previous human study, the motifs RxxSxxxs and (R)xxSxsP(MV)R (Supplemental Fig. S1) bear unique specificity determinants not previously observed in any large-scale study of plants, fungi, or animals (Reiland et al., 2009; Schwartz et al., 2009).
CONCLUSION
We have used IMAC-phosphopeptide enrichment and decision tree-driven MS/MS to identify 3,404 nonredundant sites of in vivo phosphorylation in M. truncatula Jemalong A17 roots. As this is, to our knowledge, the first large-scale plant phosphoproteomic study to utilize ETD, comparison of this data set and previously published work from other species has revealed phosphorylation motifs not previously observed in plants. Furthermore, multiple sites of phosphorylation were identified on several key proteins involved in initiating rhizobial symbiosis, including SKL, NUP133, and IPD3. We have created an open-access database (http://www.phospho.medicago.wisc.edu) to help make this, and future large-scale M. truncatula proteomic data sets, easily accessible. The protein phosphorylation sites identified provide insight into cell signaling in M. truncatula roots as well as plants in general. In addition, the novel plant phosphorylation motifs could lead to future discovery of novel kinase-substrate pairs. For any kinase (from M. truncatula or other plant species) found to phosphorylate a substrate protein on one of the identified motifs, other proteins with sequences containing the same motifs could be considered candidate substrates.
MATERIALS AND METHODS
Plant Growth, Protein Isolation, Digestion, Peptide Fractionation, and Phosphopeptide Enrichment
Seeds of Medicago truncatula Jemalong A17 were harvested, scarified, and germinated (Garcia et al., 2006). The seedlings were placed on an aeroponic system (Barker et al., 2006) and grown in nitrogen-free modified Fahraeus medium (Catoira et al., 2000) for 14 to 15 d at 22°C and 24 h of 130 to 200 μmol m−2 s−1 light. The roots, however, grew in the dark. Proteins were isolated from whole-cell lysate of root tissue or membrane-enriched fractions by previously described procedures (Gallardo et al., 2006; Lefebvre et al., 2007) modified for phosphoproteomics with the addition of a variety of phosphatase inhibitors. A 10-mg protein sample was subjected to limited digestion with trypsin for 45 min, and the resulting peptides were desalted and separated by SCX (Swaney et al., 2009). In separate experiments, 2.5-mg protein samples were digested with LysC, AspN, GluC, or ArgC, and the resulting peptides were desalted. The peptides were enriched for phosphopeptides by IMAC (Swaney et al., 2009). Further details are provided in Supplemental Materials and Methods S1.
Nano-HPLC, MS, Phosphosite Localization, Motif Analysis, and Sequence Alignment
Nano-LC-MS/MS analysis was performed on an ETD-enabled LTQ Orbitrap (Thermo Fisher Scientific; McAlister et al., 2007). While a phosphopeptide-optimized decision tree algorithm employing both ETD and CAD was utilized for most of the runs (Swaney et al., 2008), several nano-LC-MS/MS analyses for the LysC-generated peptides utilized only ETD. OMSSA was used to search the tandem MS data against a concatenated target-decoy database (Elias and Gygi, 2007) consisting of M. truncatula protein sequences from a variety of sources (Fig. 3A). FDR was determined using software written in house, which determined an optimal precursor mass accuracy threshold of ±2 ppm after correcting for systematic mass error (i.e. drift; Elias and Gygi, 2007; Swaney et al., 2009). The localization software, termed Phosphinator, compared each OMSSA-identified phosphopeptide sequence with the corresponding tandem MS spectrum in the Thermo Scientific LC-MS/MS raw data file. Phosphinator then determined whether the phosphoryl group could be localized. Motif analysis for data sets from M. truncatula, human (Swaney et al., 2009), and all Arabidopsis (Arabidopsis thaliana) peptides from the PhosPhAt 2.2 database (Heazlewood et al., 2008) was performed using version 2.0 of motif-x (Schwartz and Gygi, 2005). Sequence alignments were performed using version 5.05 of T-COFFEE (Poirot et al., 2003). Further details are provided in Supplemental Materials and Methods S1.
Database Development
All phosphopeptides identified at 1% FDR (Supplemental Table S1) were deposited into a newly created open-access M. truncatula phosphoproteomic database that can be accessed at http://www.phospho.medicago.wisc.edu. The phosphopeptides are organized by the proteins to which they map, with accession numbers indicated. Localized phosphorylation sites are indicated by underlined green lowercase letters (s, t, or y), whereas nonlocalized sites from ambiguous positional isomers are indicated with red lowercase letters without underlining (s, t, or y).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. All pLOGos of motifs extracted using motif-x version 2.0.
Supplemental Table S1. All phosphopeptides identified in M. truncatula roots.
Supplemental Materials and Methods S1. Sample preparation, data collection, and data analysis.
Supplementary Material
Acknowledgments
We gratefully acknowledge Graeme McAlister for helpful instruction on the instrumentation methodology utilized; Maegen Howes-Podoll and Heather Burch for assistance with plant growth and membrane purification; Gheorghe Craciun and Yuerong Zhu for Web site development; and Ryan Lynch and A.J. Bureta for figure preparation. We thank the International Medicago Genome Annotation Group for early access to the Mtr 3.0 gene predictions.
This work was supported by the National Science Foundation (grant nos. 0701846 and 0747990), the National Institutes of Health (grant no. R01GM080148), the Beckman Foundation, and Eli Lilly and Company.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Joshua J. Coon (jcoon@chem.wisc.edu).
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Gherardi M, Huguet T, et al (2006) The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DG, Pfaff T, Moreau D, Groves E, Ruffel S, Lepetit M, Whitehand S, Maillet F, Nair RM, Journet EP (2006) Growing M. truncatula: choice of substrates and growth conditions. In U Mathesius, ed, The Medicago truncatula Handbook. The Samuel Roberts Noble Foundation, Ardmore, OK, http://www.noble.org/MedicagoHandbook/
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci USA 106 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA 104 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF (2004) Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. Int J Mass Spectrom 236 33–42 [Google Scholar]
- de la Fuente van Bentem S, Anrather D, Roitinger E, Djamei A, Hufnagl T, Barta A, Csaszar E, Dohnal I, Lecourieux D, Hirt H (2006) Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res 34 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Hirt H (2009) Protein tyrosine phosphorylation in plants: more abundant than expected? Trends Plant Sci 14 71–76 [DOI] [PubMed] [Google Scholar]
- Duby G, Boutry M (2009) The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Arch 457 645–655 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4 207–214 [DOI] [PubMed] [Google Scholar]
- Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 20 301–305 [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al (2008) The Pfam protein families database. Nucleic Acids Res 36 D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Lesignor C, Darmency M, Burstin J, Thompson R, Rochat C, Boutin J, Kuester H, Buitink J, Leprince O, et al (2006) Seed biology of Medicago truncatula. In U Mathesius, ed, The Medicago truncatula Handbook. The Samuel Roberts Noble Foundation, Ardmore, OK, http://www.noble.org/MedicagoHandbook/
- Garcia J, Barker DG, Journet EP (2006) Seed storage and germination. In U Mathesius, ed, The Medicago truncatula Handbook. The Samuel Roberts Noble Foundation, Ardmore, OK, http://www.noble.org/MedicagoHandbook/
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH (2004) Open mass spectrometry search algorithm. J Proteome Res 3 958–964 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX (2008) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36 D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC (2007) Exploring the role of protein phosphorylation in plants: from signalling to metabolism. Biochem Soc Trans 35 28–32 [DOI] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Shukla VK, Chrispeels MJ, Larsson C, Kjellbom P (1998) Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell 10 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher NL, Zubarev RA, Bush K, Furie B, Furie BC, McLafferty FW, Walsh CT (1999) Localization of labile posttranslational modifications by electron capture dissociation: the case of gamma-carboxyglutamic acid. Anal Chem 71 4250–4253 [DOI] [PubMed] [Google Scholar]
- Kersten B, Agrawal GK, Durek P, Neigenfind J, Schulze W, Walther D, Rakwal R (2009) Plant phosphoproteomics: an update. Proteomics 9 964–988 [DOI] [PubMed] [Google Scholar]
- Kersten B, Agrawal GK, Iwahashi H, Rakwal R (2006) Plant phosphoproteomics: a long road ahead. Proteomics 6 5517–5528 [DOI] [PubMed] [Google Scholar]
- Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol 3 339–348 [DOI] [PubMed] [Google Scholar]
- Laugesen S, Messinese E, Hem S, Pichereaux C, Grat S, Ranjeva R, Rossignol M, Bono JJ (2006) Phosphoproteins analysis in plants: a proteomic approach. Phytochemistry 67 2208–2214 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Furt F, Hartmann MA, Michaelson LV, Carde JP, Sargueil-Boiron F, Rossignol M, Napier JA, Cullimore J, Bessoule JJ, et al (2007) Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol 144 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 [DOI] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H (2006) Phosphorylation and subsequent interaction with 14-3-3 proteins regulate plastid glutamine synthetase in Medicago truncatula. Planta 223 558–567 [DOI] [PubMed] [Google Scholar]
- Loque D, Lalonde S, Looger LL, von Wiren N, Frommer WB (2007) A cytosolic trans-activation domain essential for ammonium uptake. Nature 446 195–198 [DOI] [PubMed] [Google Scholar]
- Ludewig U, Neuhauser B, Dynowski M (2007) Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett 581 2301–2308 [DOI] [PubMed] [Google Scholar]
- Macek B, Mann M, Olsen JV (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol 49 199–221 [DOI] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S (2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 78 2113–2120 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298 1912–1934 [DOI] [PubMed] [Google Scholar]
- McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JE, Zabrouskov V, et al (2008) A proteomics grade electron transfer dissociation-enabled hybrid linear ion trap-orbitrap mass spectrometer. J Proteome Res 7 3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ (2007) Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem 79 3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Forbes AJ, Miller LM, Kelleher NL (2005) Detection and localization of protein modifications by high resolution tandem mass spectrometry. Mass Spectrom Rev 24 126–134 [DOI] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rouge P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20 912–921 [DOI] [PubMed] [Google Scholar]
- Miyahara A, Hirani TA, Oakes M, Kereszt A, Kobe B, Djordjevic MA, Gresshoff PM (2008) Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. J Biol Chem 283 25381–25391 [DOI] [PubMed] [Google Scholar]
- Molina H, Horn DM, Tang N, Mathivanan S, Pandey A (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA 104 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndassa YM, Orsi C, Marto JA, Chen S, Ross MM (2006) Improved immobilized metal affinity chromatography for large-scale phosphoproteomics applications. J Proteome Res 5 2789–2799 [DOI] [PubMed] [Google Scholar]
- Nikolakaki E, Drosou V, Sanidas I, Peidis P, Papamarcaki T, Iakoucheva LM, Giannakouros T (2008) RNA association or phosphorylation of the RS domain prevents aggregation of RS domain-containing proteins. Biochim Biophys Acta 1780 214–225 [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem 272 952–960 [DOI] [PubMed] [Google Scholar]
- Nita-Lazar A, Saito-Benz H, White FM (2008) Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics 8 4433–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59 519–546 [DOI] [PubMed] [Google Scholar]
- O'Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA (2002) Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem 277 16102–16115 [DOI] [PubMed] [Google Scholar]
- Peck SC (2006) Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J 45 512–522 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, Lichtenzveig J, Gish JC, Nam YW, Engstrom E, Xu K, Sckisel G, Pereira M, et al (2008) The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J 55 580–595 [DOI] [PubMed] [Google Scholar]
- Piggee C (2009) Phosphoproteomics: miles to go before it's routine. Anal Chem 81 2418–2420 [DOI] [PubMed] [Google Scholar]
- Poirot O, O'Toole E, Notredame C (2003) Tcoffee@igs: a Web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res 31 3503–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V (2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7 1019–1030 [DOI] [PubMed] [Google Scholar]
- Prayitno J, Rolfe BG, Mathesius U (2006) The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol 142 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson WR, Clark K, Young JC, Sussman MR (2004) An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 168 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Chou MF, Church GM (2009) Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol Cell Proteomics 8 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23 1391–1398 [DOI] [PubMed] [Google Scholar]
- Singh RJ, Chung GH, Nelson RL (2007) Landmark research in legumes. Genome 50 525–537 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D, Caudwell B, Cohen PT, Cohen P (1993) The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem J 296 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, McAlister GC, Coon JJ (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods 5 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ (2007) Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem 79 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Thomson JA, Coon JJ (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA 106 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA 101 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Larsen MR (2009) Analytical strategies for phosphoproteomics. Proteomics 9 1451–1468 [DOI] [PubMed] [Google Scholar]
- Trapphoff T, Beutner C, Niehaus K, Colditz F (2009) Induction of distinct defense-associated protein patterns in Aphanomyces euteiches (Oomycota)-elicited and -inoculated Medicago truncatula cell-suspension cultures: a proteome and phosphoproteome approach. Mol Plant Microbe Interact 22 421–436 [DOI] [PubMed] [Google Scholar]
- Uehlein N, Fileschi K, Eckert M, Bienert GP, Bertl A, Kaldenhoff R (2007) Arbuscular mycorrhizal symbiosis and plant aquaporin expression. Phytochemistry 68 122–129 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Larrainzar E, Glinski M, Gonzalez EM, Arrese-Igor C, Weckwerth W (2008) Absolute quantification of Medicago truncatula sucrose synthase isoforms and N-metabolism enzymes in symbiotic root nodules and the detection of novel nodule phosphoproteins by mass spectrometry. J Exp Bot 59 3307–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J, Premsler T, Sickmann A (2008) Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics 8 4466–4483 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Cociorva D, Liao L, Zabrouskov V (2006) Performance of a linear ion trap-orbitrap hybrid for peptide analysis. Anal Chem 78 493–500 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Parniske M (2005) Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J Biol Chem 280 9203–9209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.