Abstract
The color of tomato fruit is mainly determined by carotenoids and flavonoids. Phenotypic analysis of an introgression line (IL) population derived from a cross between Solanum lycopersicum ‘Moneyberg’ and the wild species Solanum chmielewskii revealed three ILs with a pink fruit color. These lines had a homozygous S. chmielewskii introgression on the short arm of chromosome 1, consistent with the position of the y (yellow) mutation known to result in colorless epidermis, and hence pink-colored fruit, when combined with a red flesh. Metabolic analysis showed that pink fruit lack the ripening-dependent accumulation of the yellow-colored flavonoid naringenin chalcone in the fruit peel, while carotenoid levels are not affected. The expression of all genes encoding biosynthetic enzymes involved in the production of the flavonol rutin from naringenin chalcone was down-regulated in pink fruit, suggesting that the candidate gene underlying the pink phenotype encodes a regulatory protein such as a transcription factor rather than a biosynthetic enzyme. Of 26 MYB and basic helix-loop-helix transcription factors putatively involved in regulating transcription of genes in the phenylpropanoid and/or flavonoid pathway, only the expression level of the MYB12 gene correlated well with the decrease in the expression of structural flavonoid genes in peel samples of pink- and red-fruited genotypes during ripening. Genetic mapping and segregation analysis showed that MYB12 is located on chromosome 1 and segregates perfectly with the characteristic pink fruit color. Virus-induced gene silencing of SlMYB12 resulted in a decrease in the accumulation of naringenin chalcone, a phenotype consistent with the pink-colored tomato fruit of IL1b. In conclusion, biochemical and molecular data, gene mapping, segregation analysis, and virus-induced gene silencing experiments demonstrate that the MYB12 transcription factor plays an important role in regulating the flavonoid pathway in tomato fruit and suggest strongly that SlMYB12 is a likely candidate for the y mutation.
Plants produce pigments to provide color to their flowers and fruit for attracting pollinators and seed dispersers. In crop plants, color is regarded as one of the most important consumer traits in fruit such as tomato (Solanum lycopersicum), pepper (Capsicum annuum), apple (Malus domestica), and strawberry (Fragaria spp.). In flowers and fruit, these colors are due mainly to carotenoid and flavonoid pigments.
Carotenoids are lipid-soluble 40-carbon isoprenoids, and more than 700 naturally occurring carotenoids have been identified (Britton et al., 2004). Carotenoids are essential for plant life, since they provide important photoprotective functions during photosynthesis in chloroplasts and serve as precursors for the phytohormone abscisic acid (Grotewold, 2006). Beyond their essential biological activities, carotenoids also accumulate in chromoplasts of flowers and fruit, where they function as yellow- to red-colored pigments (Young and Frank, 1996; Tanaka et al., 2008). Carotenoids play important roles in human nutrition and health, since they are the precursors for vitamin A, are lipophilic antioxidants, and have anticancer properties (Basu and Imrhan, 2007; Rao and Rao, 2007; Singh and Goyal, 2008).
Flavonoids are a large group of polyphenolic compounds. Based on their core structure, the aglycone, they can be grouped into different classes: chalcones, flavanones, aurones, flavonols, and anthocyanins (Fig. 1). More than 6,000 different flavonoids have been identified in nature (http://www.metabolome.jp/software/FlavonoidViewer/). This diversity is due to combinatorial modification by decorating enzymes such as glycosyl, malonyl, acyl, and methyl transferases. Besides their role as pigments to attract pollinators and seed dispersers, flavonoids are involved in many other aspects of plant growth and development, such as pollination, pathogen resistance, and protection against damage from ultraviolet light (Harborne, 1986; Harborne and Williams, 2000). Furthermore, they are good hydrophilic antioxidants and are increasingly recognized as health-protecting components in the human diet (Arts and Hollman, 2005; Hooper and Cassidy, 2006; Lotito and Frei, 2006; Fraga, 2007; Tapas et al., 2008). The anthocyanins are the widely dispersed red, purple, and blue pigments in many flowers and fruit, but several other flavonoid classes function as yellow pigments (e.g. chalcones and aurones) or copigments (e.g. flavonols; Davies, 2004; Tanaka et al., 2008).
Figure 1.
Schematic overview of the flavonoid pathway in tomato. PAL, Phe ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; HCT, cinnamoyl-CoA shikimate/quinate transferase; C3H, p-coumaroyl ester 3-hydroxylase; HQT, hydroxycinnamoyl-CoA quinate transferase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone-3-hydroxylase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid-3′5′-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; 3GT, flavonoid-3-O-glucosyltransferase; RT, flavonoid 3-O-glucoside-rhamnosyltransferase; AAC, anthocyanin acyltransferase; 5GT, flavonoid-5-glucosyltransferase.
The red color of ripe tomato fruit is due mainly to the accumulation of the carotenoid all-trans-lycopene, which is produced during fruit ripening. In addition to lycopene, tomato fruit contain significant levels of other carotenoids, such as β-carotene, phytoene, violaxanthin, and lutein. Mutants in the carotenoid pathway have an altered carotenoid composition, resulting in different fruit colors, such as orange (tangerine, beta) or yellow (r) fruit (Lewinsohn et al., 2005).
Besides carotenoids, flavonoids play a role in determining the color of tomato fruit (Schijlen et al., 2008; Bovy et al., 2010). With the exception of specific genotypes (Willits et al., 2005), flavonoids accumulate predominantly in the fruit peel, since the flavonoid pathway is not active in the fruit flesh due to a lack of expression of flavonoid biosynthetic genes in this tissue (Muir et al., 2001; Bovy et al., 2002; Colliver et al., 2002). One of the most abundant flavonoids in tomato fruit peel is the yellow-colored naringenin chalcone. It accumulates in the cuticle upon ripening and is responsible for the yellow color that develops in the peel at breaker stage, preceding the production of lycopene (Hunt and Baker, 1980). In addition, the flavonols quercetin-3-rutinoside (rutin) and kaempferol-3-rutinoside also accumulate in the peel of ripening tomato fruit. Up to 70 different flavonoids have been identified in tomato fruit, mainly consisting of different conjugated forms (i.e. different glycosides) of naringenin chalcone, quercetin, and kaempferol (Moco et al., 2006; Iijima et al., 2008).
Fruit of domesticated tomatoes do not accumulate anthocyanins, due to a lack of expression of the genes required for the formation of these compounds (Bovy et al., 2002). However, several closely related wild species do contain anthocyanins in their fruit, and this trait has recently been introgressed into the cultivated tomato through interspecific crosses with Solanum chilense and Solanum lycopersicoides. The introgressed genes, Anthocyanin fruit (Jones et al., 2003; Mes et al., 2008) and Aubergine (Mes et al., 2008), respectively, led to stimulation of anthocyanin pigmentation in tomato fruit peel (Gonzali et al., 2009).
Expression analysis of genes encoding flavonoid biosynthetic enzymes in red-fruited tomatoes revealed that chalcone synthase (CHS), flavanone-3-hydrolase (F3H), and flavonol synthase (FLS) transcript levels increase during ripening, peak at the turning stage, and then decrease slightly at the red stage. In contrast, the transcript levels of chalcone isomerase (CHI) remain low and even decrease upon ripening (Bovy et al., 2002). These expression data suggest that CHI is a rate-determining step in the production of flavonols in tomato and explain the accumulation of the CHI substrate naringenin chalcone during fruit ripening (Bovy et al., 2002). Indeed, overexpression of the petunia (Petunia hybrida) CHI gene in tomato results in up to 70-fold increase in flavonols in the fruit peel, together with a decrease in naringenin chalcone (Muir et al., 2001). These fruit do not show the characteristic yellow color at breaker/turning stage but rather develop a more dull, pinkish red color during the ripening process (Bovy et al., 2007). A similar change in fruit color was observed in CHS RNA interference fruit, which lack the accumulation of both naringenin chalcone and rutin (Schijlen et al., 2007). These results suggest that naringenin chalcone is required to give tomato its typical orange-red color and that a lack of naringenin chalcone leads to pink-colored fruit.
In Asia, pink-colored tomatoes are very popular for consumption. The pink trait was first described in 1925 in fruit with a transparent epidermis lacking a yellow pigment (Lindstrom, 1925; Fig. 2). Genetic studies revealed that pink fruit result from the monogenic, recessive y (yellow) locus present on chromosome 1, while red-colored fruit have the dominant Y allele (Lindstrom, 1925; Rick and Butler, 1956). The Y gene has not yet been identified or isolated, and its function remains to be elucidated. In this paper, we show that the development of pink tomatoes is due to a ripening-dependent down-regulation of the flavonoid pathway, resulting in a lack of naringenin chalcone accumulation in the fruit peel. Furthermore, we demonstrate that the MYB transcription factor SlMYB12 plays an important role in regulating the production of naringenin chalcone in tomato fruit and provide strong evidence that the pink fruit phenotype is likely caused by a mutation affecting the gene encoding SlMYB12. Our results suggest that this gene is a likely candidate for the Y locus.
Figure 2.
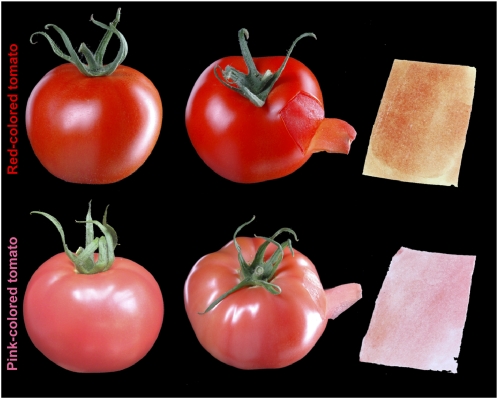
Phenotypic analysis of S. lycopersicum ‘Moneyberg’ (red tomato) and S. chmielewskii IL1b (pink tomato). Whole fruit (left and middle) and peel (right) of red (top) and pink (bottom) ripe tomato fruit are shown.
RESULTS
Biochemical Analysis of Flavonoids and Carotenoids in Pink Tomatoes
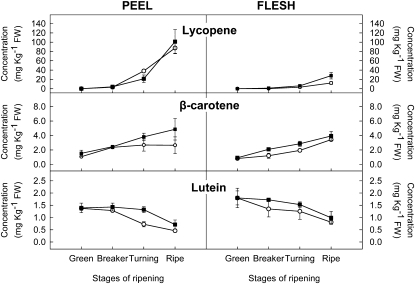
A phenotypic evaluation of a population of 55 introgression lines (ILs) derived from a cross between S. lycopersicum ‘Moneyberg’ and the wild species Solanum chmielewskii (LA1480) revealed the presence of three lines with pink-colored fruit. These pink lines all contained a homozygous introgression of S. chmielewskii DNA on chromosome 1, spanning an overlapping region of maximum 28.6 centimorgan (cM) between 18.5 and 47.1 cM (flanked by Conserved Ortholog Set II [COSII] markers C2_At5g06370 and C2_At4g15520; Fig. 3). To get insight into the biochemical basis for the pink fruit color and the levels and composition of flavonoids, phenylpropanoids and carotenoids were determined in the peel and flesh of pink fruit of TKM5U0436 (hereafter called IL1b) and red fruit of the recurrent parent cv Moneyberg (controls) at four different ripening stages. HPLC-photo diode array (PDA) analysis of peel extracts of red-colored ripe fruit revealed the presence of the main tomato flavonoids naringenin chalcone and rutin (Muir et al., 2001). In the peel of Moneyberg fruit, naringenin chalcone levels increased upon ripening and peaked at turning stage, reaching a maximum concentration of approximately 850 mg kg−1 fresh weight. Rutin levels in peel remained fairly constant during ripening or slightly decreased in control fruit. The flesh of these fruit contained only trace amounts of flavonoids, in agreement with earlier observations (Bovy et al., 2002). In contrast to Moneyberg fruit, peel extracts of the pink IL1b fruit did not show the ripening-dependent increase in naringenin chalcone, while levels of rutin were comparable to those observed in Moneyberg fruit (Fig. 4).
Figure 3.
Schematic overview of tomato chromosome 1 showing the locations of 14 COSII markers (whose names begin with C2_At) and a simple sequence repeat (SSR) marker. Numbers near the marker names indicate the genetic distances in cM from the top of the chromosome, based on the tomato-EXPEN 2000 mapping population (http://sgn.cornell.edu/). At the right part of chromosome 1, the total genetic length is mentioned. The S. chmielewskii IL population comprised four lines with single homozygous introgressions on chromosome 1 (black bars). The borders of the bars are arbitrarily drawn midway between markers positive and negative for the introgressed S. chmielewskii segment. The genetically mapped interval of the locus underlying the pink fruit phenotype is indicated by inward-facing arrows.
Figure 4.
Phenylpropanoid and flavonoid contents in peel and flesh of fruit from tomato cv Moneyberg (black squares) and IL1b (white circles) at four different stages of ripening. Values represent averages of three biological replicates. FW, Fresh weight.
Chlorogenic acid is the main phenylpropanoid in tomato fruit. In both peel and flesh of Moneyberg and IL1b fruit, chlorogenic acid levels were comparable and showed the same developmental pattern (Fig. 4), indicating that chlorogenic acid levels were not affected in pink tomato fruit.
The predominant carotenoids in red-colored ripe Moneyberg tomatoes were lycopene, β-carotene, and lutein. Levels of lycopene and β-carotene increased during ripening, while lutein levels decreased slightly. At later stages of ripening, lycopene levels were approximately 3.5-fold higher in peel than in flesh, while lutein and β-carotene levels were similar in both tissues at all developmental stages. Interestingly, peel and flesh extracts of IL1b tomatoes revealed no major differences in the levels and distribution of these three major carotenoids compared with the Moneyberg control (Fig. 5).
Figure 5.
Carotenoid contents in peel and flesh of fruit from tomato cv Moneyberg (black squares) and IL1b (white circles) at four different stages of ripening. Values represent averages of three biological replicates. FW, Fresh weight.
These results suggest that the pink phenotype is due to the absence of ripening-induced accumulation of the yellow-colored flavonoid naringenin chalcone in the fruit peel rather than to a change in carotenoid composition. This idea was confirmed by the analysis of a collection of commercial tomato materials, consisting of red and pink fruit of two segregating breeding populations (Supplemental Fig. S1) and five pink accessions (data not shown).
Expression Analysis of Phenylpropanoid and Flavonoid Pathway Genes
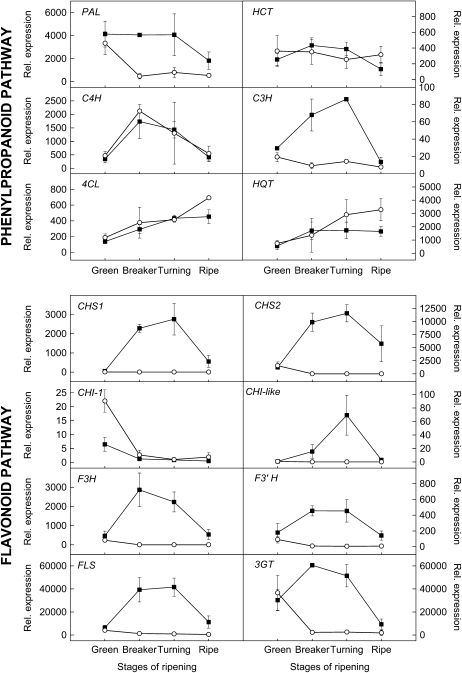
To gain insight into the molecular basis underlying the pink phenotype, the same Moneyberg and IL1b samples used for biochemical analysis were analyzed for expression of several structural genes involved in phenylpropanoid and flavonoid biosynthesis by quantitative real-time reverse transcription-PCR (qRT-PCR). Since the flavonoid pathway is only active in the fruit peel, these gene expression analyses were restricted to this tissue. In Moneyberg, transcript levels of the major Phe ammonia-lyase (PAL) gene expressed in tomato fruit were high and remained constant until the turning stage but decreased in red-ripe fruit. In contrast, PAL levels decreased strongly upon ripening of the pink IL1b fruit (Fig. 6). The other two phenylpropanoid genes involved in the production of the immediate flavonoid precursor 4-coumaroyl-CoA, those encoding cinnamate 4-hydroxylase (C4H) and 4-coumarate:CoA ligase (4CL), showed no differences between Moneyberg and IL1b fruit in the four ripening stages analyzed. In addition, three genes involved in the phenylpropanoid pathway branch leading to chlorogenic acid were studied, those encoding cinnamoyl CoA shikimate/quinate transferase (HCT), p-coumaroyl ester 3-hydroxylase (C3H), and hydroxycinnamoyl CoA quinate transferase (HQT). In Moneyberg fruit, all three genes showed a ripening-dependent increase in expression, with a peak at breaker/turning stage. In IL1b fruit, HCT and HQT expression patterns were similar to those observed in Moneyberg fruit. In contrast, C3H expression was clearly down-regulated in IL1b.
Figure 6.
Relative expression of phenylpropanoid and flavonoid genes in peel of Moneyberg (black squares) and IL1b (white circles) tomatoes at different stages of ripening. Expression levels were determined by qRT-PCR and expressed relative to the expression of the ubiquitin gene. Values represent averages of three biological replicates, each with two technical replicates.
All tested flavonoid genes, except for CHI-1, showed the same pattern of expression in Moneyberg fruit peel: levels of CHS1, CHS2, CHI-like, F3H, flavonoid-3′-hydroxylase (F3′H) , FLS, and flavonoid-3-O-glucosyltransferase (3GT) transcripts increased upon ripening, peaked at breaker or turning stage, and then slightly decreased in red-stage fruit. The expression of CHI-1, the closest homolog to the functionally characterized petunia CHI gene, was low and even decreased upon ripening. Levels of the second candidate CHI gene, a CHI-like gene, were also low compared with those of the other flavonoid pathway genes, but this gene clearly followed the ripening-dependent induction pattern. The very strong induction and high expression levels of both CHS genes at breaker/turning stage compared with the relatively low levels of both candidate CHI genes most likely lead to the accumulation of the CHS reaction product naringenin chalcone in the peel of the fruit. Unlike the observations in red-fruited Moneyberg tomatoes, the ripening-dependent induction of all the above-mentioned phenylpropanoid and flavonoid genes was absent in the peel of the pink IL1b tomatoes (Fig. 6). This lack of gene induction is, most likely, the cause of the low levels of naringenin chalcone observed in the peel of these fruit, resulting in the pink phenotype.
The MYB12 Transcription Factor Is Associated with the Pink Phenotype
The observation that the expression of many flavonoid genes was affected in the pink tomato line IL1b suggests that the gene underlying the pink phenotype encodes a regulatory protein of the flavonoid pathway rather than a biosynthetic enzyme. In plants, flavonoid biosynthesis is regulated by MYB-type transcription factors, sometimes operating in concert with basic helix-loop-helix (bHLH) transcription factors, and genes encoding flavonoid-related transcription factors have been cloned from an increasing number of plant species such as maize (Zea mays), petunia, and Arabidopsis (Arabidopsis thaliana; Schijlen et al., 2004). The phylogenetic tree of all Arabidopsis transcription factor genes (Stracke et al., 2001) was used to select Arabidopsis MYB transcription factors putatively involved in regulating genes in the phenylpropanoid and/or flavonoid pathway. These genes, belonging to subgroups 1 to 12 and 15, were used to identify the most closely related genes in tomato encoding MYB proteins using BLAST searches in Tomato Gene Index (http://compbio.dfci.harvard.edu/tgi/) and Sol Genomics Network (www.sgn.cornell.edu) databases. In this way, a total of 17 genes encoding R2R3MYB transcription factor in tomato were selected as potential candidate genes for the pink phenotype (Supplemental Table S1). A similar approach was followed to identify candidate bHLH-type transcription factor genes. In this case, the phylogenetic tree of Arabidopsis bHLH genes (Heim et al., 2003) was used to select Arabidopsis bHLH genes putatively involved in flavonoid biosynthesis (subgroup 3), and subsequently, these genes were used to identify their most closely related tomato homologs. In total, nine tomato bHLH genes were selected for further analysis (Supplemental Table S2).
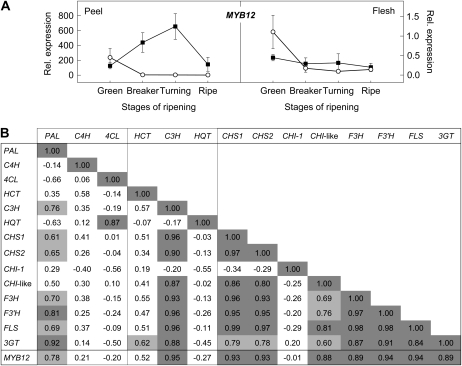
To determine the possible role of these transcription factors in regulating the production of flavonoids in red and pink tomatoes, we analyzed their expression in peel samples of pink- and red-fruited genotypes from two segregating populations, harvested at three ripening stages, using qRT-PCR (data not shown). Subsequently, the observed gene expression patterns were compared with those of the structural flavonoid genes (Supplemental Table S3). Of the 26 selected transcription factor genes, seven did not show any significant expression in tomato fruit. Correlation analysis of the remaining 19 transcription factor genes revealed that the expression of the AtMYB12 homolog TC199266 (SlMYB12) was highly correlated with the expression of the structural flavonoid genes tested, suggesting that this gene is a potential regulator of flavonol biosynthesis in tomato fruit. To confirm these results, the expression of the MYB12 gene was analyzed in more detail in Moneyberg and IL1b fruit. As shown in Figure 7A, MYB12 expression levels increased in Moneyberg fruit upon ripening, with a peak at breaker/turning stage, while in IL1b fruit, MYB12 expression was clearly down-regulated from breaker stage onward. Correlation analysis confirmed that the expression of the structural genes closely followed that of MYB12 (Fig. 7B), suggesting that MYB12 regulates the production of flavonoids in tomato fruit through activation of transcription of the genes encoding enzymes of the pathway. MYB12, therefore, was a good candidate for the gene underlying the pink phenotype of the analyzed lines.
Figure 7.
A, Relative expression of MYB12 in peel and flesh of red (black squares) and pink (white circles) tomatoes harvested at different stages of ripening, determined by qRT-PCR. Values represent means of three biological replicates ± sd. B, Correlation analysis of the structural phenylpropanoid and flavonoid gene expression profiles with the expression levels of the MYB12 gene.
Isolation and Mapping of the MYB12 Gene
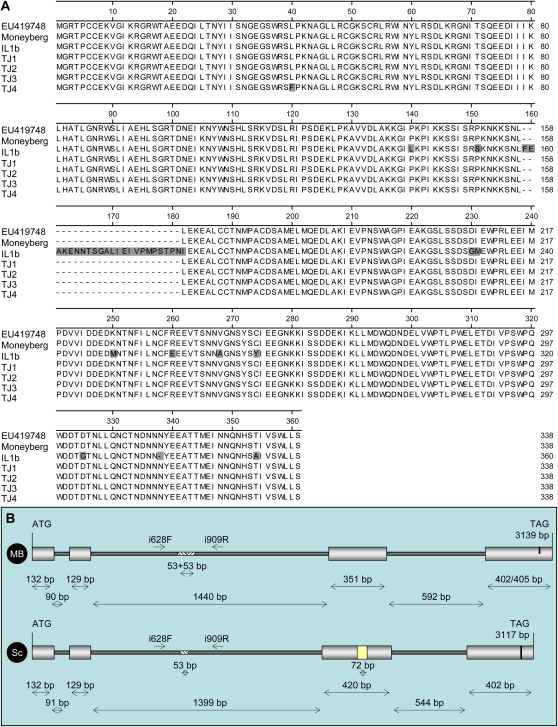
Full-length MYB12 cDNAs were isolated from green fruit samples of Moneyberg and IL1b, then sequenced and compared with the published SlMYB12 sequence (accession no. EU419748; Luo et al., 2008). As shown in Supplemental Figure S2, the Moneyberg (accession no. FN555126) and IL1b (accession no. FN555127) cDNA sequences shared 97.9% identity and were highly similar to the published SlMYB12 sequence (100% and 97.9%, respectively). Moneyberg and IL1b nucleotide sequences differed at several positions, but most striking was a 72-bp insertion in the third exon of the IL1b cDNA sequence. To check whether these single nucleotide polymorphisms and this insertion in the coding region were critical determinants of the pink phenotype, the deduced amino acid sequences of SlMYB12, Moneyberg (accession no. CBG76063.1), and IL1b (accession no. CBG76064.1) were compared with the deduced amino acid sequences of four other SlMYB12 alleles obtained from commercial tomato cultivars with pink-colored fruit (Fig. 8A). Compared with the two red alleles (EU419748 and Moneyberg), the IL1b amino acid sequence contains 11 amino acid substitutions, one amino acid deletion, and a 23-amino acid insertion. Although these differences may reflect the evolutionary distance between S. lycopersicum and S. chmielewskii, we cannot exclude the possibility that they may also affect the function of the MYB12 protein. In contrast, however, the deduced amino acid sequence of the pink MYB12 alleles obtained from commercial sources was identical to the red Moneyberg allele, except for one amino acid substitution (L→F) in cv TJ4. This suggested that deregulated MYB12 gene expression, observed in all pink genotypes tested, rather than aberrant MYB12 function is the primary cause of the pink phenotype.
Figure 8.
A, Alignment of deduced amino acid sequences of SlMYB12 alleles, including sequences of tomato cv Moneyberg (accession no. CBG76063.1), IL1b (accession no. CBG76064.1), the published sequence EU419748, and four pink tomato cultivars (TJ1, TJ2, TJ3, and TJ4). Amino acids different in one of the three sequences are boxed in gray. B, Schematic overview of the genomic structures of the SlMYB12 gene in Moneyberg (MB) and S. chmielewskii (Sc). The MYB12 gene consists of three exons (large boxes, light gray) and three introns (small boxes, dark gray). The duplicated sequence (53 bp) in the second intron is shown as a hatched box. The yellow box represents the additional 72 nucleotides detected in the S. chmielewskii sequence. Primers used to map the gene are indicated as i628F and i909R.
To analyze the genomic structure of the MYB12 gene, genomic clones were isolated from S. lycopersicum ‘Moneyberg’ and S. chmielewskii leaves using a genome-walking strategy. The MYB12 gene consisted of three introns (Fig. 8B). The second intron revealed the largest variation between the two lines, including a 53-bp duplication present in the Moneyberg sequence. This region was used to map the MYB12 gene in the S. chmielewskii IL population, using primers surrounding the duplication, resulting in PCR products of 338 bp for the Moneyberg allele and of 283 bp for the S. chmielewskii allele. As shown in Figure 9, the MYB12 gene mapped on three ILs, each carrying a homozygous S. chmielewskii introgression on chromosome 1, including IL1b, which together define an overlapping region of maximum 28.6 cM (Fig. 3). The MYB12 gene segregated perfectly with the characteristic pink fruit color in the IL population and appeared to be a perfect marker for the selection of plants with pink-colored fruit in several segregating breeding populations from diverse S. lycopersicum backgrounds, as demonstrated for one F2 population in Table I.
Figure 9.
Mapping of the MYB12 gene in the IL population derived from a cross between cultivated tomato S. lycopersicum ‘Moneyberg’ (MB) and the wild species S. chmielewskii (Sc). Ch., Chromosome. M, S. lycopersicum ‘Moneymaker’. Asterisks indicate pink tomatoes.
Table I.
Segregation of SlMYB12 in an F2 population segregating for red and pink fruit color
Values represent the number of individuals with the respective genotype.
| Phenotype | Homozygous Pink | Heterozygous | Homozygous Red |
|---|---|---|---|
| Red | 0 | 33 | 16 |
| Pink |
18 |
0 |
0 |
Virus-Induced Gene Silencing of SlMYB12 Leads to Pink Tomato Fruit Lacking Naringenin Chalcone
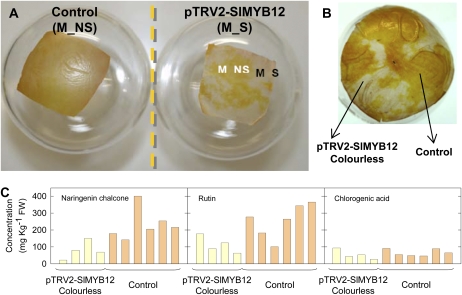
Our results thus far suggested that the pink fruit color is due to a developmentally regulated suppression of SlMYB12 gene expression. To demonstrate the direct involvement of SlMYB12 in the establishment of the pink phenotype, we used virus-induced gene silencing (VIGS) to down-regulate SlMYB12 gene expression in tomato fruit by Agrobacterium tumefaciens-mediated infiltration of S. lycopersicum ‘Moneymaker’ fruit with recombinant virus containing either a portion of the SlMYB12 gene or an empty vector (Orzaez et al., 2006). Fruit infiltrated with the control vector turned completely red, similar to the untreated Moneymaker fruit, while fruit infiltrated with the SlMYB12 VIGS construct developed pink-colored sectors in the peel regions where the infection of Agrobacterium suspension occurred. Visual inspection of the fruit epidermis revealed that these sectors were transparent and clearly lacked the accumulation of naringenin chalcone (Fig. 10, A and B). Quantification of phenylpropanoid and flavonoid levels in these silenced sectors, however, was difficult, since it was hard to separate the irregular silenced sectors from nonsilenced sectors in the same fruit, based on the subtle change in color from red to pink. Nevertheless, biochemical analysis of these sectors revealed a clear decrease in the levels of both naringenin chalcone and rutin in silenced compared with nonsilenced sectors. However, no changes in the concentration of chlorogenic acid were observed between these sectors (Fig. 10C). To gain more contrast between silenced and nonsilenced sectors, the same VIGS experiment was carried out in purple Delila and Rosea1 (Del/Ros1) S. lycopersicum ‘MicroTom’ fruit (Butelli et al., 2008; Orzaez et al., 2009), with silencing vectors that included either Del/Ros1 plus SlMYB12 or Del/Ros1 only (as a control; Supplemental Fig. S3). The phenotypic and biochemical analysis showed that pink-colored Del/Ros1-SlMYB12-silenced peel sectors contained significantly less naringenin chalcone and rutin than red-colored Del/Ros1-silenced peel sectors, in full agreement with the results obtained in the Moneymaker background. These results lead to the conclusion that silencing of the tomato SlMYB12 gene phenocopies the pink phenotype.
Figure 10.
SlMYB12 gene silencing in the cultivated tomato Moneymaker (M). A, Peel sections of fruit agroinjected with pTRV1 + pTRV2-SlMYB12 (right) or pTRV1 alone as control (left). M_S, Silenced sectors showing a colorless phenotype; M_NS, nonsilenced sectors. B, Peel of fruit agroinjected with pTRV1 + pTRV2-SlMYB12, showing the colorless silenced sectors and the colored nonsilenced sectors (control). C, Effect of SlMYB12 gene silencing in Moneymaker tomato fruit. Contents of naringenin chalcone, rutin, and chlorogenic acid in peel of four independent silenced tomato fruit and in peel of six control fruit agroinjected with pTRV1 alone are shown. FW, Fresh weight.
DISCUSSION
We have analyzed the biochemical and molecular bases underlying the pink tomato fruit color. Analysis of fruit obtained from various tomato backgrounds segregating for pink and red fruit revealed that pink fruit lack the ripening-dependent accumulation of the yellow-colored naringenin chalcone, one of the major flavonoids in tomato fruit peel. This leads to transparent peel and pink fruit color, a phenotype described in 1925 by Lindstrom, caused by the y mutation. Levels of the most abundant tomato phenylpropanoid, chlorogenic acid, and the main tomato carotenoids, lycopene, β-carotene, and lutein, were unaffected in the pink fruit analyzed, in peel or in flesh, suggesting that the gene responsible for the pink phenotype, and very likely for the known y mutation, only affects the flavonoid pathway.
In peel of red fruit, all genes in the flavonoid pathway leading to the flavonol rutin, except CHI-1, were strongly induced upon ripening. The accumulation of naringenin chalcone in peel of red-colored tomato fruit is most likely the result of the ripening-dependent increase in CHS expression, in combination with a concomitant decrease in CHI-1 expression. This creates an imbalance in the flux through the pathway, leading to the accumulation of the CHS product naringenin chalcone, due to a rate-limiting amount of CHI enzyme. Indeed, overexpression of the petunia CHI gene, the closest homolog of the tomato CHI-1 gene, in tomato fruit resulted in a significant increase in the flux through the pathway, with a concomitant decrease in naringenin chalcone (Muir et al., 2001) and a pink-colored phenotype (Colliver et al., 2002). In peel of pink fruit, however, the expression of all tested flavonoid genes remains low throughout fruit ripening; as a result, naringenin chalcone does not accumulate. The observation that all flavonoid biosynthetic genes in the pathway, except CHI-1, were affected in the pink line IL1b and all other pink genotypes tested (data not shown) suggests that the candidate gene for the pink phenotype encodes a flavonoid regulator rather than a biosynthetic enzyme.
Flavonoid pathways are regulated by a class of transcription factors belonging to the R2R3MYB family in plants. Some of these R2R3MYB transcription factors act in a complex composed of MYB, bHLH, and WD40 repeat proteins (Ramsay and Glover, 2005). We used a bioinformatic approach to select candidate MYB and bHLH transcription factor genes that might be involved in regulating the flavonoid pathway in tomato (Stracke et al., 2001; Heim et al., 2003). A total of 26 candidate genes encoding R2R3MYB and bHLH transcription factors were selected, and their expression in peel of ripening red and pink genotypes was determined using qRT-PCR. Expression of one transcription factor gene, MYB12, was correlated strongly with the expression of all biosynthetic flavonoid genes in red- and pink-colored tomato fruit, suggesting that this gene may be the direct regulator of the flavonoid pathway in tomato fruit peel.
SlMYB12 is the closest homolog of the Arabidopsis MYB12 gene (80% amino acid sequence identity), which regulates the production of flavonols in Arabidopsis seedlings (Mehrtens et al., 2005). Similarly, the MYB12 ortholog from grape (Vitis vinifera) is involved in regulating flavonol biosynthesis, since its reduced expression has been related to a drastically reduced flavonol content in grape fruit after shading and leaf-removal treatments (Matus et al., 2009). AtMYB12 does not need a bHLH partner to activate its target genes and lacks the conserved amino acid signatures for interaction with bHLH proteins (Zimmermann et al., 2004; Mehrtens et al., 2005). Indeed, these motifs are absent in SlMYB12 as well (data not shown). In addition to AtMYB12, two other MYB transcription factor genes (AtMYB11 and AtMYB111) are involved in regulating flavonol biosynthesis in Arabidopsis. Through differential expression, each of these genes regulates flavonol production in different parts of the Arabidopsis seedling (Stracke et al., 2007). In addition to MYB12, we identified a MYB111 (accession no. AI771790) homolog in tomato as well. This EST showed no significant expression in tomato fruit (Supplemental Table S3) and may be involved in controlling flavonol production in other parts of the tomato plant.
Three strategies were employed to determine whether MYB12 is the gene underlying the pink phenotype. First, the segregation of the MYB12 gene was followed in the S. chmielewskii IL population and in several commercial breeding populations segregating for pink fruit color. The SlMYB12 marker appeared to segregate perfectly with the pink fruit color in all populations tested, suggesting that SlMYB12 is the gene underlying the pink phenotype or lies very close to the pink locus. Second, we mapped the genetic position of MYB12 to a region of maximum 28.6 cM on the short arm of chromosome 1, flanked by COSII markers C2_At5g06370 (at 18.5 cM) and C2_At4g15520 (at 47.1 cM). Although the segregation and mapping results suggest that all pink lines analyzed originate from the same genetic locus, most likely Y, allelism tests should provide more conclusive data showing that the pink lines tested in this study are allelic to the original y mutant. Third, the pink fruit color was phenocopied by transient VIGS-mediated silencing of SlMYB12 gene expression. In the Moneymaker background, silenced sectors in ripe fruit displayed the typical pink color and had a transparent peel. Biochemical analysis of flavonoids revealed that both naringenin chalcone and rutin levels were significantly lower in silenced sectors of the fruit peel than in nonsilenced sectors. In parallel, VIGS experiments were carried out in a purple transgenic Microtom background, in which the Antirrhinum anthocyanin regulators Del and Ros1 were expressed in the fruit under control of the E8 promoter, leading to accumulation of high levels of purple anthocyanins (Butelli et al., 2008). These plants improve the efficiency and visual monitoring of VIGS experiments (Orzaez et al., 2009), since silencing of the Del/Ros1 transgenes leads to easily recognizable red-colored cosilenced sectors within the purple background. VIGS of Del/Ros1 (control) or SlMYB12 in combination with Del/Ros1 resulted in fruit with red (control) or pink (SlMYB12) silenced sectors. Biochemical analysis revealed that pink sectors showed significantly decreased levels of naringenin chalcone and rutin compared with red sectors. Since the Del/Ros1 and SlMYB12 transcription factors to a large extent activate the same genes in the flavonoid pathway, we could not exclude the possibility that silencing of Del/Ros1 influences the silencing effect of SlMYB12. However, it appeared to be very easy to use this system to monitor the effects of even a flavonoid-related transcription factor, since the results of the Moneymaker and Microtom VIGS experiments were comparable.
It has been shown recently that overexpression of the AtMYB12 gene in tomato fruit leads to a strong induction of flavonoid and phenylpropanoid gene expression and highly increased levels of flavonols and hydroxycinnamic acids, in particular chlorogenic acid, in both fruit peel and flesh (Luo et al., 2008). In contrast, chlorogenic acid levels were unaffected in peel and flesh of pink IL1b fruit compared with Moneyberg or in VIGS-silenced SlMYB12 fruit. Gene expression analysis of IL1b and Moneyberg fruit revealed that, except for C3H, the genes required for the synthesis of chlorogenic acid (HCT and HQT) are not primary targets of SlMYB12 regulation, suggesting that the activation of the pathway to chlorogenic acid in fruit overexpressing AtMYB12 most likely reflects a dosage effect of the AtMYB12 protein, through recruitment of new targets, probably through lower affinity binding sites in the promoters of the HCT and HQT genes. A similar dose-dependent response of target genes has been described for AtMYB4 in Arabidopsis (Jin et al., 2000) and for c-MYB activity in human (Andersson et al., 1999). Interestingly, of all target genes tested, those showing lower responses to AtMYB12 overexpression in tomato (C4H and 4CL) were unaffected in their expression levels in the pink tomatoes, supporting the idea that these and the HCT and HQT genes represent lower affinity targets for regulation than the core genes of flavonol synthesis (PAL, CHS, CHI, F3H, and FLS).
Comparison of the deduced amino acid sequences encoded by the MYB12 genes from EU419748, Moneyberg, and IL1b tomato lines revealed high homology (97.9%) between the sequences, although the encoded proteins differed at several positions. The differences between the predicted proteins of the EU419748/Moneyberg and IL1b lines all reside in the region encoding the hypervariable C-terminal domain of R2R3MYB proteins, all maintain the open reading frame of the encoded protein, and all are consistent with the levels of sequence variation found between orthologous R2R3MYB proteins from different species found by others (Di Stilio et al., 2009). No differences were found in the deduced amino acid sequences of three SlMYB12 alleles obtained from independent pink S. lycopersicum cultivars and the SlMYB12 sequence in Moneyberg and EU419748, suggesting that deregulated MYB12 gene expression, rather than aberrant MYB12 function, is the primary cause of the pink phenotype. Several examples of cis-acting mutations leading to deregulated expression of Myb transcription factor genes have been reported. In white grape cultivars, expression of the grape VvmybA1a gene is abolished due to a retrotransposon insertion in the promoter region, leading to a loss of anthocyanin pigmentation (Kobayashi et al., 2004). In contrast, a rearrangement in the upstream region of the apple MYB10 gene led to high and deregulated MYB10 expression and concomitant induction of the anthocyanin pathway, resulting in apple fruit with red fruit flesh (Espley et al., 2009).
In conclusion we have shown that the pink fruit color of all the lines analyzed in this study is due to the absence of the yellow-colored flavonoid naringenin chalcone. This is caused by a deregulated expression of MYB12, a transcription factor gene regulating the flavonoid pathway leading to flavonols such as rutin. Genetic mapping, segregation analysis, and VIGS results suggest strongly that the MYB12 gene is a likely candidate of the locus leading to pink fruit, probably the Y locus. Complementation of pink genotypes by a wild-type MYB12 allele, however, should provide final proof for this hypothesis. Since MYB12 gene expression is affected in pink genotypes, it is tempting to speculate that this is due to a mutation in a cis-element affecting gene expression rather than gene function. Indeed, sequence analysis of the MYB12 gene did not reveal any mutations in the coding sequences of three independent pink alleles of MYB12 in the S. lycopersicum background. Although several single nucleotide polymorphisms and insertions/deletions specific for the pink allele were found in introns and the promoter region of the MYB12 gene in IL1b compared with the sequence in the Moneyberg background, we were unable to pinpoint the causative mutation. This issue will be addressed in future experiments. From an applied perspective, it is clear that MYB12 is located at or near the locus underlying the pink fruit phenotype and can be used as an efficient genetic marker to select for the pink phenotype in future breeding programs.
MATERIALS AND METHODS
Plant Material
A population of 55 ILs derived from a cross between the cultivated tomato Solanum lycopersicum ‘Moneyberg’ and the wild species Solanum chmielewskii (LA1840) was grown to maturity in a greenhouse, and fruit were analyzed phenotypically. Fruit of IL1b (TKM5U0436), containing a homozygous introgression of the S. chmielewskii genome on chromosome 1 in the Moneyberg background, showed a characteristic pink color compared with the red commercial tomato fruit. Six plants of IL1b and Moneyberg were subsequently grown to maturity under greenhouse conditions in Wageningen, The Netherlands. Fruit were harvested at four stages of ripening, and the fruit peel was carefully separated from the rest of the fruit (the flesh tissue) using a scalpel. Both tissues were immediately frozen in liquid nitrogen, ground to a fine frozen powder using an analytical electric mill, and stored at −80°C until use. Each replicate consisted of at least six fruit of the same ripening stage obtained from two different plants.
For biochemical and molecular analyses, Enza Zaden Research and Development provided fruit obtained from individual genotypes of two breeding populations segregating for the pink phenotype (population 35025, two pink- and one red-fruited genotype; population 35037, one pink- and one red-fruited genotype) and from an additional set of five pink varieties. Samples consisted of a pool of at least six fruit per genotype, collected at three stages of ripening. Samples were separated into peel and flesh tissues, immediately frozen in liquid nitrogen, ground, and stored at −80°C until analysis.
The sequences of different MYB12 alleles were determined from tomato cv Moneyberg, IL1b, and four different pink S. lycopersicum cultivars (TJ1, TJ2, TJ3, and TJ4).
For segregation analysis, an F2 population consisting of 67 plants segregating for the pink and red fruit color was analyzed with a MYB12-specific marker.
Flavonoid and Carotenoid Extraction and HPLC Analysis
Flavonoid extraction and HPLC-PDA and liquid chromatography-quadrupole time of flight-mass spectrometry analyses were carried out according to Bino et al. (2005). The detected flavonoid compounds were identified using authentic standards and accurate mass liquid chromatography-mass spectrometry analysis (Moco et al., 2006).
Carotenoids were extracted as described previously by López-Ráez et al. (2008) and analyzed by HPLC-PDA according to Bino et al. (2005).
RNA Isolation and qRT-PCR Gene Expression Analysis
Total RNA was isolated from 150 mg of peel or flesh of tomato fruit using 1.5 mL of Trizol reagent (Invitrogen) according to the manufacturer's instructions. Before cDNA synthesis, total RNA was treated with DNase-I Amplification Grade (Invitrogen) and purified with an RNeasy Mini Kit (Qiagen). An aliquot of 1 μg of total RNA was used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad Laboratories) in a 20-μL final volume according to the manufacturer. Expression levels of each gene were measured in duplicate reactions, performed with the same cDNA pool, in the presence of fluorescent dye (iQ SYBR Green Supermix) using an iCycler iQ instrument (Bio-Rad Laboratories) with specific primer pairs (Supplemental Tables S1, S2, and S4). The constitutively expressed mRNAs encoding ubiquitin or glyceraldehyde 3-phosphate dehydrogenase were used as internal references in case of Moneyberg and IL1b or segregating breeding populations, respectively. Expression levels were determined relative to the internal reference and multiplied by a factor of 1,000. Calculations of each sample were carried out according to the comparative Ct (threshold cycle) method.
DNA Markers and Assays
A set of PCR-based markers consisting of 130 COSII markers (Wu et al., 2006) and three simple sequence repeats (Frary et al., 2005), previously mapped in the tomato genome and covering all 12 tomato chromosomes, were used to genotype the population of 55 ILs (Prudent et al., 2009). Sequences of the primers are available on the Sol Genomics Network Web site (http://www.sgn.cornell.edu).
For the COSII markers, amplicon size differences between the two parents were detected in 12% of the cases and were used to genotype the IL population directly; in the other cases, the amplicons were digested with different restriction enzymes (TaqI, HinfI, AluI, DraI, RsaI, and MseI) to identify polymorphisms. Where no polymorphisms were detected, single-band amplicons were purified and sequenced. Amplicon sequences were aligned and examined for polymorphisms using the program CAPSdesigner (http://www.sgn.cornell.edu/tools/caps_designer/caps_input.pl). Thereafter, the IL population was genotyped via cleaved-amplified polymorphic sequence assays (Konieczny and Ausubel, 1993).
Cloning, Sequencing, Mapping, and Segregation Analysis of the MYB12 Transcription Factor
MYB12 full-length cDNA sequences were amplified from green-stage fruit peel of Moneyberg and IL1b tomatoes using the SMART RACE cDNA amplification kit (Clontech Laboratories). MYB12 genomic DNA was amplified using the GenomeWalker kit (Clontech Laboratories). The amplified sequences of MYB12 full-length cDNA and genomic DNA were cloned into pGEM-T-Easy vector (Promega) and sequenced.
The MYB12 gene was mapped using the S. chmielewskii IL population, with a PCR-based marker, using primers i628F (forward; 5′-CACAATAATTTGGTGCTCCGATCTAAC-3′) and i909R (reverse; 5′-ATATTAAATTTATCACACGAACAACAGC-3′) in the second intron of the MYB12 gene.
VIGS of the SlMYB12 Gene
Wild-type S. lycopersicum ‘Moneymaker’ and transgenic tomato plants accumulating high levels of anthocyanins in the fruit (Butelli et al., 2008) were used for VIGS of the SlMYB12 gene (Orzaez et al., 2006, 2009).
Two vectors for VIGS of SlMYB12 were produced: one designed for cosilencing of both the color-monitoring Del/Ros1 module and SlMYB12, to be used in the Del/Ros1 background, and the other containing only the SlMYB12 module, to be used in the Moneymaker background (Orzaez et al., 2009). To do this, a 234-bp SlMYB12 fragment between oligonucleotides P07DEC04MYBF2 (5′-CCTAAAGCCGTAGTTGATTTGGC-3′) and P07DEC03MYBR2 (5′-CTGAACTAAGGGCTTCCCTTGGC-3′) was first cloned into pCR8/GW-TOPO and subsequently transferred by LR recombination into the pTRV2-Del/Ros1-GW destination vector to produce pTRV2-Del/Ros1-SlMYB12 or into pTRV2-GW (Liu et al., 2002) to produce pTRV2-SlMYB12.
All plant inoculations were performed using Agrobacterium tumefaciens strain C58 cultures harboring the different pTRV constructs. Coinfiltration experiments were carried out as described previously by Orzaez et al. (2009). When the Del/Ros1 background was used, anthocyanin-depleted samples, corresponding to Del/Ros1 silencing, were collected from pTRV2-Del/Ros1-SlMYB12- or pTRV2-Del/Ros1-agroinjected fruit. Peel sectors from different fruit were pooled and frozen in liquid nitrogen. When the Moneymaker background was used, SlMYB12-silenced sectors were separated from nonsilenced ones on the basis of the pink phenotype of the silenced sectors.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FN555126, FN555127, CBG76063.1, and CBG76064.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Concentration of flavonoids and carotenoids in peel of two F2 populations segregating for pink and red tomato fruit.
Supplemental Figure S2. Sequence alignment of tomato MYB12 genes.
Supplemental Figure S3. SlMYB12 gene silencing in purple transgenic Microtom tomatoes expressing the Del/Ros1 genes.
Supplemental Table S1. Overview of MYB transcription factor genes analyzed for expression in red and pink tomato fruit.
Supplemental Table S2. Overview of bHLH transcription factor genes analyzed for expression in red and pink tomato fruit.
Supplemental Table S3. Correlation analysis of expression levels of MYB and bHLH transcription factor and flavonoid genes in several S. lycopersicum breeding populations segregating for pink and red fruit.
Supplemental Table S4. Oligonucleotides of phenylpropanoid and flavonoid genes used for SYBR Green RT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Keygene for kindly providing the S. chmielewskii IL population as well as Dr. Sjaak van Heusden, Mr. Paul Dijkhuis, and Mrs. Fien Meijer-Dekens for excellent greenhouse management and plant cultivation and for providing genomic DNA of this population.
This work was supported by the European Union as part of the EU-SOL project (grant no. PL 016214–2) and by the Centre for BioSystems Genomics, provided under the auspices of the Netherlands Genomics Initiative.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Arnaud Bovy (arnaud.bovy@wur.nl).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andersson KB, Berge T, Matre V, Gabrielsen OS (1999) Sequence selectivity of c-Myb in vivo: resolution of a DNA target specificity paradox. J Biol Chem 274 21986–21994 [DOI] [PubMed] [Google Scholar]
- Arts ICW, Hollman PCH (2005) Polyphenols and disease risk in epidemiologic studies. Am J Clin Nutr 81 317S–325S [DOI] [PubMed] [Google Scholar]
- Basu A, Imrhan V (2007) Tomatoes versus lycopene in oxidative stress and carcinogenesis: conclusions from clinical trials. Eur J Clin Nutr 61 295–303 [DOI] [PubMed] [Google Scholar]
- Bino RJ, De Vos CHR, Lieberman M, Hall RD, Bovy A, Jonker HH, Tikunov Y, Lommen A, Moco S, Levin I (2005) The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol 166 427–438 [DOI] [PubMed] [Google Scholar]
- Bovy A, De Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, Schijlen E, Hall RD (2007) Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy AG, Gomez-Roldan V, Hall RD (2010) Strategies to optimize the flavonoid content of tomato fruit. In C Santos-Buelga, MT Escribano-Bailon, V Lattanzio eds, Recent Advances in Polyphenols Research, Vol II. Wiley-Blackwell Publishing, Oxford (in press)
- Britton G, Liaaen-Jensen S, Pfander H (2004) Carotenoids Handbook. Birkhauser Verlag, Basel, p 186
- Butelli E, Titta L, Giorgio M, Mock H-P, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26 1301–1308 [DOI] [PubMed] [Google Scholar]
- Colliver S, Bovy A, Collins G, Muir S, Robinson S, De Vos CHR, Verhoeyen ME (2002) Improving the nutritional content of tomatoes through reprogramming their flavonoid biosynthetic pathway. Phytochem Rev 1 113–123 [Google Scholar]
- Davies KM (2004) An introduction to plant pigments in biology and commerce. In KM Davies, ed, Plant Pigments and Their Manipulation. Annual Plant Reviews, Vol 14. Blackwell Publishing/CRC Press, Oxford/Boca Raton, FL, pp 1–22
- Di Stilio V, Martin V, Schulfer A, Connelly C (2009) An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol 183 718–728 [DOI] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga CG (2007) Plant polyphenols: how to translate their in vitro antioxidant actions to in vivo conditions. IUBMB Life 59 308–315 [DOI] [PubMed] [Google Scholar]
- Frary A, Xu Y, Liu J, Mitchell S, Tedeschi E, Tanksley S (2005) Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theor Appl Genet 111 291–312 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Mazzucato A, Perata P (2009) Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci 14 237–241 [DOI] [PubMed] [Google Scholar]
- Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57 761–780 [DOI] [PubMed] [Google Scholar]
- Harborne JB (1986) Nature, distribution and function of plant flavonoids. Prog Clin Biol Res 213 15–24 [PubMed] [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55 481–504 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 735–747 [DOI] [PubMed] [Google Scholar]
- Hooper L, Cassidy A (2006) A review of the health care potential of bioactive compounds. J Sci Food Agric 86 1805–1813 [Google Scholar]
- Hunt GM, Baker EA (1980) Phenolic constituents of tomato fruit cuticles. Phytochemistry 19 1415–1419 [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, Suzuki T, Suzuki H, Okazaki K, Kitayama M, et al (2008) Metabolite annotations based on the integration of mass spectral information. Plant J 54 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Trancriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Mes P, Myers JR (2003) Characterization and inheritance of the Anthocyanin fruit (Aft) tomato. J Hered 94 449–456 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304 982. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4 403–410 [DOI] [PubMed] [Google Scholar]
- Lewinsohn E, Sitrit Y, Bar E, Azulay Y, Ibdah M, Meir A, Yosef E, Zamir D, Tadmor Y (2005) Not just colors: carotenoid degradation as a link between pigmentation and aroma in tomato and watermelon fruit. Trends Food Sci Technol 16 407–415 [Google Scholar]
- Lindstrom EW (1925) Inheritance in tomatoes. Genetics 10 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gomez-Roldan V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Becard G, Mulder P, et al (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178 863–874 [DOI] [PubMed] [Google Scholar]
- Lotito SB, Frei B (2006) Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: cause, consequence, or epiphenomenon? Free Radic Biol Med 41 1727–1746 [DOI] [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Paul B, Weisshaar B, Martin C (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J 56 316–326 [DOI] [PubMed] [Google Scholar]
- Matus JT, Loyola R, Vega A, Peña-Neira A, Bordeu E, Arce-Johnson P, Alcalde JA (2009) Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J Exp Bot 60 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes PJ, Boches P, Myers JR, Durst R (2008) Characterization of tomatoes expressing anthocyanin in the fruit. J Am Soc Hortic Sci 133 262–269 [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, De Groot J, Van Beek TA, Vervoort J, De Vos CHR (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, Ric De Vos CH, Van Tunen AJ, Verhoeyen ME (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19 470–474 [DOI] [PubMed] [Google Scholar]
- Orzaez D, Medina A, Torre S, Fernandez-Moreno JP, Rambla JL, Fernandez-del-Carmen A, Butelli E, Martin C, Granell A (2009) A visual reporter system for VIGS in tomato fruit based on anthocyanin accumulation. Plant Physiol 150 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaez D, Mirabel S, Wieland WH, Granell A (2006) Agroinjection of tomato fruits: a tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol 140 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent M, Causse M, Génard M, Tripodi P, Grandillo S, Bertin N (2009) Genetic and physiological analysis of tomato fruit weight and composition: influence of carbon availability on QTL detection. J Exp Bot 60 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10 63–70 [DOI] [PubMed] [Google Scholar]
- Rao AV, Rao LG (2007) Carotenoids and human health. Pharmacol Res 55 207–216 [DOI] [PubMed] [Google Scholar]
- Rick CM, Butler L (1956) Cytogenetics of the tomato. Adv Genet 8 267–382 [Google Scholar]
- Schijlen EGWM, Beekwilder J, Hall RD, van der Meer IM (2008) Boosting beneficial phytochemicals in vegetable crop plants. CAB Reviews 3 1–21 [Google Scholar]
- Schijlen EGWM, De Vos CHR, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, Van Tunen AJ, Bovy AG (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijlen EGWM, De Vos CHR, Van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65 2631–2648 [DOI] [PubMed] [Google Scholar]
- Singh P, Goyal GK (2008) Dietary lycopene: its properties and anticarcinogenic effects. Compr Rev Food Sci Food Saf 7 255–270 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54 733–749 [DOI] [PubMed] [Google Scholar]
- Tapas AR, Sakarkar DM, Kakde RB (2008) Flavonoids as nutraceuticals: a review. Trop J Pharm Res 7 1089–1099 [Google Scholar]
- Willits MG, Kramer CM, Prata RTN, De Luca V, Potter BG, Stephens JC, Graser G (2005) Utilization of the genetic resources of wild species to create a nontransgenic high flavonoid tomato. J Agric Food Chem 53 1231–1236 [DOI] [PubMed] [Google Scholar]
- Wu F, Mueller LA, Crouzillat D, Petiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single copy, orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AJ, Frank HA (1996) Energy transfer reactions involving carotenoids: quenching of chlorophyll fluorescence. J Photochem Photobiol B 36 3–15 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.