Abstract
The functional protein phosphatase type 2C from beechnut (Fagus sylvatica; FsPP2C1) was a negative regulator of abscisic acid (ABA) signaling in seeds. In this report, to get deeper insight on FsPP2C1 function, we aim to identify PP2C-interacting partners. Two closely related members (PYL8/RCAR3 and PYL7/RCAR2) of the Arabidopsis (Arabidopsis thaliana) BetV I family were shown to bind FsPP2C1 in a yeast two-hybrid screening and in an ABA-independent manner. By transient expression of FsPP2C1 and PYL8/RCAR3 in epidermal onion (Allium cepa) cells and agroinfiltration in tobacco (Nicotiana benthamiana) as green fluorescent protein fusion proteins, we obtained evidence supporting the subcellular localization of both proteins mainly in the nucleus and in both the cytosol and the nucleus, respectively. The in planta interaction of both proteins in tobacco cells by bimolecular fluorescence complementation assays resulted in a specific nuclear colocalization of this interaction. Constitutive overexpression of PYL8/RCAR3 confers ABA hypersensitivity in Arabidopsis seeds and, consequently, an enhanced degree of seed dormancy. Additionally, transgenic 35S:PYL8/RCAR3 plants are unable to germinate under low concentrations of mannitol, NaCl, or paclobutrazol, which are not inhibiting conditions to the wild type. In vegetative tissues, Arabidopsis PYL8/RCAR3 transgenic plants show ABA-resistant drought response and a strong inhibition of early root growth. These phenotypes are strengthened at the molecular level with the enhanced induction of several ABA response genes. Both seed and vegetative phenotypes of Arabidopsis 35S:PYL8/RCAR3 plants are opposite those of 35S:FsPP2C1 plants. Finally, double transgenic plants confirm the role of PYL8/RCAR3 by antagonizing FsPP2C1 function and demonstrating that PYL8/RCAR3 positively regulates ABA signaling during germination and abiotic stress responses.
It is well established that the plant hormone abscisic acid (ABA) plays a key role in different plant developmental processes, including seed formation, dormancy, and germination (Bewley, 1997; González-Guzmán et al., 2002; Finkelstein et al., 2008), as well as in plant responses to several abiotic stresses such as drought, salt, and cold (Hugouvieux et al., 2001; Finkelstein et al., 2002; Nambara and Marion-Poll, 2005; Christmann et al., 2006). ABA acts through a complex signaling cascade to induce changes in gene expression, thus affecting multiple physiological processes (Skriver and Mundy, 1990; Cutler and McCourt, 2005; Pei and Kuchitsu, 2005). Numerous ABA-regulated genes have been identified, and genome-scale analyses indicate that more than 2,900 genes are responsive to ABA in Arabidopsis (Arabidopsis thaliana; Nemhauser et al., 2006). Several ABA signaling components, some of them associated with germination and involved in the removal of sensitivity to ABA function, have been characterized, including guanine nucleotide-binding proteins (G proteins), kinases, phosphatases, protein degradation pathways, transcription factors, and secondary messengers (Finkelstein et al., 2002; López-Molina et al., 2003; Nishimura et al., 2004, 2007; Verslues and Zhu, 2005; Zhang et al., 2005; Pandey et al., 2006; Sáez et al., 2006; Yoine et al., 2006; Holdsworth et al., 2008).
Evidence suggests that ABA signaling in both seeds and guard cells involves heterotrimeric G proteins such as the Arabidopsis Gα subunit (GPA1), the Gβ subunit (AGB1), and the candidate G protein-coupled receptor (GCR1). Results from knockout mutants lacking one or more of these components showed that GPA1, AGB1, and GCR1 each negatively regulates ABA signaling in seed germination and early seedling development, leading to hypersensitivity to ABA and reduced seed dormancy (Pandey et al., 2006). Recently, Pandey et al. (2009) have reported the identification of two novel membrane proteins from Arabidopsis, GTG1 and GTG2 (GPCR-type G proteins), that interact with GPA1 but also have intrinsic GTP-binding and GTPase activities. GTG1 and GTG2 bind ABA in vitro, and double mutants lacking both proteins exhibit ABA hyposensitivity. They suggest that GTG proteins are a new type of G protein that function as membrane-localized receptors of ABA and mediate ABA responses in vivo.
Most recently, an ABA-specific-binding protein, ABAR/CHLH (for putative abscisic acid receptor/Mg-chelatase H subunit), that mediates ABA signaling as a positive regulator in Arabidopsis, has been found to specifically bind ABA using a newly developed ABA-affinity chromatography technique and a [3H]ABA-binding assay, supporting that ABAR/CHLH may act as an intracellular ABA receptor (Wu et al., 2009).
A pivotal role for phosphorylation and dephosphorylation processes in ABA signaling through kinases and phosphatases is well supported by biochemical and genetic evidence (Leung and Giraudat, 1998; Campalans et al., 1999; Finkelstein et al., 2002). The snrk2.2/snrk2.3 double mutant of two protein kinases, namely SNF1-RELATED PROTEIN KINASE2.2 (SnRK2.2) and SnRK2.3, showed strong ABA insensitivity for germination in comparison with wild-type and single mutant seeds (Fujii et al., 2007).
Moreover, protein phosphatases type 2C (PP2Cs) were identified as major components of the ABA signaling pathway (Koornneef et al., 1989, 2002; Leung et al., 1994, 1997; Meyer et al., 1994; Rodríguez et al., 1998a, 1998b). Arabidopsis abi mutants abi1 and abi2 with dominant mutations (Koornneef et al., 1989, 2002) are closely related members of the PP2C family (Schweighofer et al., 2004). These mutants have greatly reduced phosphatase activity; however, knockout mutations in these loci show weak hypersensitivity to ABA and hyperdormancy phenotype (Merlot et al., 2001). Currently, several PP2Cs are known to regulate ABA signaling in Arabidopsis: ABI1, ABI2, PP2CA, HAB1, and AHG1 (Nishimura et al., 2007). Expression of many of these protein phosphatases is induced by ABA, acting as negative regulators of the hormone signaling pathway and providing a negative feedback mechanism to attenuate the ABA response (Gosti et al., 1999; Merlot et al., 2001; Tahtiharju and Palva, 2001; Sáez et al., 2004, 2006; Kuhn et al., 2006; Yoshida et al., 2006b). Studies on knockout/knockdown and double mutants in several PP2C family members have demonstrated the pivotal role of these PP2Cs in ABA signaling and the existence of some redundancy (Sáez et al., 2006; Yoshida et al., 2006b; Nishimura et al., 2007). Recently, the PYR1-PYL/RCAR family of proteins has been reported to function as ABA sensors by binding and inhibiting PP2Cs, which triggers a reversible protein phosphorylation cascade to mediate ABA responses (Ma et al., 2009; Park et al., 2009b). The individual functions of the different PYL/RCAR members in ABA signaling are now emerging, and PYL5/RCAR8 has been described as an intracellular ABA receptor modulating stress responses (Santiago et al., 2009).
Although most of the research is focused on Arabidopsis, important progress is also being made in other plant species. These findings may help to answer questions about the fundamental mechanisms controlling plant growth, development, and stress responses and the role that protein phosphorylation/dephosphorylation plays in these processes.
We previously reported the cloning of FsPP2C1, a functional PP2C specifically expressed in beechnut (Fagus sylvatica), that was up-regulated upon ABA addition in seeds and early seedlings (Lorenzo et al., 2001). Overexpression of FsPP2C1 in Arabidopsis conferred a reduced degree of dormancy compared with wild-type seeds as well as ABA insensitivity in both seed germination and different abiotic stresses. In addition, FsPP2C1 transgenic plants showed ABA-resistant early root growth and diminished induction of the ABA response genes RAB18 and KIN2, but no effect on the stomatal closure regulation was observed, which proved that FsPP2C1 acts as a negative regulator of ABA signaling (González-García et al., 2003), as described for other Arabidopsis PP2Cs such as ABI1, ABI2, and HAB1.
These PP2Cs may have different substrates/partners inside the cells, but only a few have been identified (Christmann et al., 2006), and the specific interactions and mechanisms have not been demonstrated yet. These substrates include transcription factors involved in the drought/stress response (Himmelbach et al., 2002), protein kinases such as OST1/SnRK2.6 (Yoshida et al., 2006a), K+ channels (Cherel et al., 2002), calcineurin B-like calcium sensors (Guo et al., 2002), a glutathione peroxidase (Miao et al., 2006), and a stress-induced photoprotective plastid protein (Yang et al., 2006).
To get deeper insight into the function of this PP2C from Fagus in ABA signaling and ABA-regulated responses, we started a search for putative FsPP2C1 cell substrates or interacting proteins by yeast two-hybrid analysis. We have found at least one protein that strongly interacts with FsPP2C1, named PYL8/RCAR3, and this interaction is interestingly localized in the nucleus. This FsPP2C1-interacting partner belongs to the BetV I subfamily described by Radauer et al. (2008), whose members have been identified as PP2C inhibitors (Ma et al., 2009; Park et al., 2009b; Santiago et al., 2009). Overexpression of PYL8/RCAR3 produces hypersensitivity to ABA in seed germination and increased tolerance to water stress in vegetative tissues, suggesting an important role for this protein in ABA signaling and ABA-regulated genes involved in dormancy, germination, and stress responses.
Characterization and function of these proteins will serve for a better understanding of the PP2C regulatory mechanisms involved in ABA signaling and in the ABA-mediated mechanisms controlling seed dormancy, germination, and stress responses.
RESULTS
Identification of FsPP2C1-Interacting Proteins by Yeast Two-Hybrid Screening
We have previously reported the isolation and functional characterization of FsPP2C1 as a plant type 2C protein phosphatase, with all the conserved features of the catalytic domain of these proteins (Lorenzo et al., 2001), which acts as a negative regulator of ABA responses in seeds (González-García et al., 2003). Our results suggest that this PP2C from beech, which is a foreign protein in Arabidopsis tissues, possesses at least its corresponding signaling components (substrate, activator, or partner) in this plant model species.
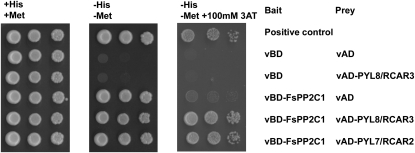
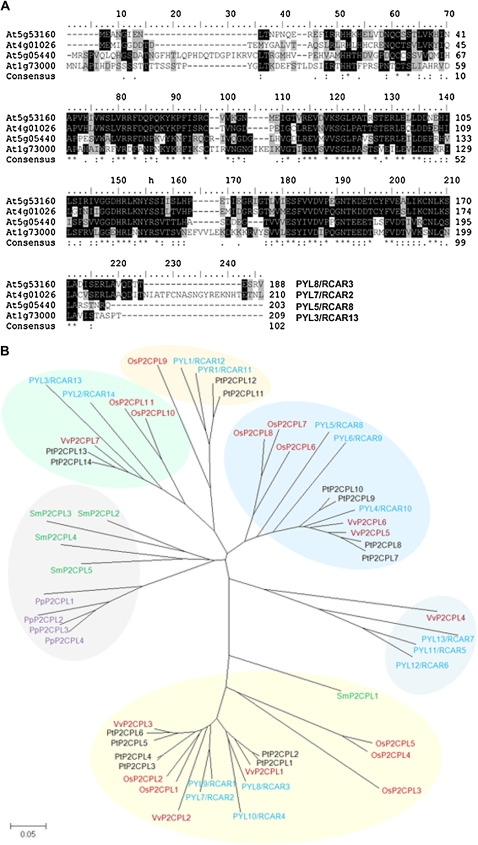
In a search of FsPP2C1-interacting proteins (PP2C-IPs) in Arabidopsis, a yeast two-hybrid screening was performed using FsPP2C1 as bait. As a result, we have identified three different proteins (PYL8/RCAR3, PYL7/RCAR2, and PP2C-IP3) showing interaction with FsPP2C1 (Table I). Two of the PP2C-IPs are closely related members of an Arabidopsis gene family, namely At5g53160 (PYL8/RCAR3) and At4g01026 (PYL7/RCAR2; Fig. 1). In particular, five clones belonging to the same PYL8/RCAR3 were identified, explaining a strong interaction. BLAST searches in Arabidopsis using the amino acid sequence encoded by both genes as query revealed high similarity with the PYR1-PYL/RCAR gene family recently identified (Ma et al., 2009; Park et al., 2009b) and composed of 14 members. All members encode small proteins, ranging between 165 and 221 amino acid residues (approximately 20 kD), characterized by the presence of the well-conserved BetV I family signature in many members of the family (Fig. 2A), as revealed by protein domain analysis using the InterPro database (http://www.ebi.ac.uk/interpro). This signature is present in different plant allergens, and specifically, the BetV I family corresponds to particularly potent allergens from white birch (Betula verrucosa) pollen.
Table I.
FsPP2C1-interacting proteins obtained by yeast two-hybrid assay in Arabidopsis
| Clone | Arabidopsis Genome Initiative No. | Sequence | No. of Clones |
|---|---|---|---|
| PYL8/RCAR3 | At5g53160 | −46…691 | 5 |
| PYL7/RCAR2 | At4g01026 | 24…556 | 1 |
| PP2C-IP3 |
At2g23380 (CLF) |
6…983 |
1 |
Figure 1.
Yeast two-hybrid assay for FsPP2C1-interacting proteins. Two-hybrid interaction test of FsPP2C1 (whole protein) with two different proteins, At5g53160 (PYL8/RCAR3) and At4g01026 (PYL7/RCAR2). Left, control plate. Middle and right, test plates (−His,−Met and −His,−Met,+3-aminotriazole [3AT], respectively). For the constructs, the FsPP2C1 was fused to the Gal4 DNA-binding domain (BD) and At5g53160 (PYL8/RCAR3) and At4g01026 (PYL7/RCAR2) were fused to the Gal4 activation domain (AD).
Figure 2.
A, Sequence alignment of PYL8/RCAR3 (At5g53160) protein with other members of the BetV I family in Arabidopsis. Positions with identical amino acid residues are highlighted in black, while similar amino acid residues are colored in gray. The consensus sequence is represented by asterisks, colons, and dots. B, Phylogenetic tree of the plant PYR-PYL/RCAR BetV I family reported to the updated database (http://www.phytozome.net/) with high similarity to Arabidopsis. Accession numbers are shown in Supplemental Table S1. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (Tamura et al., 2007), and the tree has been built using the minimum evolution method. The clades within the family are shaded in different colors.
The phylogenetic relationship between PYL8/RCAR3 and other closely related plant protein PYR-PYL/RCARs is shown in Figure 2B. A phylogenetic tree was constructed using the 14 Arabidopsis PYR-PYL/RCARs and homologs from the genome sequence databases of five lower and higher plants (Physcomitrella patens, Selaginella moellendorfii, Populus trichocarpa, Vitis vinifera, and Oryza sativa; Fig. 2B; Supplemental Table S1). The phylogenetic tree reveals the presence of ancient gene duplication events that predate the speciation of dicots, monocots, and mosses, leading to the six distinct clades indicated in Figure 2B. The data available after recent sequencing projects have revealed the conservation of PYR-PYL/RCAR in lower land plants such as the moss P. patens and in the lycophyte S. moellendorfii. Different PYR-PYL/RCAR subgroups can be identified, and proteins from all plant species analyzed are present in at least one subgroup, suggesting the probable existence of PP2C-IP homologs in all considered genomes as well as in the common ancestor of all the species examined. One of the clades contains exclusively sequences from the lower plants P. patens and S. moellendorfii, suggesting that this lineage of plants contains a copy of BetV I-containing proteins with specialized functions. Additionally, three clades can be clearly distinguished, which include most of the PYR-PYL/RCARs from higher plants where duplications are present and two other clades including Arabidopsis and one or two more higher species.
The high identity found in the BetV I domain and the evolutionary conservation among these PYR-PYL/RCARs support a regulatory function for this domain. Moreover, the interaction of two members of this family with the ABA-related plant type 2C protein phosphatase FsPP2C1 suggests that they might have a role in the ABA signal transduction pathway, but it has not been investigated yet.
Subcellular Localization of the FsPP2C1 and PYL8/RCAR3 Proteins
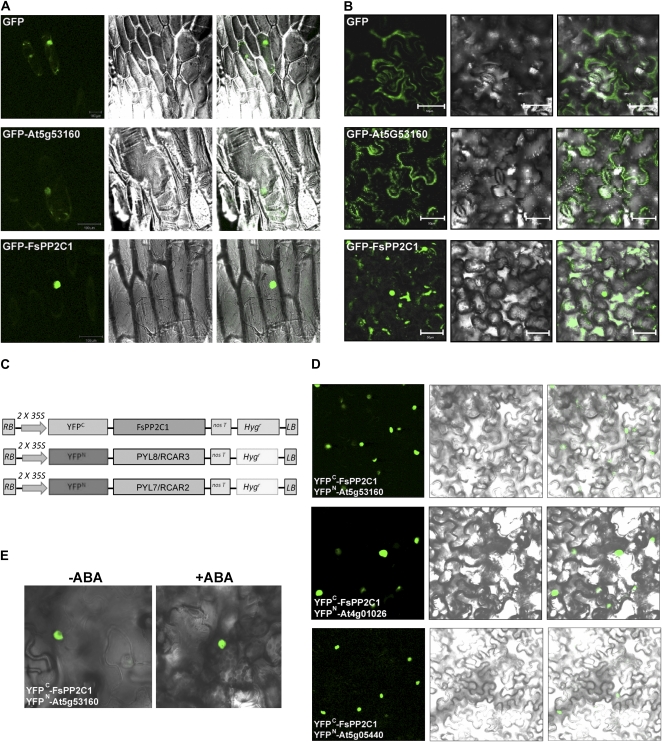
To ascertain the potential site of action of FsPP2C1 and its interacting partner within the cell, the subcellular localization of these proteins was investigated using N-terminal GFP fusions of full-length FsPP2C1 and PYL8/RCAR3. As depicted in Figure 3A, transient expression of these constructs in epidermal onion cells showed that the PYL8/RCAR3 cellular expression pattern was similar to that of the native GFP, demonstrating that the full-length protein appeared in both the cytoplasm and the nucleus. Furthermore, FsPP2C1 was mostly localized in the nucleus. The exact localization of this protein was corroborated with confocal microscopy slides of cell sections through the nucleus and showing the expression of GFP-FsPP2C1. Interestingly, C-terminal GFP fusions of FsPP2C1 altered the subcellular localization of the native protein in tobacco (Nicotiana benthamiana) and Arabidopsis cells, indicating the presence of signals in this end that might be relevant for its localization pattern (Supplemental Fig. S1).
Figure 3.
Subcellular localization of FsPP2C1, PYL8/RCAR3 (At5g53160), and the protein interactions between FsPP2C and PYL/RCAR proteins determined in leaf epidermis of tobacco plants. A, Transiently transformed epidermal onion cells. Confocal microscopy slides of cell sections show the exact localization of the GFP-FsPP2C1 and GFP-At5g53160 proteins in epidermal onion cells. B, Agroinfiltration analysis in tobacco leaves. Constructs delivered in both cases correspond to N-terminal GFP fusions of full-length FsPP2C1 (35S:GFP-FsPP2C1) and At5g53160 (35S:GFP-At5g53160). Nuclear and cytosolic distribution of the GFP protein alone is shown as a control (35S:GFP). C, Constructs used in the transformation of tobacco. LB, Left border; RB, right border. D, BiFC detection of protein-protein interactions in subcellular locations. The interactions between YFPn-At5g53160 and YFPc-FsPP2C1 (top), YFPn-At4g01026 and YFPc-FsPP2C1 (middle), and YFPn-At5g05440 and YFPc-FsPP2C1 (bottom) are shown. E, Confocal microscopy images showing the exact localization of the FsPP2C1-At5g53160 interaction in the nucleus in the absence and presence of ABA.
In a similar way, agroinfiltration analysis of the same constructs delivered in tobacco leaves confirmed the same subcellular localization for FsPP2C1, highlighting the nuclear pattern and the localization for the FsPP2C1-interacting partner (PYL8/RCAR3) appearing in the whole cell (Fig. 3B).
In Planta Interaction between FsPP2C1 and PYL/RCAR Proteins
Bimolecular fluorescence complementation (BiFC) assays were used to detect the interaction between FsPP2C1 and the PYL/RCAR proteins PYL8/RCAR3 (At5g53160) and PYL7/RCAR2 (At4g01026) in plant cells. With this purpose, FsPP2C1 was translationally fused to the C-terminal 84-amino acid region of the yellow fluorescent protein (YFPC) in the pYFPC43 vector (derived from pMDC43; Curtis and Grossniklaus, 2003), which generated a YFPC-FsPP2C1 fusion protein (Fig. 3C). Moreover, At5g53160 and At4g01026 were translationally fused to the N-terminal 155-amino acid region of the yellow fluorescent protein (YFPN) in the pYFPN43 vector, which generated the YFPN-At5g53160 and YFPN-At4g01026 fusion proteins, respectively (Fig. 3C). The corresponding constructs were codelivered into tobacco leaf cells by Agrobacterium tumefaciens infiltration, and as a result, fluorescence was observed in the nucleus of tobacco cells (Fig. 3, D and E), in agreement with the previous finding in the two-hybrid assay and subcellular localization. Similar results were obtained when another member of the PYR1-PYL/RCAR family of interacting partners, PYL5/RCAR8 (At5g05440), was used for BiFC assays (Fig. 3D), showing the specificity of the FsPP2C1-PYL/RCAR family interaction.
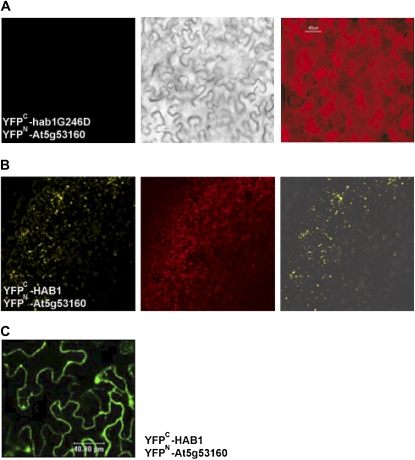
Additionally, we corroborated the feasibility of these PP2C-interacting partners by analyzing the interaction between PYL8/RCAR3 and members of the Arabidopsis clade A PP2Cs to whom FsPP2C1 is the closest ortholog. Therefore, we used HAB1 and the mutated version hab1G246D, corresponding to the same mutation found in the dominant abi1-1 and abi2-1 mutants (Fig. 4). Interestingly, we were able to also demonstrate the interaction between PYL8/RCAR3 and HAB1, while the mutation hab1G246D was able to abolish this interaction (Fig. 4, A and B). These in vivo targeting experiments also determined the subcellular localization of the interaction in both the nucleus and the cytosol (Fig. 4C), which is quite similar to that described for HAB1 (Sáez et al., 2008).
Figure 4.
Subcellular localization of the protein interaction between Arabidopsis HAB1 and PYL8/RCAR3 (At5g53160) determined in leaf epidermis of tobacco plants. A, No BiFC detection of protein-protein interactions was found between the mutated version of HAB1 (YFPc-hab1G246D) and YFPn-At5g53160. B, The interaction between YFPn-At5g53160 and YFPc-HAB1 is shown. C, Confocal microscopy image showing the exact localization of the HAB1-At5g53160 interaction in the cell.
The specific nuclear localization of the FsPP2C1-PYL8/RCAR3 and the rest of the FsPP2C1-PYL/RCAR interactions suggest an interesting function for these complexes in ABA signaling and ABA-regulated gene expression.
Generation and Characterization of 35S:PYL8/RCAR3 Transgenic Lines
Previously, we had shown that FsPP2C1 was strongly up-regulated by ABA treatment, specifically expressed in ABA-treated dormant seeds, and that this expression was negatively correlated with germination (Lorenzo et al., 2001). The expression of the PP2C-interacting partner PYL8/RCAR3 (At5g53160) displayed a pattern largely opposite to that of FsPP2C1 and was down-regulated by ABA. Gene expression analysis using Genevestigator software (http://www.genevestigator.ethz.ch; Zimmermann et al., 2004) revealed that expression of PYL8/RCAR3 was strongly down-regulated by salt and osmotic stress as well as by ABA treatment (Supplemental Fig. S2A). In contrast with most of the other PYR-PYL/RCAR members, PYL7/RCAR2 (At4g01026) seems to behave differently in the gene expression profiles from Genevestigator shown in Supplemental Figure S2A, since an increased expression in response to ABA, salt, osmotic, or drought stress is observed. Furthermore, using the eFP browser from BAR (http://bar.utoronto.ca), we compared the transcription levels of PYL8 (At5g53160) versus PYL7 (At4g01026) in different plant tissues and developmental stages (Supplemental Fig. S2, B and C), showing a different expression pattern mainly in seeds (dry and imbibed) and seedlings, which may indicate different functions for some members of this new family.
Taking into account that homologous genes of the same family usually present functional redundancy to varying degrees, we have chosen a gain-of-function approach to provide genetic evidence on the role of the PYL8/RCAR3 gene in ABA signaling by generating 35S:PYL8/RCAR3 transgenic lines to further analyze the ABA responses in them (Fig. 5).
Figure 5.
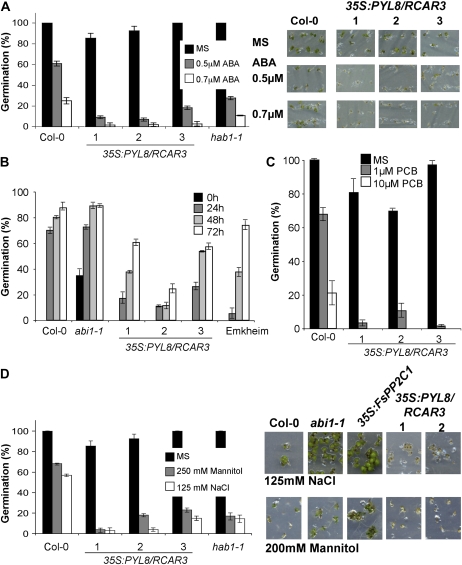
Seed-specific phenotypes of 35S:PYL8/RCAR3 plants. A, ABA-hypersensitive inhibition of germination in 35S:At5g53160 lines as compared with wild-type plants. Percentage of seed germination and early seedling growth in the presence of the indicated ABA concentrations are shown. Approximately 200 seeds of Col-0 and three independent 35S lines (1–3) were sown and scored 8 d later. Photographs show wild-type (Col-0) and three 35S:At5g53160 independent lines (1–3) in medium lacking or supplemented with 0.5 and 0.7 μm ABA 8 d after sowing. B, Dormancy assay of PYL8/RCAR3-overexpressing seeds. Germination percentage was determined at 5 d after seed stratification at 4°C for 0 h (black bars), 24 h (dark gray bars), 48 h (light gray bars), and 72 h (white bars). Three independent PYL8/RCAR3 transgenic lines in wild-type (Col-0) background were used, as well as the abi1-1 mutant and wild-type plants with different degrees of dormancy (Col-0 and Emkheim). C, Germination assay in the presence of the GA biosynthesis inhibitor paclobutrazol (PCB). Germination rating is represented as the percentage of seeds that germinated and developed green cotyledons in the presence of 1 and 10 μm paclobutrazol. Three independent 35S:PYL8/RCAR3 transgenic lines in wild-type (Col-0) background were used (1–3), as well as the indicated wild-type plants. Seeds were scored 5 d after sowing. D, Stress germination assays. Percentage of seed germination and early seedling growth in MS medium or medium supplemented with either 250 mm mannitol or 125 mm NaCl are shown. Seeds were scored 4 d after sowing. Photographs show wild-type (Col-0), abi1-1, 35S:FsPP2C1 (González-García et al., 2003), and two 35S:At5g53160 independent lines (1 and 2) in medium supplemented with 125 mm NaCl or 200 mm mannitol. Data are averages ± se from three independent experiments.
To this end, the coding region of PYL8/RCAR3 was fused to the cauliflower mosaic virus 35S promoter in the binary vector pBIN121, and the construct was used to transform Arabidopsis plants (Columbia [Col-0] background; Supplemental Fig. S3A).
Most of the 35S:PYL8/RCAR3 T1 kanamycin-resistant lines recovered showed an ABA-hypersensitive phenotype to different degrees in seed germination, as compared with untransformed plants or plants transformed with the empty vector (data not shown). For each line, T2 seeds were plated on selection medium to test the segregation ratio of kanamycin resistance. Finally, six T3 homozygous lines were selected, and Southern-blot analysis displayed double and single insertions of the 35S:PYL8/RCAR3 transgene (Supplemental Fig. S3B). Expression levels of the 35S:PYL8/RCAR3 transgene in three different lines analyzed by quantitative (Q)-PCR presented in Supplemental Figure S3C showed substantially higher (3- to 8-fold) expression compared with wild-type plants, as expected. Out of the T3 homozygous lines obtained, three lines with a single insertion were selected for further analysis.
Constitutive Expression of PYL8/RCAR3 in Arabidopsis Confers ABA Hypersensitivity in Seeds
The specific interaction of PYL/RCAR with FsPP2C1, which is up-regulated by ABA in beech seeds (Lorenzo et al., 2001), prompted us to test whether constitutive expression of PYL8/RCAR3 in Arabidopsis would affect ABA sensitivity in seeds.
Seed germination of PYL8/RCAR3-overexpressing transgenic plants in medium supplemented with ABA is shown in Figure 5A. At 8 d poststratification, radicle emergence and development of green and expanded cotyledons was mostly inhibited in 35S:PYL8/RCAR3 seeds at 0.5 μm ABA (and also under 0.7 μm ABA), while control Col-0 seeds showed nearly 70% germination in 0.5 μm ABA. To further substantiate this result, we compared the germination of ABA-hypersensitive mutants (hab1) and wild-type seeds with that of 35S:PYL8/RCAR3 transgenics in medium supplemented with ABA (Fig. 5A). The addition of 0.5 μm ABA decreased the percentage of germination in hab1-1 to 25%, similar to the germination observed in 35S:PYL8/RCAR3 transgenic lines. Clearly, under a concentration not completely inhibitory for the wild type (0.5 μm ABA), PYL8/RCAR3-overexpressing lines reached less than 20% germination. 35S:PYL8/RCAR3 seeds were unable to germinate and grow under this low ABA concentration; therefore, they showed enhanced sensitivity to ABA.
Mature Arabidopsis seeds exhibit primary dormancy when freshly released from the mother plant, which means that seeds are unable to germinate under the appropriate environmental conditions unless they are treated with dormancy-breaking treatments such as stratification or gibberellins (GAs; Koornneef and Karssen, 1994). To determine the degree of dormancy of 35S:PYL8/RCAR3 seeds, we compared the germination percentage of the seeds harvested at the same time after different cold treatment periods (0, 24, 48, and 72 h) with that of two wild-type plants with different degrees of dormancy (Col-0 and Emkheim) and with ABA-related mutants that produce nondormant seeds (abi1-1). As a result, all of the PYL8/RCAR3 transgenic lines exhibited an enhanced degree of dormancy compared with the Col-0 wild type, very similar to that of the Emkheim ecotype but significantly different from the abi mutants (Fig. 5B). In the absence of stratification at 4°C, PYL8/RCAR3 transgenic seeds were unable to germinate, reaching 10% to 25% germination after 1 d of treatment, suggesting that this protein may be related to ABA-regulated dormancy.
To fully characterize the hypersensitive phenotype of 35S:PYL8/RCAR3 seeds, an additional seed germination assay was carried out in the presence of paclobutrazol, a well-known inhibitor of GA biosynthesis. GAs are antagonistic to ABA; therefore, seeds with reduced sensitivity to ABA (or diminished ABA levels) show paclobutrazol-resistant germination (Koornneef et al., 1998). In contrast to the wild type, 35S:PYL8/RCAR3 seeds were unable to germinate and develop green cotyledons in medium supplemented with 1 and 10 μm paclobutrazol, indicating that they are more dependent on GA biosynthesis at this developmental stage (Fig. 5C).
These results demonstrate that 35S:PYL8/RCAR3 seeds exhibit an enhanced degree of seed dormancy and sensitivity to inhibition of germination by exogenous ABA and paclobutrazol, suggesting that PYL8/RCAR3 might be involved as a positive regulator in ABA responsiveness in seeds.
35S:PYL8/RCAR3 Seeds Are Hypersensitive to Salt and Osmotic Stresses
It has been suggested previously that ABA regulates many different stress responses. To test whether the ABA sensitivity induced by overexpression of PYL8/RCAR3 during germination is also effective against other ABA-mediated stresses that increase ABA levels, we analyzed the seed germination response of PYL8/RCAR3 transgenic lines in the presence of low concentrations of NaCl and mannitol (Fig. 5D) and compared it with that of wild-type plants.
Seed germination under 250 mm mannitol and 125 mm NaCl leads to a severe delay in radicle emergence and further growth arrest in 35S:PYL8/RCAR3 individuals; in contrast, wild-type seeds were able to germinate and develop green cotyledons under such conditions (Fig. 5D). These results indicate that PYL8/RCAR3-expressing seeds are neither osmotolerant nor resistant to low salt concentration in this germination assay.
Effect of PYL8/RCAR3 Overexpression in Vegetative Tissues
To determine the extent to which PYL8/RCAR3 overexpression could affect whole plant phenotypes, we analyzed ABA sensitivity in vegetative tissues of the ABA-related mutant abi1-1, three 35S:PYL8/RCAR3 transgenic lines, and wild-type plants.
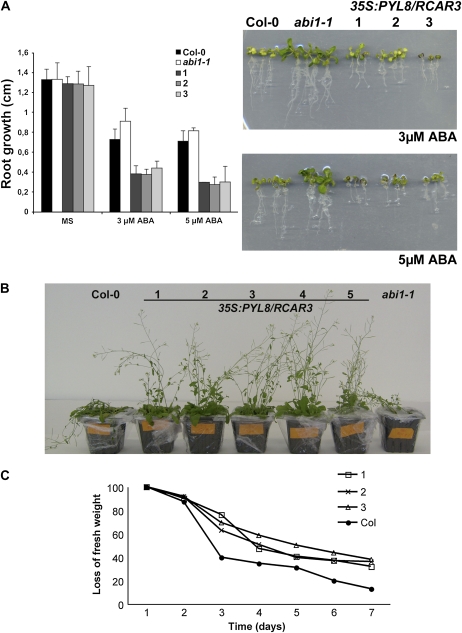
ABA has an inhibitory effect on root growth and, consequently, ABA-insensitive mutants and FsPP2C1-overexpressing plants are resistant to this ABA-mediated process (Himmelbach et al., 1998; González-García et al., 2003). Twelve-day-old 35S:PYL8/RCAR3 seedlings grown in the presence of 10 and 30 μm ABA showed an enhanced inhibition of root growth compared with wild-type plants and to a greater extent with that of abi mutants (Fig. 6A). When grown in the absence of ABA, the PYL8/RCAR3-overexpressing plants did not display any visible phenotypic alteration.
Figure 6.
Vegetative phenotypes of 35S:PYL8/RCAR3 plants. A, Root growth inhibition assay for scoring ABA hypersensitivity. Growth of Col-0 and abi1-1 plants and three independent 35S:PYL8/RCAR3 lines (1–3) in medium lacking (MS) or supplemented with 3 and 5 μm ABA is shown. Data are averages ± se from three independent experiments (n = 20 seedlings per experiment). Photographs show representative seedlings 7 d after the transfer of 5-d-old seedlings from MS to plates supplemented with 3 and 5 μM ABA. B, Whole plant transpiration assay. Photograph showing the drought phenotype of five independent 35S:PYL8/RCAR3 lines (1–5) compared with wild-type and abi1-1 plants. C, Loss of fresh weight measured in whole plants of either Col-0 or three independent 35S:PYL8/RCAR3 lines (1–3). Data values represent one of three independent experiments with similar results, and se in all the genotypes and time points is less than 5%.
ABA triggers stomatal closure and consequent reduction in water loss under drought conditions. The Arabidopsis ABA-insensitive mutants abi1-1 and abi2-1 are impaired in the ABA-induced stomatal closure and, therefore, in their ability to limit transpiration upon drought. On the contrary, PYL8/RCAR3 overexpression did affect stomatal regulation, since 35S:PYL8/RCAR3 plants showed lower rates of transpiration than wild-type plants under drought conditions (not watered after 7 d), which lost approximately 80% fresh weight after 7 d (Fig. 6, B and C).
Altered Expression of ABA-Responsive Genes in PYL8/RCAR3 Transgenics
To examine whether the enhancement in ABA sensitivity in transgenic plants was accompanied by altered expression of ABA-responsive genes, we compared the expression of several previously reported ABA-inducible genes, RAB18, Δ1-pyrroline-5-carboxylate synthase (P5CS1), RD29A, and RD29B, in 35S:PYL8/RCAR3 with that of Col-0 and abi1-1 (Fig. 7). RAB18 (Lang and Palva, 1992; Parcy et al., 1994) is an ABA-inducible gene whose expression is drastically inhibited both in abi1-1 and abi2-1 mutants (Leung et al., 1997), and accumulation of P5CS1 mRNA is induced by drought, salinity, and ABA and is reduced by approximately 50% in the abi1-1 mutant compared with wild-type plants (Strizhov et al., 1997). Similarly, RD29A and RD29B are cold-, drought-, and ABA-inducible genes that contain ABA-responsive elements in their promoters. RD29A contains in addition the dehydration-responsive element, which is involved in a rapid response to conditions of dehydration or high salt (Yamaguchi-Shinozaki and Shinozaki, 1994).
Figure 7.
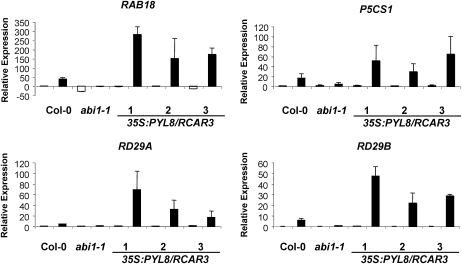
Expression of the ABA-regulated RAB18, P5CS1, RD29A, and RD29B genes in 35S:PYL8/RCAR3 plants (1–3) compared with the wild type (Col-0 transgenic plants) and abi1-1 controls. mRNA levels of the indicated genes were determined by Q-PCR analysis using total RNAs isolated from mock-treated plants (white bars) or 10 μm ABA-treated plants (black bars) for 3 h.
Regarding this effect, 35S:PYL8/RCAR3 plants behave opposite to the abi1-1 mutant, as they showed a high induction (3- to 5-fold) in the expression of RAB18, P5CS1, RD29A, and RD29B upon ABA treatment from that detected in wild-type plants.
Taken together, these results indicate that expression of PYL8/RCAR3 in vegetative tissues increases ABA-responsive gene expression and is also linked to ABA-related physiological responses such as enhanced dormancy level, stress-resistant germination, inhibition of root growth, and desiccation tolerance.
Overexpression of Both PYL8/RCAR3 and FsPP2C1 Show Enhanced Response to ABA
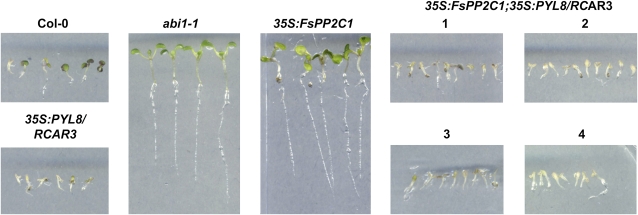
To further assess the real function of PYL8/RCAR3 in planta and its relevance in the ABA signal transduction pathway, we generated double transgenic lines that overexpressed both FsPP2C1 and PYL8/RCAR3. Functional analysis of these plants has also shown an enhanced response to ABA during seed germination and early seedling growth, similar to PYL8/RCAR3-overexpressing lines and greatly different from the ABA insensitivity displayed by the gain-of-function 35S:FsPP2C1 lines or the dominant abi1-1 mutant (Fig. 8). These results confirm that PYL8/RCAR3 is able to antagonize FsPP2C1 function in the plant.
Figure 8.
Seed-specific phenotypes of double transgenic 35S:FsPP2C1;35S:PYL8/RCAR3 plants. ABA-hypersensitive inhibition of germination in 35S:FsPP2C1;35S:PYL8/RCAR3 lines as compared with wild-type plants is shown. Seed germination and early seedling growth in the presence of 0.5 μm ABA concentration are also shown. Approximately 200 seeds of Col-0 and four independent 35S lines (1–4) were sown and scored 8 d later. Photographs show wild-type (Col-0) and four 35S:At5g53160 independent lines (1–4) in medium supplemented with 0.5 μm ABA 8 d after sowing. [See online article for color version of this figure.]
DISCUSSION
ABA is one of the main factors controlling seed formation, dormancy, and germination (Koornneef and Karssen, 1994; Bewley, 1997) and abiotic stress responses (López-Molina et al., 2001; González-Guzmán et al., 2002). Genetic screens used to find new components of ABA signaling associated with germination have recently identified many loci that are involved in the removal of sensitivity to ABA function (Nishimura et al., 2004, 2007; Zhang et al., 2005; Pandey et al., 2006; Sáez et al., 2006; Yoine et al., 2006). These loci encode diverse functions, including those associated with protein degradation pathways (López-Molina et al., 2003; Zhang et al., 2005) and kinase/phosphatase components of signaling pathways.
PP2Cs are key enzymes in ABA signaling, since Arabidopsis PP2C mutants show altered responses to ABA-regulated processes. We have previously isolated and characterized a PP2C from beechnut, FsPP2C1, with an important role in ABA-mediated signal transduction in both seed dormancy/germination and different abiotic stress responses (Lorenzo et al., 2001). Although there is much evidence on the function of PP2Cs in ABA signaling, studies on their substrates/partners, and therefore the way they act into the cell, are very scarce. Our previous published results (González-García et al., 2003) revealed that 35S:FsPP2C1 plants behave as abi1-1 and abi2-1 mutants, as they showed a severe reduction in the expression of RAB18 upon ABA treatment. Taking these results together, we conclude that FsPP2C1 functions as a negative regulator of ABA signaling in Arabidopsis plant tissues (even as a foreign protein), and we reasoned that the PP2C from beech might possess at least the corresponding signaling components (substrate, activator, or partner) in this plant model species to achieve its function.
With these premises, we started the search for putative cell substrates of FsPP2C1 by yeast two-hybrid analysis (Table I; Fig. 1). Thus, we have identified some proteins interacting with FsPP2C1 (PP2C-IPs) and characterized one of them, PYL8/RCAR3. PYL8/RCAR3 belongs to the BetV I family (Fig. 2A), characterized by the presence of a well-conserved signature found in different plant allergens and particularly potent in the case of white birch pollen. BetV I is a representative of the group 10 family of pathogenesis-related (PR) proteins, and other PR-10 members also display the BetV I signature (Iturriaga et al., 1994; Srivastava et al., 2006). However, we have found that this structural motif is also present in many members of the Arabidopsis family hereby identified, which are not related to PR-10 proteins, as shown by amino acid sequence alignments (Fig. 2B). Further NMR analysis confirmed earlier predictions of IgE-binding epitopes of BetV I and revealed a protein structure composed of six antiparallel β-strands and three α-helices (Faber et al., 1996; Markovic-Housley et al., 2003; Spangfort et al., 2003), which defines a hydrophobic cavity involved in the binding of lipid molecules (Radauer et al., 2008).
Recently, Ma et al. (2009) identified a 14-member protein family, named RCAR, in a yeast two-hybrid screen for plant proteins that interact with PP2Cs that negatively regulate ABA signaling. RCAR proteins bind ABA and block the phosphatase activity of PP2Cs in an ABA-dependent manner in vivo. The ABA affinity of the RCAR-PP2C protein complex is much higher than that of RCAR, which is consistent with a heteromeric receptor complex. Concomitantly, Park et al. (2009b) identified the same 14-member family of proteins, named PYR1 and PYL (for PYR-like), which are functionally redundant and mediate multiple ABA responses in vivo. Furthermore, using biochemical and genetic approaches, they found that ABA promotes the interaction of PYR1 with group A PP2Cs, leading to inhibition of PP2C enzymatic activity. In light of these results, both teams conclude that PYR1, PYLs, and RCARs form a large subfamily of cytosolic ABA receptors belonging to the BetV I family that act by binding and inhibiting type 2C protein phosphatases, thus triggering a reversible protein phosphorylation cascade that mediates ABA responses (Ma et al., 2009, Park et al., 2009b).
Studies of orthologous genes and functional tests in heterologous systems have shown that the ABA signal transduction pathway is mostly conserved among evolutionarily distant plant species (for review, see Finkelstein et al., 2002; González-García et al., 2003). The phylogenetic analysis described here shows that the PYR-PYL/RCAR gene family has been subjected to several gene duplication events and that some of these duplication events are ancient, predating the radiation of the major groups of land plants. Such gene duplication events were often followed by diversification of protein function; thus, the presence of the ancient clades shown in Figure 2B suggests that some members of the BetV I family have distinct functions. The phylogenetic analysis will be useful for determining orthology relationships among the genes and identifying targets for future functional analyses.
The site of action of PP2Cs in ABA responses is unknown. In a recent report, Moes et al. (2008) described the nuclear localization of ABI1 wild-type and mutant proteins and the requirement of a functional nuclear localization sequence in order to regulate ABA sensitivity and ABA-dependent gene expression, and they suggested that ABI1 reprograms sensitivity toward ABA in the nucleus. Similarly, Sáez et al. (2008) confirmed the interaction of HAB1 and SWI3B (an Arabidopsis homolog of the yeast SWI3 subunit of SWI/SNF chromatin-remodeling complexes) in the nucleus and suggested a direct involvement of HAB1 in the regulation of ABA-induced transcription. Remarkably, one of the ABI1 and HAB1 ortholog genes in beech (FsPP2C1) is also localized mainly in the nucleus (Fig. 3, A and B), although the consensus sequence differs from that described in the C-terminal extension of ABI1 (Lorenzo et al., 2001). The N-terminal extension of the PP2Cs has been suggested to facilitate the interaction with different substrates (Rodríguez, 1998) and N-terminal deletions of ABI1 and AtPP2CA, leading to enhanced PP2C activity, which suggests a regulatory function for this domain (Sheen, 1998). Interestingly, evidence that the AtPP2CA C-terminal (catalytic) domain is directly involved in the interaction with ATK2 (Cherel et al., 2002) and ATK3 (Vranova et al., 2001) has been provided.
BiFC analysis provides in planta evidence for the interaction of FsPP2C1 and several members of the PYR-PYL/RCAR family (Fig. 3, D and E). While FsPP2C1 subcellular location is mainly in the nucleus, PYL8/RCAR3 is localized in both nucleus and cytosol; however, the BiFC assay clearly identified a nuclear colocalization of both proteins. Interestingly, the interaction between FsPP2C1 and PYL7/RCAR2 and PYL5/RCAR8 is also present mostly in the nucleus. It is noteworthy that most of the targets previously identified for clade A PP2Cs were not nuclear proteins (Cherel et al., 2002; Guo et al., 2002; Ohta et al., 2003; Miao et al., 2006; Yang et al., 2006; Yoshida et al., 2006a). However, it is supposed that the interaction of ABI1 with the transcription factor ATHB6 must be nuclear (Himmelbach et al., 2002).
Although FsPP2C1 and the corresponding PP2C orthologs belonging to clade A (Schweighofer et al., 2004) have been described as negative regulators of ABA signaling, they are strongly induced by ABA as a sort of negative feedback mechanism (Lorenzo et al., 2001; Merlot et al., 2001; González-García et al., 2003). Conversely, expression of PYL8/RCAR3 was strongly down-regulated by salt and osmotic stress as well as ABA treatment (Supplemental Fig. S2A). Therefore, expression of clade A PP2Cs and most PYR-PYL/RCARs mirrors an opposite pattern that would fit into the negative feedback regulatory mechanism, taking into account that PYR-PYL/RCAR proteins inhibit the activity of clade A PP2Cs (Ma et al., 2009; Park et al., 2009b; Santiago et al., 2009; Szostkiewicz et al., 2009).
In contrast with most of the other PYR-PYL/RCAR members, PYL7/RCAR2 (At4g01026) displays an opposite gene expression profile (Supplemental Fig. S2A), with increased expression in response to ABA, salt, osmotic, or drought stress. A comparison of the transcription levels of PYL8 (At5g53160) versus PYL7 (At4g01026) in different plant tissues and developmental stages (Supplemental Fig. S2, B and C) highlights the different expression patterns in seed development, dry and imbibed seeds, and seedlings, which may indicate different functions for some members of this new family.
The features of PYL8/RCAR3 (i.e. interaction with ABA-regulated PP2Cs and ABA down-regulation) made it a suitable candidate potentially involved in aspects of ABA signaling in seeds and, consequently, as a regulator of seed dormancy. Genetic evidence was necessary to assess whether PYL8/RCAR3 functions as a positive or a negative regulator of seed dormancy, germination, and stress responses. In this study, we used a gain-of-function approach in Arabidopsis to investigate PYL8/RCAR3 function (Supplemental Fig. S3).
Overexpression of PYL8/RCAR3 in Arabidopsis results in hypersensitivity to ABA in seeds. Based on these results, we conclude that PYL8/RCAR3 is a positive regulator of ABA signaling, since constitutive expression in Arabidopsis leads to enhanced seed responses to ABA and, consequently, to enhanced dormancy (Fig. 5).
Unlike abi mutants, seeds of 35S:PYL8/RCAR3 transgenic plants displayed enhanced dormancy (Fig. 5B), which is indicative of a higher responsiveness to endogenous ABA in seeds (Gosti et al., 1999). Indeed, whereas the germination of wild-type Arabidopsis seeds was not suppressed by 0.5 μm ABA, PYL8/RCAR3-overexpressing transgenic seeds were unable to display radicle emergence and develop green and expanded cotyledons under these conditions (Fig. 5A), as the ABA-hypersensitive mutant hab1-1 does (Sáez et al., 2006). Taken together, these data are consistent with a role of PYL8/RCAR3 as a positive regulator of ABA signaling in seeds. In addition to down-regulation by ABA, PYL8/RCAR3 expression also decreased after unfavorable conditions, such as salt and osmotic stresses, suggesting a role for this gene during the stress response (Supplemental Fig. S2).
The reliability of the physiological role of PYL8/RCAR3 is also reinforced by the germination assay in the presence of the GA biosynthesis inhibitor paclobutrazol (Fig. 5C). GAs and ABA play antagonistic roles in seed germination, and apparently, ABA-insensitive (or ABA-defective) individuals need less GA during germination (Léon-Kloosterziel et al., 1996). Clearly, the GA requirement was enhanced in 35S:PYL8/RCAR3 seeds compared with wild-type seeds (Fig. 5C), suggesting that expression of PYL8/RCAR3, by the promotion of ABA signaling, negatively regulates seed germination.
Another important role of ABA is the prevention of seed germination under unfavorable water conditions (González-Guzmán et al., 2002). Compelling evidence has shown that osmotic stress delays seed germination and arrests early seedling development. Several studies show that ABA plays an inhibitory role in these processes, since both ABA-insensitive and ABA-deficient mutants are able to bypass them (Werner and Finkelstein, 1995; Léon-Kloosterziel et al., 1996; López-Molina et al., 2001; González-Guzmán et al., 2002), while ABA-hypersensitive mutants are not (Sáez et al., 2006). As previously observed with hab1 (Sáez et al., 2004), PYL8/RCAR3-expressing lines were unable to germinate and carry out early growth in medium with the organic solute mannitol (200 mm) as well as low salt concentration (125 mm NaCl) compared with the germinative ability of wild-type plants (Fig. 5D). These results indicate that PYL8/RCAR3 produces a general hypersensitivity to osmotic stress and are in agreement with the finding that PYL8/RCAR3 transcripts are highly affected by water deficit in Arabidopsis (Supplemental Fig. S2). Thus, PYL8/RCAR3 overexpression in Arabidopsis promotes the inhibition of seed germination under low water potential conditions and contributes to the stress response.
The role of ABA in the promotion of stomatal closure is a well-characterized response in vegetative tissues, reducing water loss by transpiration during drought. Several Arabidopsis mutants are impaired in the ABA-induced stomatal closure in Arabidopsis (abi1, abi2, gca2, and ost1) and therefore in their ability to limit transpiration upon drought (Pei et al., 2000; Mustilli et al., 2002). By measuring the loss of total water, we confirmed that the transpiration rate of PYL8/RCAR3-overexpressing plants, and consequently the stomatal closure, differs from that of wild-type plants (Fig. 6). Indeed, after 7 d under drought conditions (no-water conditions), 35S:PYL8/RCAR3 plants remained green and turgid, whereas the abi1-1 mutant and the wild type (Col-0) showed wilting. These results indicate that ABA responsiveness by PP2C-IP1 overexpression is also maintained in adult plants. Further analysis of ABA responsiveness in vegetative tissues indicated that inhibition of early root growth by 3 to 5 μm ABA was enhanced in 35S:PYL8/RCAR3 seedlings, demonstrating once again that PYL8/RCAR3-overexpressing seedlings are more sensitive to ABA at this stage of development. The inhibitory effect of high ABA concentrations on root growth has been attributed to activation of the ethylene response pathway (Beaudoin et al., 2000; Ghassemian et al., 2000).
The activation of the ABA signal in 35S:PYL8/RCAR3 plants is further sustained by the enhanced induction of the ABA-responsive genes RAB18, P5CS1, RD29A, and RD29B (Fig. 7). RAB18 is a dehydrin only found in ABA-treated plants and accumulates in Arabidopsis dry seeds (Nylander et al., 2001). abi1-1 and abi2-1 mutants show impaired induction of this gene (Leung et al., 1997) that is induced in antisense AtPP2CA plants (Tahtiharju and Palva, 2001). P5CS1 is a stress-regulated protein whose transcriptional regulation and gene expression are induced by drought, salinity, and ABA; also, it is reduced by approximately 50% in the abi1-1 mutant compared with wild-type plants (Strizhov et al., 1997). RD29A and RD29B are cold-, drought-, and ABA-inducible genes that contain ABA-responsive elements in their promoters. RD29A contains in addition the dehydration-responsive element, which is involved in a rapid response to conditions of dehydration or high salt (Yamaguchi-Shinozaki and Shinozaki, 1994).
It is widely accepted that ABA responses depend on coordinated interactions between positive and negative regulators, which are required for the proper control of this complex signaling pathway that operates in a cell. Many transcriptional factors of ABA-inducible genes are known, including ABA-responsive element-binding proteins (ABA-INSENSITIVE5 [ABI5]/ABF/AREB/AtbZIP family), ABI3/VP1/B3, ABI4/APETALA2, MYC, MYB, and HD-ZIP domain proteins (Giraudat et al., 1992; Suzuki et al., 1997; Finkelstein et al., 1998; Choi et al., 2000; Finkelstein and Lynch, 2000; Uno et al., 2000; Bensmihen et al., 2002; Himmelbach et al., 2002; Abe et al., 2003). Most of these transcriptional factors play a positive role in ABA signaling, but some of them function as repressors of the ABA response (Himmelbach et al., 2002; Pandey et al., 2005; Song et al., 2005). AtSAT32 is involved in both salinity tolerance and ABA signaling as a positive regulator in Arabidopsis (Park et al., 2009a).
In this work, another member of the PYR-PYL/RCAR group of proteins directly interacting with clade A PP2Cs and with a role as a positive regulator of ABA signaling has been clearly identified. Functional evidence using double transgenic plants (Fig. 8) confirms the role of PYL8/RCAR3 by antagonizing FsPP2C1 function in the presence of ABA and supports a role of PYL8/RCAR3 as a positive regulator of ABA signaling in seeds and abiotic stress responses. PYL8/RCAR3 overexpression leads to both enhanced seed dormancy, as compared with untransformed plants, and inhibition of germination under low concentrations of ABA or osmoticum medium. This ABA-hypersensitive phenotype is also relevant at the molecular level, and together with the ABA-repressed expression of PYL8/RCAR3 transcripts, it suggests that PYL8/RCAR3 positively regulates key physiological processes such as germination and abiotic stress through the modulation of ABA signaling. A recent report by Szostkiewicz et al. (2009) identified RCAR3 as an interactor of ABI1 and ABI2 able to repress the activity of both PP2Cs, fully supporting the results presented in this work. In addition, Santiago et al. (2009) have reported the function of another member of the PYR1-PYL/RCAR ABA receptor family, PYL5/RCAR8, in the activation of the ABA signaling pathway, also corroborating a role as a positive regulator of ABA signaling.
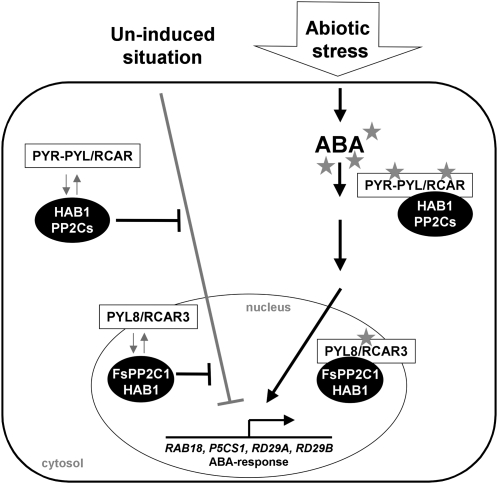
The hypothetical model shown in Figure 9 takes into account that PP2Cs are negative regulators of ABA signaling and that the interaction with this new family of PP2C partners (PYR1-PYL/RCAR) would lead to modification of ABA perception or inactivation of PP2C activity, as reported by Ma et al. (2009), Park et al. (2009b), Santiago et al. (2009), and Szostkiewicz et al. (2009). In terms of functional interaction, a new window is opened, since differences in hormone sensitivity and selectivity have been found between different PYR1-PYL/RCAR members in the ABA receptor complexes (Szostkiewicz et al., 2009). Obviously, other PYL/RCARs of the 14 members belonging to the BetV I family may interact directly with the rest of Arabidopsis clade A PP2Cs (nine members) described so far (i.e. ABI1, ABI2, HAB1, HAB2, and AtPP2CA) to form different perception complexes in relation to the plethora of tissue, developmental, and environmental ABA responses. Finally, in the phenomenon of beechnut life, the putative function of the FsPP2C1-PYL/RCAR complexes may contribute to promote the transition from seed dormancy to germination during the early weeks of stratification, mainly due to changes in ABA perception/sensitivity during these key developmental processes.
Figure 9.
Schematic model describing the function of the PP2C-PYL/RCAR interaction in the control of the ABA signaling pathway. In the absence of ABA, PYR-PYL/RCAR proteins may interact with PP2Cs, but these PP2Cs remain active. Upon ABA perception, PYR-PYL/RCAR proteins interact with PP2Cs, maintaining these PP2Cs as inactive and subsequently activating the transcription of ABA-responsive genes (including PP2Cs). The specific colocalization of FsPP2C1-PYL8/RCAR3 in the nucleus is shown.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) wild-type and transgenic plants used throughout this work were in the Col-0 ecotype. They were routinely grown in a growth chamber under 40% humidity, 22°C temperature, and 16-h-light/8-h-dark photoperiod at 80 to 100 μE m−2 s−1 in pots containing a 1:3 vermiculite:soil mixture. For in vitro culture, seeds were surface sterilized in 75% bleach solution and 0.01% Triton X-100 for 5 min and, finally, washed three times in sterile distilled water. Stratification of the seeds was conducted during 3 d at 4°C, unless otherwise indicated. Afterward, seeds were sown on Murashige and Skoog (1962; MS) plates containing solid medium composed of MS basal salts and 2% Glc, solidified with 0.7% agar, and adjusted to pH 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled-environment growth chamber.
Yeast Two-Hybrid Screening
Bait cloning and yeast two-hybrid screening were performed by Hybrigenics (http://www.hybrigenics.com/services). FsPP2C1 was cloned in an inducible yeast two-hybrid vector optimized by Hybrigenics. The bait construct was transformed in the L40ΔGAL4 yeast strain (Fromont-Racine et al., 1997). An Arabidopsis 1-week-old seedling random-primed cDNA library, transformed into the Y187 yeast strain and containing 10 million independent fragments, was used for mating. High mating efficiency was obtained using a specific mating method (Legrain and Selig, 2000; Legrain, 2002). The screen was first performed on a small scale to adapt the selective pressure to the intrinsic property of the bait. The bait was found to autoactivate the yeast two-hybrid system, and 100 mm 3-aminotriazole was found to be the optimal concentration to reduce background colonies. Then, the full-scale screen was performed in conditions ensuring a minimum of 50 million interactions tested, in order to cover five times the primary complexity of the yeast-transformed cDNA library (Rain et al., 2001). A total of 123 million interactions were actually tested with FsPP2C1. After selection on medium lacking Leu, Trp, and His, seven positive clones were picked, and the corresponding prey fragments were amplified by PCR and sequenced at their 5′ and 3′ junctions. Sequences were then filtered and contiged as described previously (Formstecher et al., 2005) and compared with the latest release of the GenBank database using BLASTN (Altschul et al., 1997). A predicted biological score (PBS) was applied to assess the reliability of each interaction, as described previously (Formstecher et al., 2005). Briefly, the PBS relies on two different levels of analysis. First, a local score takes into account the redundancy and independence of prey fragments as well as the distributions of reading frames and stop codons in overlapping fragments. Second, a global score takes into account the interactions found in all the screens performed at Hybrigenics using the same library. In addition, potential false positives are flagged by a specific “E” PBS. This is done by discriminating prey proteins containing “highly connected” domains, previously found several times in screens performed on libraries derived from the same organism. The PBS scores have been shown to positively correlate with the biological significance of interactions (Rain et al., 2001; Wojcik et al., 2002).
GFP Fusions and Transient Expression Assays
The coding regions of FsPP2C1 and PYL8/RCAR3 were obtained and fused to the C-terminal end of the GFP in the pMDC43 vector using the Gateway technology (Invitrogen).
Following sequence verification of the inserts by DNA sequencing, GFP fusion and control constructs were transiently expressed into epidermal onion (Allium cepa) cells by particle bombardment. DNA absorption to gold particles and bombardment using a helium-driven particle accelerator (PDS-1000/He; Bio-Rad) were performed according to the manufacturer's recommendations. Five micrograms of plasmid was used per transformation, and all target materials were bombarded twice.
Agroinfiltration Analysis
FsPP2C1 was translationally fused to the YFPC in the pYFPC43 vector (derived from pMDC43; Curtis and Grossniklaus, 2003), and At5g53160, At4g01026, and At5g05440 were translationally fused to the YFPN in the pYFPN43 vector using the Gateway technology and primers listed in Supplemental Table S2. The constructs generated were used to transform the C58C1 (pGV2260) Agrobacterium tumefaciens strain (Deblaere et al., 1985) and delivered into leaf cells of tobacco (Nicotiana benthamiana) by Agrobacterium infiltration as described previously (Voinnet et al., 2003), together with the C58C1 (pCH32) Agrobacterium strain to avoid gene silencing. Basically, Agrobacterium strains were resuspended in 10 mm MgCl2, 10 mm MES, pH 5.6, and 200 μm acetosyringone. After infiltration with 2 mL using a syringe, plants were maintained during 3 to 4 d under greenhouse conditions.
Fluorescence Microscopy
The fluorescence photographs of cells expressing the GFP and YFP reporter genes under the control of the 35S promoter were obtained using a Zeiss Axiovert 200 confocal microscope and a Bio-Rad Radiance 2100 laser scanning confocal imaging system with LaserSharp version 5 Image software acquisition. For GFP and YFP detection, the excitation source was an argon ion laser at 488 nm and detection filters ranged from 515 to 530 nm and 500 to 540 nm, respectively.
Vector Construction and Plant Transformation
To generate the constructs, the coding region of the PYL8/RCAR3 cDNA was amplified by PCR using the primers from Supplemental Table S2 and cloned into the pGEM-T vector (Invitrogen). The PYL8/RCAR3 clone was excised from the pGEM-T vector using SacI-XbaI double digestion and subcloned into the doubly digested pBIN121 vector. The T-DNA region of the pBIN121-PYL8/RCAR3 construct was transferred to Agrobacterium C58C1 (pGV2260; Deblaere et al., 1985) by electroporation. Arabidopsis plants (Col-0 ecotype) were transformed by the floral dip method (Clough and Bent, 1998). Seeds were harvested and plated on kanamycin selection medium (50 μg mL−1) to identify T1 transgenic plants. Approximately 100 of the T2 seeds were plated on MS kanamycin agar plates, and transgenic lines with a 3:1 (resistant:sensitive) segregation ratio were selected. Southern-blot analysis was performed as described by González-García et al. (2003) to select lines carrying a single T-DNA copy. T3 progeny homozygous for kanamycin resistance were used for further studies.
Generation of Double Transgenic Lines
The generation of transgenic lines simultaneously overexpressing both FsPP2C1 and PYL8/RCAR3 was performed by transferring pollen from 35S:PYL8/RCAR3 lines to emasculated flowers from different FsPP2C1-overexpressing lines and subsequent selection of the F2 siblings on plates containing 50 μm kanamycin. Coexpression of FsPP2C1 and PYL8/RCAR3 in double transgenic plants was verified by PCR analysis using the primer 35S-F (5′-ACGCACAATCCCACTATCCTTCG-3′) and those indicated in Supplemental Table S2. Additionally, F3 progeny were analyzed in 0.5 and 3 μm ABA and properly genotyped in accordance with ABA sensitivity.
Germination Assays
To measure ABA sensitivity, seeds were plated on solid medium composed of MS basal salts, 2% Glc, and different concentrations of ABA (0.5 and 0.7 μm). For the dormancy assay, seed lots to be compared were harvested on the same day from individual plants grown in identical environmental conditions and stratified during 0, 1, 2, and 3 d at 4°C. Each value represents the average germination percentage of 80 to 100 seeds with the se values of at least three replicates.
To determine sensitivity to inhibition of germination by low osmoticum and salt, the medium was supplemented with 250 mm mannitol and 125 mm NaCl, respectively. To measure paclobutrazol sensitivity, seeds were plated on medium containing 1 and 10 μm paclobutrazol. The percentage of seeds that had germinated and developed fully green expanded cotyledons was determined in all assays after 7 d of sowing.
Root Growth and Transpiration Assays
The root growth assay for scoring ABA sensitivity was done by measuring root growth after 7 d of transferring of 5-d-old seedlings onto vertical MS plates containing 3 and 5 μm ABA. For the transpiration assays, the loss of fresh weight of whole 4-week-old plants at the same developmental stage was measured at room temperature after the indicated periods of time.
Q-PCR Analysis
Seeds from wild-type (Col-0), 35S:PYL8/RCAR3, and abi1-1 plants were grown in 125-mL flasks containing 20 mL of MS solution and 2% Glc. The flasks were shaken at 130 rpm under cool fluorescent light. After 10 d, seedlings were mock treated or treated with 10 μm ABA during 3 h. Plant material was collected and frozen in liquid nitrogen. Total RNA for quantitative reverse transcription (RT)-PCR was extracted from Arabidopsis 10-d-old frozen seedlings using Tri-reagent as described by the manufacturer (Ambion). Extracted RNAs were then subjected to RT. One microgram of DNase-treated total RNA was used for RT with a SuperScript Kit (Roche). The RT product was used for quantitative PCR using the primers designed from the RAB18, P5CS1, RD29A, and RD29B genes with the aid of Primer Express software 1.0 (Applied Biosystems) and listed in Supplemental Table S2. The actin gene β-ACT8 was used as a control. Primers were described previously (Sáez et al., 2006).
Real-time PCR was performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Amplification was carried out with Brilliant SYBR Green QPCR MasterMix (Stratagene) according to the manufacturer's instructions.
The thermal profile for SYBR Green real-time PCR was 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
To generate the standard curves, cDNA isolated from Arabidopsis seedlings was serially diluted by a factor of 10 and aliquots of the dilutions were used in standard real-time PCRs. Each value determination was repeated three times to ensure the slope of the standard curves and to determine the sd. The concentration of unknown samples was calculated with the ABI PRISM 7000 SDS software, which created threshold cycle values and extrapolated relative levels of PCR product from the standard curve. The expression of all genes was normalized against the expression of the endogenous control gene (Actin). All experiments were repeated at least three times and yielded similar results.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ277743 and At5g53160.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of C-terminal GFP fusions of FsPP2C1 in tobacco and Arabidopsis cells.
Supplemental Figure S2. Expression pattern of PYL8 in the context of PYR-PYL/RCAR genes.
Supplemental Figure S3. Generation and molecular analysis of 35S:PYL8/RCAR3 transgenic lines.
Supplemental Table S1. Gene identification numbers of the 14 Arabidopsis PYR-PYL/RCARs in the phylogenetic tree (Fig. 2B), including the PYR-PYL/RCAR homologs of several lower and higher plants.
Supplemental Table S2. Oligonucleotides used in the different constructs (transgenic plants and BiFC) and for Q-PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Pedro L. Rodríguez and his research group for providing material and technical assistance in the BiFC experiments and also for their stimulating discussions. The help and comments of Mike Thon in the phylogenetic analysis are also acknowledged. Seeds from the Emkheim ecotype were kindly provided by M. Koornneef and L. Bentsink.
This work was supported by the Ministerio de Educación y Ciencia of Spain (grant nos. BIO2005–08473, CSD2007–00057 [TRANSPLANTA], and BIO2008–04698 to O.L. and grant no. BFU2006–07622/BFI to D.R.), by the Junta de Castilla y León (grant no. SA065A07 to O.L. and grant no. SA073A08 to D.R.), by a Researcher Training Fellowship (FPI) from the Ministerio de Educación y Ciencia (Spain) to A.M., and by a Marie Curie European Reintegration Grant (grant no. FP7–PEOPLE–ERG–2008 to L.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Oscar Lorenzo (oslo@usal.es).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Messeguer R, Goday A, Pagès M (1999) Plant responses to drought, from ABA signal transduction events to the action of the induced proteins. Plant Physiol Biochem 37 327–340 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, McCourt P (2005) Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol 138 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB (2002) Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275 1723–1730 [DOI] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol (Stuttg) 8 314–325 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber C, Lindemann A, Sticht H, Ejchart A, Kungl A, Susani M, Frank RW, Kraft D, Breitenbach M, Rosch P (1996) Secondary structure and tertiary fold of the birch pollen allergen Bet v 1 in solution. J Biol Chem 271 19243–19250 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59 387–415 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16 277–282 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Rodríguez D, Nicolás C, Rodríguez PL, Nicolás G, Lorenzo O (2003) Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol 133 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3 233–244 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Iten M, Grill E (1998) Signalling of abscisic acid to regulate plant growth. Philos Trans R Soc Lond B Biol Sci 353 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179 33–54 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487 [DOI] [PubMed] [Google Scholar]
- Iturriaga EA, Leech MJ, Barratt DH, Wang TL (1994) Two ABA-responsive proteins from pea (Pisum sativum L.) are closely related to intracellular pathogenesis-related proteins. Plant Mol Biol 24 235–240 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ (1998) Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5 33–36 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM (1989) In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol 90 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM (1994) Seed dormancy and germination. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 313–334
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Palva ET (1992) The expression of a rab-related gene, RAB18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20 951–962 [DOI] [PubMed] [Google Scholar]
- Legrain P (2002) Protein domain networking. Nat Biotechnol 20 128–129 [DOI] [PubMed] [Google Scholar]
- Legrain P, Selig L (2000) Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett 480 32–36 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10 655–661 [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49 199–222 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Rodríguez D, Nicolás G, Rodríguez PL, Nicolás C (2001) A new protein phosphatase 2C (FsPP2C1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiol 125 1949–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068 [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H (2003) Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J Mol Biol 325 123–133 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455 [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54 806–819 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco. Physiol Plant 15 473–497 [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56 165–185 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50 935–949 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Murayama M, Asami T, Shinozaki K, Hirayama T (2004) Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol 45 1485–1499 [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45 263–279 [DOI] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu JK (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136 136–148 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Chung MS, Koh HS, Lee DJ, Ahn SJ, Kim CS (2009. a) Isolation and functional characterization of the Arabidopsis salt-tolerance 32 (AtSAT32) gene associated with salt tolerance and ABA signaling. Physiol Plant 135 426–435 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al (2009. b) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K (2005) Early ABA signaling events in guard cells. J Plant Growth Regul 24 296–307 [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734 [DOI] [PubMed] [Google Scholar]
- Radauer C, Lackner P, Breiteneder H (2008) The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol 8 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, et al (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409 211–215 [DOI] [PubMed] [Google Scholar]
- Rodríguez PL (1998) Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol Biol 38 919–927 [DOI] [PubMed] [Google Scholar]
- Rodríguez PL, Benning G, Grill E (1998. a) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421 185–190 [DOI] [PubMed] [Google Scholar]
- Rodríguez PL, Leube MP, Grill E (1998. b) Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol 38 879–883 [DOI] [PubMed] [Google Scholar]
- Sáez A, Apostolova N, González-Guzmán M, González-García MP, Nicolás C, Lorenzo O, Rodríguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37 354–369 [DOI] [PubMed] [Google Scholar]
- Sáez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodríguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL (2008) HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL (2009) Modulation of drought resistance by the abscisic acid-receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60 575–588 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9 236–243 [DOI] [PubMed] [Google Scholar]
- Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangfort MD, Mirza O, Ipsen H, Van Neerven RJ, Gajhede M, Larsen JN (2003) Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by x-ray crystallography and site-directed mutagenesis. J Immunol 171 3084–3090 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Rahman MH, Shah S, Kav NN (2006) Constitutive expression of the pea ABA-responsive 17 (ABR17) cDNA confers multiple stress tolerance in Arabidopsis thaliana. Plant Biotechnol J 4 529–549 [DOI] [PubMed] [Google Scholar]
- Strizhov N, Abraham E, Okresz L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 12 557–569 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E (2009) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J (in press) [DOI] [PubMed]
- Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26 461–470 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33 375–379 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Vranova E, Tahtiharju S, Sriprang R, Willekens H, Heino P, Palva ET, Inze D, Van Camp W (2001) The AKT3 potassium channel protein interacts with the AtPP2CA protein phosphatase 2C. J Exp Bot 52 181–182 [PubMed] [Google Scholar]
- Werner WE, Finkelstein R (1995) Arabidopsis mutants with reduced response to NaCl and osmotic stress. Physiol Plant 93 659–666 [Google Scholar]
- Wojcik J, Boneca IG, Legrain P (2002) Prediction, assessment and validation of protein interaction maps in bacteria. J Mol Biol 323 763–770 [DOI] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, Liu ZQ, Du SY, Mei C, Zhao CX, Wang XF, Shang Y, Jiang T, et al (2009) The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol 150 1940–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sulpice R, Himmelbach A, Meinhard M, Christmann A, Grill E (2006) Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc Natl Acad Sci USA 103 6061–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K (2006) The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J 47 49–62 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006. a) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006. b) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.