Abstract
Processing bodies (PBs) are specialized cytoplasmic foci where mRNA turnover and translational repression can take place. Stress granules are related cytoplasmic foci. The CCCH tandem zinc finger proteins (TZFs) play pivotal roles in gene expression, cell fate specification, and various developmental processes. Human TZF binds AU-rich elements at the 3′ untranslated region and recruits decapping, deadenylation, and exonucleolytic enzymes to PBs for RNA turnover. Recent genetic studies indicate that plant TZFs are involved in gene regulation and hormone-mediated environmental responses. It is unknown if plant TZFs can bind RNA and be localized to PBs or stress granules. The Arabidopsis (Arabidopsis thaliana) AtTZF1/AtCTH/AtC3H23 was identified as a sugar-sensitive gene in a previous microarray study. It is characterized by a TZF motif that is distinct from the human TZF. Higher plants such as Arabidopsis and rice (Oryza sativa) each have a gene family containing this unique TZF motif. Here, we show that AtTZF1 can traffic between the nucleus and cytoplasmic foci. AtTZF1 colocalizes with markers of PBs, and the morphology of these cytoplasmic foci resembles that of mammalian PBs and stress granules. AtTZF1-associated cytoplasmic foci are dynamic and tissue specific. They can be induced by dark and wound stresses and are preferentially present in actively growing tissues and stomatal precursor cells. Since AtTZF1 can bind both DNA and RNA in vitro, it raises the possibility that AtTZF1 might be involved in DNA and/or RNA regulation.
In eukaryotic cells, gene expression is determined not only by transcription but also by posttranscriptional events that control RNA processing, transport, stability, localization, and, ultimately, translation (Garneau et al., 2007). After being exported from the nucleus, mRNAs can be translated or degraded in the cytoplasm. When mRNAs fail to pass the quality control (Doma and Parker, 2007; Shyu et al., 2008) or translation is repressed (Valencia-Sanchez et al., 2006), mRNA-protein complexes are temporarily stored in cytoplasmic foci, known as processing bodies (PBs; Sheth and Parker, 2006; Anderson and Kedersha, 2008; Balagopal and Parker, 2009), the location where microRNA-mediated mRNA decay also takes place (Liu et al., 2005; Eulalio et al., 2007; Parker and Sheth, 2007; Franks and Lykke-Andersen, 2008). Depending on the cellular condition, mRNAs in the PBs can either reenter into the normal translation cycle (Brengues et al., 2005) or exit to the mRNA decay pathway where deadenylation, decapping, and 5′ to 3′ exonucleolytic decay take place (Sheth and Parker, 2003; Coller and Parker, 2005). While a number of proteins involved in mRNA decay have been found in PBs (Newbury et al., 2006; Eulalio et al., 2007; Parker and Sheth, 2007; Anderson and Kedersha, 2008), tandem zinc finger (TZF) proteins are unique in that they can recruit and activate mRNA decay enzymes (Fenger-Grøn et al., 2005; Lykke-Andersen and Wagner, 2005) as well as nucleate PB formation for silencing mRNAs that contain AU-rich elements (AREs) in the 3′ untranslated regions (UTRs; Franks and Lykke-Andersen, 2007; Stoecklin and Anderson, 2007).
TZF proteins are characterized by two identical C-x8-C-x5-C-x3-H motifs separated by 18 amino acids (Blackshear et al., 2005). The zinc ion is coordinated by three Cys residues and one His, and each CCCH zinc finger is capable of binding to the 5′-UAUU-3′ half-site of the class II ARE 5′-UAUUUAUU-3′ (Carrick et al., 2004; Hudson et al., 2004; Barreau et al., 2005). In humans, the TZF gene family consists of ZFP36, ZFP36L1, and ZFP36L2. ZFP36 is better known as tristetraprolin (TTP) and is induced by insulin, serum, and other growth factors (Carrick et al., 2004). TTP causes mRNA degradation by binding to the AREs at the 3′ UTR of many important regulators, including tumor necrosis factor-α (TNF-α; Carballo et al., 1998), granulocyte macrophage-colony stimulating factor (Carballo et al., 2000), and possibly many other genes (Barreau et al., 2006; Lai et al., 2006).
A genome-wide analysis of CCCH zinc finger proteins has identified 68 AtC3H genes in Arabidopsis (Arabidopsis thaliana) and 67 OsC3H genes in rice (Oryza sativa; Wang et al., 2008). Of the 26 Arabidopsis genes containing the TZF motif, only two have the TZF motif with the same spacing as human TTP (hTTP; Blackshear et al., 2005). By contrast, 11 out of those 26 genes in subfamily IX contain a unique TZF motif that can only be found in plants. In this unique subfamily, the functions of most of the genes are unknown, with the exception of PEI1, AtSZF1/AtSZF2, and SOMNUS, which are associated with embryogenesis (Li and Thomas, 1998), salt stress responses (Sun et al., 2007), and light-dependent seed germination (Kim et al., 2008), respectively. Intriguingly, both AtSZF1 and SOMNUS have been shown to be localized in the nucleus, not in PBs.
TTP shuttles between the nucleus and cytoplasm (Taylor et al., 1996) via interaction with cellular trafficking factors and either the putative nuclear localization signal (NLS) or the nuclear exit signal, respectively (Phillips et al., 2002). Under stress conditions, TTP can be concentrated in discrete cytoplasmic foci, known as stress granules (SGs) and PBs. SGs and PBs are spatially, compositionally, and functionally linked (Kedersha et al., 2005; Newbury et al., 2006; Kedersha and Anderson, 2007; Anderson and Kedersha, 2008; Buchan et al., 2008; Balagopal and Parker, 2009). The roles of TTP in posttranscriptional regulation are supported by its association with proteins involved in translational repression and mRNA silencing in PBs. TTP activates mRNA decay by binding to the AREs via the TZF motif and by interacting with enzymes involved in deadenylation, decapping, and exonucleolytic activities by the N-terminal activation domain (Lykke-Andersen and Wagner, 2005). TTP nucleates PB formation to silence ARE-containing mRNAs through both translational repression and mRNA decay (Franks and Lykke-Andersen, 2007). In Caenorhabditis elegans, TZF proteins (e.g. OMA-1, OMA-2, and PIE-1) are also associated with cytoplasmic RNPs, termed P granules (Mello et al., 1996; Shimada et al., 2006). These TZF proteins are required for germline specification through differential gene expression during asymmetrical embryonic divisions. PIE-1 is a dual-function protein that inhibits transcription in nucleus and promotes NOS2 RNA maintenance and protein translation in the cytoplasm. It has been postulated that PIE-1 affects maternal RNA expression by direct binding to RNA on P granules (Tenenhaus et al., 2001).
While ARE-mediated mRNA turnover has been studied extensively in animals, it has not been investigated in depth in plants. The only evidence so far shows that a synthetic AUUUA repeat in the 3′ UTR could shorten the half-lives of reporter genes in tobacco (Nicotiana tabacum), raising the possibility that the process of ARE-mediated mRNA decay also exists in plants (Ohme-Takagi et al., 1993). Recent genome-wide study of mRNA decay rates in Arabidopsis revealed multiple AREs as determinants for short half-lives, although the exact AUUUA motif was not among those identified in silico (Narsai et al., 2007). The RNA granules (Anderson and Kedersha, 2006) were first identified in tomato (Solanum lycopersicum) cells as heat shock granules (HSGs; Nover et al., 1983, 1989), although a recent report indicates that plant HSGs are not associated with mRNAs (Weber et al., 2008). Nevertheless, this recent report has revealed that plants do contain both PBs and SGs and that they are distinct from HSGs. It appears that the components in PBs (DCP1, DCP2, and XRN4) and SGs (eIF4E and RNA-binding proteins) are conserved from humans to plants. Some earlier studies have also shown that plant-decapping complexes are present in cytoplasmic foci, resembling mammalian and yeast PBs (Xu et al., 2006; Goeres et al., 2007; Iwasaki et al., 2007).
In an effort to globally identify Glc-responsive genes in Arabidopsis (Price et al., 2004), a CCCH subfamily IX member, AtTZF1/AtC3H23/AtCTH, was identified. Here, we show that AtTZF1 can traffic between the nucleus and cytoplasmic foci. The morphology of the cytoplasmic foci resembles that of PBs and SGs in mammalian and yeast cells. AtTZF1 colocalizes with hTTP and PB/SG markers in plant cells. The cytoplasmic foci are present in specific tissues and can be induced by stresses. We further demonstrate that AtTZF1 can bind both DNA and RNA in vitro. Results of RNA mobility gel shift assays indicate that AtTZF1 does not bind to the hTTP-specific AREs, consistent with the notion that RNA-binding specificity is determined by the structure of the TZF motif.
RESULTS
AtTZF1 Belongs to a Plant-Unique Gene Family
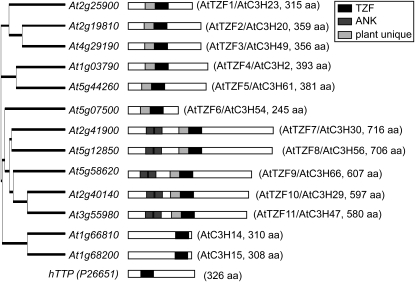
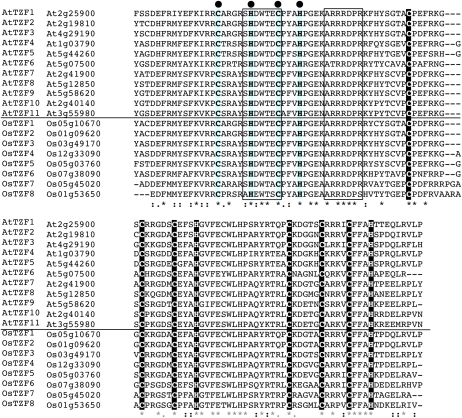
The TZF motif in AtTZF1 shares some similarities with hTTP. In hTTP, TZF is characterized by two identical C-x8-C-x5-C-x3-H motifs, separated by 18 amino acids (Blackshear et al., 2005). By contrast, AtTZF1 contains two different motifs, C-x7-C-x5-C-x3-H and C-x5-C-x4-C-x3-H, separated by 16 amino acids. A BLAST search identified 11 Arabidopsis genes (Fig. 1) and eight rice genes with similar TZF motifs, plus a stretch of extremely conserved 50 amino acids upstream of the TZF motif (Fig. 2). Neither the TZF nor the plant-unique motif could be found in any other organisms outside the plant kingdom. In the plant-unique region, an uncharacterized C-x5-H-x4-C-x3-H motif was found (Fig. 2). In addition, two other motifs, SHDWTEC and ARRRDPR, were also unique to these genes. Compared with other family members in Arabidopsis, AtTZF7 to AtTZF11 have longer C termini and extended N termini with two predicted ankyrin repeats (Puntervoll et al., 2003; Wang et al., 2008) that could be involved in protein-protein interaction.
Figure 1.
Phylogenetic analysis of the AtTZF gene family. All AtTZF genes are characterized by the C-x7-C-x5-C-x3-H-x16-C-x5-C-x4-C-x3-H motif, except that At1g03790 and At5g44260 contain the variant C-x8-C-x5-C-x3-H-x16-C-x5-C-x4-C-x3-H motif. Also shown are At1g66810 and At1g68200, which contain the TZF motif C-x8-C-x5-C-x3-H-x18-C-x8-C-x5-C-x3-H, identical to the TZF in hTTP. Nomenclatures with the AtC3H symbols (Wang et al., 2008) are included for cross-reference. ANK, Ankyrin repeat.
Figure 2.
Plant-unique TZF motif. Sequence comparison of the highly conserved CCCH TZF motif (white letters on black background) and upstream 50 amino acids of Arabidopsis AtTZF and rice OsTZF genes using the ClustalW program (http://www.ebi.ac.uk/Tools/clustalw2/index.html). A conserved C-x5-H-x4-C-x3-H motif is highlighted by black dots, and two other conserved motifs, SHDWTEC and ARRRDPR, are boxed. [See online article for color version of this figure.]
Unique Subcellular Localization of AtTZF1
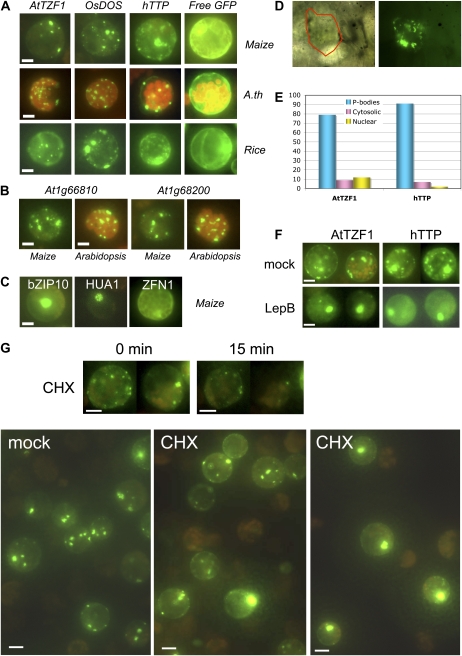
Under certain conditions, mouse TTP is associated with PBs and SGs in the cytoplasm (Kedersha et al., 2005). However, current annotations for AtTZFs in various databases consistently indicate a nuclear localization. To determine the subcellular localization of AtTZF1, we first used protoplast transient expression systems (Sheen, 2001). The GFP fusion reporter gene was expressed in maize (Zea mays), Arabidopsis, and rice protoplasts using established procedures (Jang and Sheen, 1994; Chen et al., 2006; Yoo et al., 2007). Several reporters were also tested to ensure the fidelity of the protoplast transient expression system. Consistent with the mouse counterpart, hTTP was localized in cytoplasmic foci (Fig. 3A). Interestingly, while AtTZF1 was predicted to be in the nucleus (Heazlewood et al., 2007), it was predominantly localized in the cytoplasm, often with multiple foci resembling PBs and SGs (Fig. 3A). The patterns were similar when GFP was translationally fused to either the C or the N terminus of AtTZF1 (data not shown). These cytoplasmic foci varied in size, shape, intensity, and number, depending on the conditions of individual cells. As cauliflower mosaic virus (CaMV) 35S promoter could produce artifacts due to high expression levels, PAtTZF1:AtTZF1-GFP was used to determine if cytoplasmic foci could form under native expression levels. While both the parallel (CaMV 35S:GFP) and internal (CaMV 35S:GASA6-mCherry) controls expressed at high levels, no expression could be detected for PAtTZF1:AtTZF1-GFP (Supplemental Fig. S1). This is consistent with the results of expression analyses showing that AtTZF1 expresses at the background levels in leaves (shown in Fig. 5 below). As an alternative approach to lower the expression levels, we performed protoplast transient expression analysis using reduced amounts of CaMV 35S:AtTZF1-GFP DNA. As shown in Supplemental Figure S2, there was a clear dose response between the amount of DNA and the number, size, and intensity of the cytoplasmic foci. Therefore, the cytoplasmic foci did not seem to be entirely artifacts, because even when the DNA was reduced to 25% (10 μg) of the standard level in transient transformation, numerous cells with cytoplasmic foci could still be found, albeit they were small, dim, and fewer per cell.
Figure 3.
AtTZF1 is localized in the cytoplasmic foci that resemble PBs. A, AtTZF1-GFP, rice homolog OsDOS (Os01g09620)-GFP, and hTTP (P26651)-GFP are localized in cytoplasmic foci in etiolated maize, green Arabidopsis (A.th), and etiolated rice leaf mesophyll protoplasts. By contrast, free GFP is localized in the cytoplasm in a diffusive pattern. Bar = 10 μm. B, The closest Arabidopsis homologs of hTTP, At1g66810 and At1g68200, are also predominantly localized in cytoplasmic foci. Bar = 10 μm. C, For comparison, transcription factor bZIP10 (At4g02640)-GFP is localized in the nucleus, HUA1-GFP (At3g12680; six CCCH zinc fingers) is localized in the nucleolus, and ZFN1-GFP (At3g02830; five CCCH zinc fingers) is localized in the cytoplasm. Bar = 10 μm. D, AtTZF1-GFP is localized in the cytoplasmic foci in lima bean cotyledon cells transformed via particle bombardment. A single cell is outlined in red in a corresponding transmitting bright-light image (left). E, Differential subcellular localization of AtTZF1 and hTTP in maize protoplasts. Shown are the percentages of cells (AtTZF1, 669 cells; hTTP, 376 cells) with respect to the pattern indicated. F, AtTZF1 and hTTP can shuttle between cytoplasmic foci and the nucleus. Shown are maize protoplasts that express either AtTZF1-GFP or hTTP-GFP in the absence/presence of leptomycin B (LepB), an inhibitor of nuclear export receptor. Bar = 10 μm. G, Negative effects of CHX on cytoplasmic foci formation. The number and intensity of cytoplasmic foci are reduced by the CHX treatment in 15 min (top panel). However, some cytoplasmic foci are relatively insensitive to CHX (bottom middle panel), indicating their non-PB/SG identities. By contrast, some CHX-treated cells show GFP signals in the nucleus (bottom right panel), indicating that CHX is effective in reducing AtTZF1-associated cytoplasmic foci but is ineffective when AtTZF1 is localized in the nucleus. Bar = 10 μm.
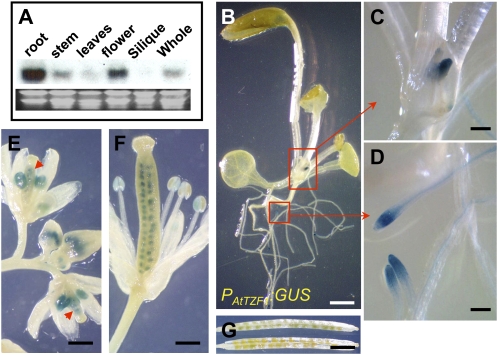
Figure 5.
Temporal and spatial expression of AtTZF1. A, RNA gel-blot analysis indicates that AtTZF1 mRNA is differentially expressed in different tissues. B, GUS staining showing differential expression of PAtTZF1:GUS in root tips and vasculature. Bar = 2 mm. C, Enlarged view of shoot apical region showing GUS stain in young leaf primordia. Bar = 200 μm. D, Enlarged view showing GUS stain in root tips and vasculature. Bar = 200 μm. E, PAtTZF1:GUS is expressed in anthers and female gametophytes (arrowheads) before pollination. Bar = 0.5 mm. F and G, Differential expression of PAtTZF1:GUS during embryo development. Shown are high levels of PAtTZF1:GUS expression in early stages of embryo development (F; bar = 0.5 mm) and very low to no expression after bending-cotyledon stages (G; bar = 2.5 mm).
The 10 other members of the AtTZF gene family (data not shown) and the two closest hTTP homologs, At1g66810 and At1g68200, were also localized in cytoplasmic foci (Fig. 3B). A rice homolog of AtTZF1, OsDOS (Os01g09620), was reported to be nucleus localized in root tips (Kong et al., 2006). Although OsDOS could be localized in the nucleus in some protoplasts, it was found mainly in PB-like foci in protoplasts (Fig. 3A). The subcellular localization patterns of TZF proteins from different organisms were similar in Arabidopsis, maize, and rice protoplasts.
The protoplast transient expression systems appeared to be reliable, as the free GFP was uniformly distributed in the cytoplasm (Fig. 3A) and transcription factor bZIP10 was found in the nucleus (Fig. 3B). To further determine the specificity of these subcellular localization patterns, two other CCCH zinc finger genes were tested. Consistent with a previous report, HUA1 (At3g12680; six CCCH zinc fingers) was localized in the nucleolus (Cheng et al., 2003) and ZFN1 (At3g02830; five CCCH zinc fingers) was evenly diffused in the cytoplasm (Fig. 3C), suggesting that the PB-like cytoplasmic foci were specific to TZF genes. As protoplasts were incubated in high-mannitol solution (0.5–0.6 m) and the osmotic stress might be responsible for protein inclusion body formation, particle bombardment was used to deliver the reporter gene into intact lima bean (Phaseolus lunatus) cotyledons. Consistent with the results of protoplast systems, AtTZF1 was localized in cytoplasmic foci in lima bean cotyledons, resembling PBs or SGs (Fig. 3D).
Similar to TTP, while AtTZF1 often localized in cytoplasmic foci, it could also be localized in the nucleus with lower frequencies (Fig. 3E). In fact, it was common to find AtTZF1 in both locations within the same cells (data not shown). Compared with hTTP, AtTZF1 was localized in the nucleus with a higher frequency. To determine if AtTZF1 could shuttle between cytoplasm and nucleus, leptomycin B was used to block the perception of nuclear exit signal (Kudo et al., 1999; Ryu et al., 2007). As a result, both AtTZF1-GFP and hTTP-GFP signals were retained within nuclei in protoplasts treated with leptomycin B (Fig. 3F), implicating a nucleus-cytoplasm shuttling process. Since no predictable NLS could be found in AtTZF1 protein sequence, a sequential deletion analysis was performed to identify regions containing potential NLS. A GFP fusion of the AtTZF1 fragment without the N terminus and the TZF region was exclusively localized in cytoplasm, indicating that the NLS had been lost (P.-C. Lin, C. Hah, S.G. Kang, J. Qu, and J.-C. Jang, unpublished data). Together, these results support the notion that AtTZF1 can traffic between the nucleus and cytoplasm.
It is well documented that cycloheximide (CHX) inhibits translation elongation, thereby preventing PB and SG assembly in both mammalian and plant cells (Kedersha et al., 2005; Weber et al., 2008). The number and intensity of cytoplasmic foci were reduced in 15 min by the CHX treatment (Fig. 3G), suggesting that they might be PBs and/or SGs. However, some cytoplasmic foci were relatively insensitive to CHX treatment (Fig. 3G, bottom middle), indicating that AtTZF1 could be associated with non-PB/SG-type cytoplasmic bodies. Moreover, some cells showed GFP signals only in the nuclei after the CHX treatment, indicating that CHX was effective in reducing the cytoplasmic foci but ineffective when AtTZF1 was localized in or relocalized to the nucleus, and possibly associated with non-PB/SG components (Fig. 3G, bottom right).
AtTZF1 Is Colocalized with PB and SG Markers
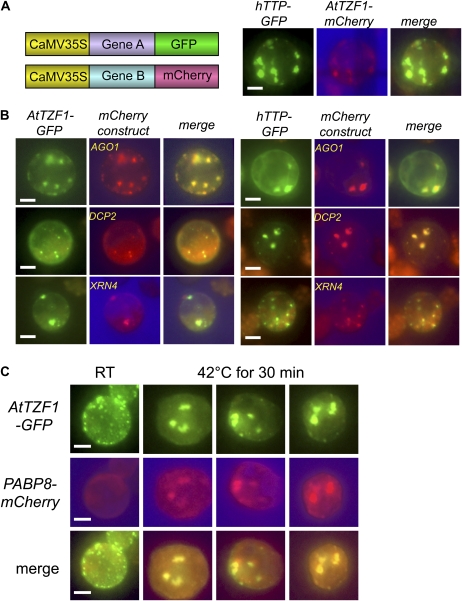
To further determine if AtTZF1-GFP was associated with PBs, colocalization experiments were conducted using both confirmed and putative Arabidopsis PB markers (Newbury et al., 2006; Eulalio et al., 2007; Kedersha and Anderson, 2007; Parker and Sheth, 2007; Anderson and Kedersha, 2008). AGO1 was chosen due to its roles in gene silencing (Baumberger and Baulcombe, 2005; Brodersen et al., 2008; Mi et al., 2008); DCP2 and XRN4 are bona fide RNA-processing enzymes and localized in PBs (Gazzani et al., 2004; Souret et al., 2004; Xu et al., 2006; Goeres et al., 2007; Iwasaki et al., 2007; Weber et al., 2008). We first demonstrated that AtTZF1 was colocalized with hTTP in cytoplasmic foci (Fig. 4A). Similar to the mammalian counterparts, plant AGO1, DCP2, and XRN4 were all localized in cytoplasmic foci resembling PBs. Remarkably, all three markers were also colocalized with AtTZF1 and hTTP, indicating their localization in PBs (Fig. 4B). To determine if AtTZF1 could also be associated with SGs, a colocalization experiment was conducted using the putative SG marker PABP8 (At1g49760; Newbury et al., 2006; Anderson and Kedersha, 2008). Unlike AtTZF1, the PABP8-GFP was randomly distributed in cytoplasm at room temperature (Fig. 4C). Since plant SGs could be induced by heat shock (Weber et al., 2008), transformed protoplasts were heat shocked at 42°C for 30 min. There were dramatic changes in subcellular localization patterns for both AtTZF1 and PABP8 after heat shock. Consequently, they were colocalized in patches of cytoplasmic granules (Fig. 4C). Together, these results indicate that AtTZF1 has a dynamic subcellular localization pattern in the nucleus, PBs, and SGs.
Figure 4.
AtTZF1 is colocalized with PB and SG markers. A, Constructs used for the colocalization studies, and colocalization results of hTTP and AtTZF1 in maize protoplasts. Bar = 10 μm. B, Both AtTZF1 and hTTP are colocalized with PB markers AGO1, DCP2, and XRN4. Bar = 10 μm. C, AtTZF1 can be localized in the SGs. Both AtTZF1 and putative SG marker PABP8 form aggregates and colocalize after heat shock at 42°C for 30 min. Bar = 10 μm. RT, Room temperature.
AtTZF1 Expression and Subcellular Localization in Intact Plants
To determine the temporal-spatial expression pattern of AtTZF1, transgenic plants expressing a promoter-reporter fusion gene (PAtTZF1:GUS) were used. Consistent with the results of RNA gel-blot analysis (Fig. 5A), PAtTZF1:GUS was differentially expressed in different organs (Fig. 5B), with the highest levels in the leaf primordia (Fig. 5C), root tips, vasculature (Fig. 5D), and flowers (Fig. 5, E and F). It appeared that the expression of AtTZF1 was confined to the young tissues. For example, the expression levels of AtTZF1 decreased along with the maturation of the anthers (Fig. 5, compare E and F) and embryos (Fig. 5, compare F and G). During embryo development, AtTZF1 was mainly expressed in the integuments of female gametophytes (Fig. 5, E and F) and completely diminished when embryos reached bending-cotyledon stages (Fig. 5G).
To determine the subcellular localization of AtTZF1, plants expressing a translational fusion gene (AtTZF1 coding sequence-GFP) driven by either native AtTZF1 promoter or by the ubiquitous CaMV35S promoter were generated. Since PAtTZF1:AtTZF1-GFP was expressed at relatively low levels (data not shown), CaMV35S:AtTZF1-GFP plants were used to further investigate the subcellular localization of AtTZF1 in intact tissues. Under the growth conditions used, PB-like AtTZF1-GFP cytoplasmic foci were occasionally found in root meristems (Fig. 6, A and B), and rarely present in the elongation zone (data not shown). Overall, the cytoplasmic foci of AtTZF1-GFP were difficult to detect in intact green organs, including cotyledons, leaves, hypocotyls, stems, and siliques. However, few distinct cytoplasmic foci could be found in the leaf protoplasts isolated from intact CaMV35S:AtTZF1-GFP plants (Fig. 6C). It was unclear if the cytoplasmic foci were induced by protoplast isolation.
Figure 6.
Subcellular localization of AtTZF1 in CaMV35S:AtTZF1-GFP plants. A, AtTZF1-associated cytoplasmic foci are most commonly found in the root meristems, where the protein is accumulated at higher levels. B, Closeup view of a root tip showing cytoplasmic foci within each cell. Bar = 50 μm. Boxed regions are shown with enlarged views. C, Cytoplasmic foci can be found in the leaf protoplasts isolated from CaMV35S:AtTZF1-GFP plants. Bar = 10 μm.
AtTZF1-Associated Cytoplasmic Foci Are Stress Inducible
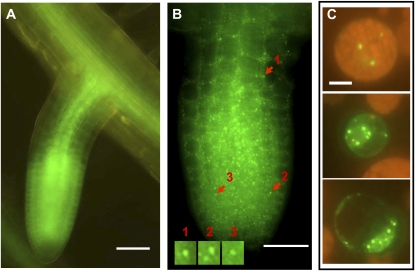
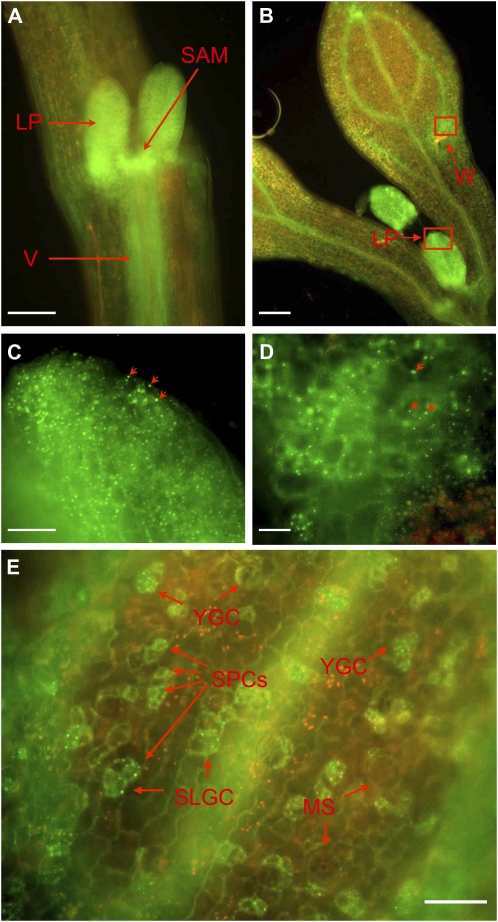
As it was difficult to visualize AtTZF1-GFP expression in green tissues, we examined its expression in dark-grown etiolated seedlings. The strong GFP signal was found not only in the roots but also in the vasculature, shoot apical meristems, and leaf primordia (Fig. 7, A and B). In contrast to green plants, cytoplasmic foci were readily visible in etiolated leaf primordia (Fig. 7C) and mesophyll cells near a mechanical wound pinched by a pair of forceps during sample handling (Fig. 7D). In etiolated cotyledons, cytoplasmic foci were especially distinct in stomatal precursor cells, including meristemoids and guard mother cells (Geisler et al., 2000), in young stomata, and in stomatal-lineage ground cells but not as apparent in mature stomata and pavement cells (Fig. 7E).
Figure 7.
AtTZF1-associated cytoplasmic foci are present in etiolated seedlings and can be induced by wounding. A, AtTZF1-GFP is highly abundant in leaf primordia (LP), shoot apical meristem (SAM), and vasculature (V). B, In contrast to the fully expanded cotyledons, where AtTZF1-GFP is localized mainly in the vasculature, it is present in all cells in the true leaf primordia. In addition, AtTZF1-GFP cytoplasmic foci can be induced in the cells surrounding a wound (W) pinched by a pair of fine-tip forceps. Bar = 100 μm. C, Enlarged view is shown from the red rectangular area in B, a leaf primordium with numerous cytoplasmic foci. Some cytoplasmic foci are indicated by arrows. Bar = 20 μm. D, Enlarged view is shown from the red rectangular area in B, showing an induction of cytoplasmic foci in the cells surrounding a wounding site (bottom right). Some cytoplasmic foci are indicated by arrows. Bar = 10 μm. E, In etiolated cotyledons, AtTZF1-associated cytoplasmic foci are especially distinct in stomatal precursor cells (SPCs), in young guard cells (YGCs), and in stomatal-lineage ground cells (SLGCs) but are not apparent in mature stomata (MS). Bar = 30 μm.
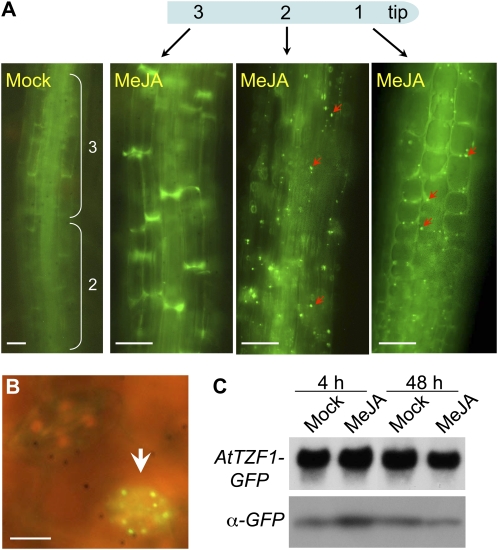
To verify the effects of wounding, light-grown green plants were treated with methyl jasmonate (MeJA), a gaseous hormone that plants use to transmit wound signals systematically (Farmer and Ryan, 1990). As predicted, MeJA enhanced the formation of cytoplasmic foci in both root meristem and elongation zones (Fig. 8A). In addition, cytoplasmic foci were preferentially induced in the stomata in the green leaves (Fig. 8B). While the mechanism of the MeJA induction is currently unknown, these results are consistent with the findings in yeast and mammals, in which SGs and PBs are induced by various stresses (Kedersha et al., 2005; Anderson and Kedersha, 2008). To determine if the induction of cytoplasmic foci was a result of gene expression change, RNA and protein gel-blot analyses were conducted. Results showed that AtTZF1-GFP mRNA abundance was similar among different samples, only slightly reduced for the 48-h MeJA-treated sample (Fig. 8C). Compared with the mock treatment, the protein level was slightly higher at 4 h but lower at 48 h in MeJA-treated plants (Fig. 8C). Thus, the induction of cytoplasmic foci was likely due to unknown posttranslational events but not to a large change in protein abundance per se.
Figure 8.
Wound hormone MeJA induces cytoplasmic foci formation. A, Cytoplasmic foci are normally found in root meristems. However, they can be found in transition (1) and elongation-differentiation (2) zones (arrowheads) after the MeJA (100 μm) treatment. Also shown are mock-treated root zone 2 and fully elongated zone 3. Bar = 50 μm. B, Cytoplasmic foci can be induced by MeJA in most stomata (arrowhead). Bar = 10 μm. C, Induction of cytoplasmic foci is unlikely a direct consequence of increased protein levels. RNA gel blot shows similar expression levels between mock- and MeJA-treated plants. The protein level is higher only in 4-h MeJA-treated samples. Images in A and B were taken from 48-h samples. [See online article for color version of this figure.]
AtTZF1 Binds Both DNA and RNA in Vitro
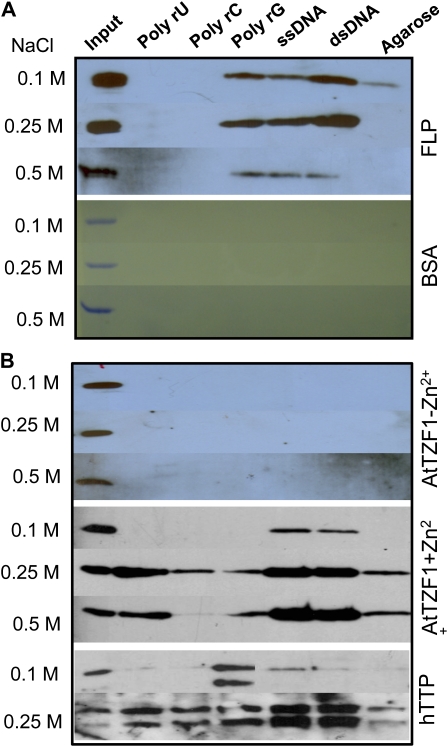
Since it was unknown if any of the unique plant TZFs could bind DNA or RNA, we performed in vitro binding assays using recombinant proteins produced in Escherichia coli. We first performed the bead-binding assays (Yang et al., 1998) using a positive control HUA1 that is known to bind both DNA and RNA. The results (data not shown) were consistent with a previous report (Li et al., 2001). We subsequently used a MYB domain-containing transcription factor (At1g14350) and found positive binding with DNA but not RNA (Fig. 9A). Finally, we used bovine serum albumin (BSA) as a negative control and showed that BSA could bind neither DNA nor RNA (Fig. 9A). Together, these results indicated that our assay conditions were reliable. Surprisingly, no binding could be detected when AtTZF1 was tested and the salt concentration ranged from 0.1 to 0.5 m (Fig. 9B). As Zn2+ might be important for binding, 50 μm ZnCl2 was added to the binding reactions. Results showed that AtTZF1 could only bind to DNA in 0.1 m NaCl and could bind ribohomopolymer U when the salt concentration increased to 0.25 and 0.5 m (Fig. 9B). Consistent with a previous report (Brewer et al., 2004), hTTP bound neither DNA nor RNA in the absence of Zn2+ (data not shown) but could bind both when Zn2+ was added to the reactions. Nucleic acid binding for hTTP was also enhanced by higher salt concentration (Fig. 9B). The binding between AtTZF1 and poly-rC or poly-rG was either weak or an artifact, as negative control agarose also showed a similar signal (Fig. 9B). The binding with poly-rG was likely nonspecific, because all tested proteins (except for BSA) could bind poly-rG (data not shown). Together, these results indicate that AtTZF1 and hTTP can bind both DNA and RNA.
Figure 9.
AtTZF1 binds both DNA and RNA in vitro. A, In vitro bead-binding assays showing that the MYB transcription factor FLP preferentially binds DNA. Binding to the poly-rG appears to be nonspecific. As a negative control, BSA binds neither DNA nor RNA. B, Results of in vitro bead-binding assays indicate that AtTZF1 preferentially binds single- and double-stranded DNA at 0.1 m NaCl and can bind ribohomopolymer U when salt concentration increases to 0.25 to 0.5 m. Poly-rG and poly-rC binding appears to be nonspecific, as the negative control (agarose) can generate similar signals. The binding requires the presence of Zn2+, as no binding is detected in the absence of ZnCl2 in the binding buffer. In contrast to AtTZF1, hTTP not only binds both single-stranded and double-stranded DNA but also binds all ribohomopolymers with similar affinities at 0.5 m NaCl. The results by poly-rG are likely false positive, because every protein tested so far can generate similar signals (data not shown). [See online article for color version of this figure.]
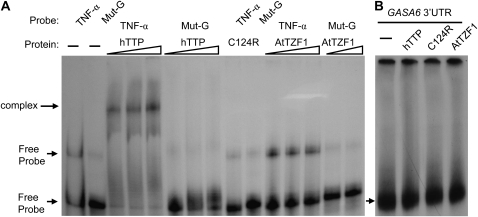
Since GASA6 was down-regulated in AtTZF1 overexpression plants (P.-C. Lin, Y. Jikumaru, C. Hah, M.C. Pomeranz, S.G. Kang, Y. Kamiya, and J.-C. Jang, unpublished data) and contains two ARE-like motifs (5′-TATTTTATTTAATTATTT-3′ and 5′-ATTTTATTTATTAATTA-3′) at the 3′ UTR, we performed RNA electrophoretic mobility shift assay (REMSA) to determine if AtTZF1 could bind AREs. As shown in Figure 10A, hTTP formed a complex with TNF-α 3′ UTR probe and caused a band shift. This shift was abolished when the A nucleotides were replaced by G nucleotides in the TNF-α 3′ UTR probe (Mut-G) or when hTTP was mutated (C124R) to abolish the RNA-binding ability (Lai et al., 2002). The band shift was not observed for the combination of AtTZF1 with either TNF-α 3′ UTR or mutated Mut-G probe. To further confirm this, a synthetic probe with multiple tandem UAUU elements (3U/4U probe) was used in EMSA. Consistent with the TNF-α 3′ UTR probe, AtTZF1 did not form a complex with 3U/4U probe (data not shown). Together, these results suggest that AtTZF1 does not bind to the typical AREs that hTTP can interact with with high affinity. Interestingly, neither hTTP nor AtTZF1 could form a complex with GASA6 3′ UTR (Fig. 10B), indicating that both the structure of the TZF motif and the RNA sequence are important for binding specificity (Blackshear et al., 2005).
Figure 10.
AtTZF1 does not bind the hTTP consensus binding site. A, REMSA shows that hTTP can bind the 3′ UTR of TNF-α with specificity, because mutation in either RNA probe (MutG) or TZF domain (C124R) can eliminate the binding (indicated by the arrow). By contrast, no RNA-protein complex can be detected when AtTZF1 is used. B, REMSA results indicate that neither hTTP nor AtTZF1 can bind the GASA6 3′ UTR. The arrowhead indicates free probe.
DISCUSSION
The mammalian CCCH-type TZF proteins control mRNA turnover via interactions with the 3′ UTR AREs (Blackshear et al., 2005) and recruitment and activation of mRNA decay enzymes in PBs (Lykke-Andersen and Wagner, 2005). These TZF proteins are also crucial for the nucleation of PBs where ARE-mediated gene silencing takes place (Franks and Lykke-Andersen, 2007). The C. elegans TZF motifs are also important for localization and translational control in the P-granules (Tenenhaus et al., 2001; Jadhav et al., 2008). In plants, although the molecular mechanisms are unknown, multiple genetic studies have shown that plant-unique TZF genes are important for both developmental and environmental responses (Li and Thomas, 1998; Kong et al., 2006; Sun et al., 2007; Kim et al., 2008; Guo et al., 2009). Based on the results of reverse genetic analyses, AtTZF1 is likely involved in GA/ABA-mediated developmental and environmental responses (P.-C. Lin, Y. Jikumaru, C. Hah, M.C. Pomeranz, S.G. Kang, Y. Kamiya, and J.-C. Jang, unpublished data). Here, we report that AtTZF1 (AtCTH/AtC3H23) can traffic between the nucleus and cytoplasmic foci, a characteristic of hTTP. The morphology of AtTZF1 cytoplasmic foci resembles that of hTTP when localized in PBs and SGs in mammalian cells. These PB/SG-like cytoplasmic foci are present in active growing cells and are induced by various stresses. AtTZF1 binds both DNA and RNA in vitro, raising the possibility that it might play a role in DNA and/or RNA regulation.
PBs in Plants
The composition and function of mammalian and yeast PBs have been under intensive investigation recently. However, except for Arabidopsis decapping enzymes (Xu et al., 2006; Goeres et al., 2007; Iwasaki et al., 2007) and XRN4 (Weber et al., 2008), the composition of plant PBs is unknown. It was shown that DCP1, DCP2, and decapping regulator VCS were localized in PB-like cytoplasmic foci, in tobacco epidermal cells, and in Arabidopsis root cells. Furthermore, DCP1 and DCP2 were colocalized with the yeast PB marker DHH1 in yeast cells (Xu et al., 2006). These results suggest that plant decapping enzymes are associated with PBs. In another report (Goeres et al., 2007), VCS and TDT (DCP2) were found to be localized in cytoplasmic foci in the root elongation zone. The latest report showed that DCP1, DCP2, and XRN4 were colocalized in the PBs (Weber et al., 2008). Here, we report that AtTZF1 may be associated with PBs, and the presence of putative PBs is affected by developmental and environmental conditions. This proposition is supported by multiple lines of evidence. (1) Arabidopsis, rice, and human TZF proteins are localized in PB-like cytoplasmic foci in Arabidopsis, maize, and rice protoplasts. (2) AtTZF1 is colocalized with hTTP in maize protoplasts. (3) Both AtTZF1 and hTTP are colocalized with plant AGO1, DCP2, and XRN4 in PB-like cytoplasmic foci. (4) CHX inhibits the formation of AtTZF1-associated cytoplasmic foci. (5) AtTZF1 is localized in PB-like cytoplasmic foci in transgenic Arabidopsis plants. (6) AtTZF1-associated cytoplasmic foci are found specifically in actively growing cells. (7) PB-like cytoplasmic foci can be induced by dark, mechanical wounding, and the stress hormone MeJA. We have also shown that the localization patterns are specific to TZF proteins, because proteins with a different number and arrangement of CCCH zinc fingers, such as HUA1 and ZFN1, are not localized in the cytoplasmic foci (Fig. 3C). In fact, all 11 genes in the AtTZF family (Fig. 1) can be localized in PB/SG-like cytoplasmic foci (M.C. Pomeranz and J.-C. Jang, unpublished data), suggesting that the conserved TZF domain and/or the 50-amino acid plant-unique motif (Fig. 2) are responsible for this unique localization pattern, as regions outside the TZF domain are less conserved in the family.
Cytoplasmic mRNA-protein complex granules include PBs, SGs, and HSGs in plants (Weber et al., 2008), polar or germinal granules in germlines of flies and worms, neuronal granules in neurons, and SGs and PBs in mammalian somatic cells (Anderson and Kedersha, 2006). In animals, while the composition varies, some conserved components are found in different granules and different organisms (Strome, 2005; Anderson and Kedersha, 2006; Barbee et al., 2006). In mammals, SGs and PBs are compositionally and functionally linked. SGs, as reflected by the name, are induced by many stresses such as heat, oxidative stress, UV irradiation, hypoxia, viral infection, and Glc starvation. The core components of SGs are stalled preinitiation complexes, small ribosomal subunits, mRNAs, and mRNA-binding proteins. SGs vary in size and shape. By contrast, PBs are more uniform; the main components in PBs are 5′ to 3′ mRNA decay machinery, activators of mRNA decay, translational regulators, microRNA-silencing machinery, mRNAs, and mRNA-binding proteins (Kedersha et al., 2005; Anderson and Kedersha, 2006, 2008; Newbury et al., 2006; Kedersha and Anderson, 2007; Franks and Lykke-Andersen, 2008; Balagopal and Parker, 2009). Similar to what we have reported here, PBs are found in actively growing cells and induced by arsenite treatment in mammalian cells. Interestingly, overexpression of mammalian TZF, TTP, or BRF1 not only induces PB and SG formation but also promotes their physical interaction (Kedersha et al., 2005). Recently, it has been shown that TTP or BRF1 is critical in inducing PB formation, hence facilitating AU-rich element-mediated decay processes (Franks and Lykke-Andersen, 2007). Here, we have shown that AtTZF1 is likely localized not only in PBs but also in SGs, as it colocalized with PABP8 (Fig. 4C), a putative counterpart of mammalian SG marker (Newbury et al., 2006). The morphology of those cytoplasmic foci in AtTZF1-GFP transgenic plants resembles that of PBs in mammals and yeast: small and uniform. However, large and irregular cytoplasmic foci in protoplasts either could be artifacts due to overexpression or they are a mixture of SGs, PBs, and unknown granules. This is also evidenced by the insensitivity of some of the cytoplasmic foci to the CHX treatments (Fig. 3G). In mammalian cells, overexpression of PB components can create abnormally large PBs that exert aberrant function and behave abnormally (Kedersha and Anderson, 2007). To verify these possibilities, work is in progress using recently identified plant SG markers (Weber et al., 2008) to colocalize AtTZF1 in different tissues under various conditions.
Developmental and Environmental Control of PB/SG Formation
AtTZF1-associated cytoplasmic foci are mostly found in regions with active cell division and expansion, such as the main root tips, lateral root primordia, the shoot apical meristem, stomatal precursor cells, and unexpanded young leaves (Figs. 6 and 7). It is noteworthy that during lateral root formation, AtTZF1 protein is accumulated at the highest levels in root meristems and gradually decreases, becoming restricted to the vasculature in the regions where cells begin to differentiate and elongate (Fig. 6A). This could be due to the differential activities exerted by the CaMV 35S promoter. Alternatively, AtTZF1 protein abundance may be controlled by tissue-specific cues. In C. elegans, the P-granule-associated TZF proteins (e.g. PIE-1) are crucial in germline specification (Strome, 2005; Strome and Lehmann, 2007). It has been demonstrated that the first zinc finger of PIE-1 is required for the degradation of PIE-1 itself in the somatic blastomeres during asymmetric division, and the second zinc finger is important for differential segregation and enrichment of PIE-1 to the germ lineage and targeting to the P-granules (Reese et al., 2000). These results highlight the possible similarity and importance of differential expression of TZF proteins in development. It will be imperative to determine if AtTZF1's abundance is controlled by specific developmental cues in Arabidopsis.
The induction of putative plant PBs and/or SGs by dark, wounding, and MeJA is consistent with the characteristics of stress-induced mammalian SGs and PBs (Kedersha et al., 2005; Anderson and Kedersha, 2006, 2008; Kedersha and Anderson, 2007; Balagopal and Parker, 2009). It has been shown recently that plant PBs and SGs can also be induced by stresses such as heat and hypoxia (Weber et al., 2008). The cytoplasmic foci induced by MeJA in stomatal cells (Fig. 8B) are noteworthy because both ABA and MeJA can trigger stomatal closure, via cytoplasmic alkalization, preceding reactive oxygen species (ROS) production and a series of actions on activities of different ion channels (Suhita et al., 2004; Munemasa et al., 2007; Saito et al., 2008). ABA- or MeJA-induced stomatal closure can be suppressed by exogenous application of inhibitor of NADPH oxidase or the NADPH oxidase double mutant atrbohD/F (Suhita et al., 2004). The stomatal hyperclosure in AtTZF1 gain-of-function plants (P.-C. Lin, Y. Jikumaru, C. Hah, M.C. Pomeranz, S.G. Kang, Y. Kamiya, and J.-C. Jang, unpublished data) and the induction of cytoplasmic foci by MeJA (Fig. 8, A and B) suggest that AtTZF1 participates in stress responses via the association with PBs and/or SGs. While plant cell growth and stress response seem to be two distinct processes, it is remarkable that ROS also play a critical role in cell growth (Foreman et al., 2003). As AtTZF1-associated cytoplasmic foci are predominantly found in actively growing or stressed cells, it raises the possibility that ROS is involved in plant PB/SG induction, as demonstrated in mammalian cells (Lian and Gallouzi, 2009). Work is in progress to test this hypothesis using pharmacological and mutant approaches.
How Does AtTZF1 Affect Gene Expression?
On the basis of phenotypic and gene expression analyses, we propose that AtTZF1 is involved in sugar/ABA/GA responses (P.-C. Lin, Y. Jikumaru, C. Hah, M.C. Pomeranz, S.G. Kang, Y. Kamiya, and J.-C. Jang, unpublished data). A plausible mechanism for AtTZF1 to control gene expression is via posttranscriptional regulation, an area that has not been addressed widely in plant sugar signaling. For example, the mRNA half-life of rice α-amylase gene is affected by sugar via destabilization signal sequences at the 3′ UTR (Chan and Yu, 1998). Furthermore, the 3′ UTR of the maize cell wall invertase gene Incw1 is likely to affect sugar-dependent mRNA metabolism and translation (Cheng et al., 1999). In fact, translational repression caused by sugar starvation appears to be a predominant mechanism in silencing genes involved in cell cycle and cell growth in plants (Nicolai et al., 2006). Unfortunately, we know nothing about the trans-acting RNA-binding factors involved in these processes. Finally, the Arabidopsis sugar response mutant low-beta-amylase1 (lba1) is caused by a mutation in UPF1, an RNA helicase involved in the nonsense-mediated mRNA decay pathway (Yoine et al., 2006). Interestingly, the animal UPF1 binds to mRNAs that contain premature termination codons and directs them to PBs for degradation (Brogna et al., 2008). It would be critical to understand whether or not AtTZF1 and LBA1 affect the expression of sugar-responsive genes via AU-rich element-mediated decay and nonsense-mediated mRNA decay, respectively, in PBs.
On the basis of gene expression analyses, we suspected that AtTZF1 might bind 3′ UTR AREs of genes down-regulated in AtTZF1 gain-of-function plants and destabilize these mRNAs (P.-C. Lin, Y. Jikumaru, C. Hah, M.C. Pomeranz, S.G. Kang, Y. Kamiya, and J.-C. Jang, unpublished data). However, it is not surprising that while AtTZF1 can bind RNA (Fig. 9B), it does not bind to either typical AREs or the candidate GASA6 3′ UTR (Fig. 10). There are several possibilities to account for the negative interaction between AtTZF1 and typical AREs. (1) The space within and between the two CCCH zinc fingers in AtTZF1 is different from the mammalian TZFs. Although it has been implicated that a single C-x8-C-x5-C-x3-H motif might be sufficient for ARE binding (Hudson et al., 2004), AtTZF1 may bind different RNA motifs. (2) AtTZF1 does not contain a typical lead-in sequence before each finger. The lead-in motif (R/KYKTEV) is critical for the TTP family to bind AREs (5′-UAUUUAUU-3′). They form hydrophobic pockets that participate in backbone hydrogen-bonding interactions, important for recognition of the U and A bases at the 5′ end of each half-site of the ARE (Hudson et al., 2004). Comparative studies indicated that variants of the lead-in sequence could recognize divergent RNA sequences other than typical AREs (Pagano et al., 2007; Farley et al., 2008). (3) AtTZF1 alone is insufficient to bind these sequences. For example, the repression of maternally transcribed hunchback (hb) mRNA in the posterior is mediated by Nanos and Pumilio during embryonic patterning of Drosophila. While Pumilio binds specifically to 3′ UTR of hb, Nanos can bind hb only when recruited by the Pumilio to the ternary complex (Sonoda and Wharton, 1999). (4) The DNA/RNA binding might be mediated through a domain outside the predicted TZF motif in AtTZF1 (e.g. the 50-amino acid regions upstream of the TZF domain with a conserved putative C-x5-H-x4-C-x3-H motif). Arabidopsis does contain two genes (At1g66810 and At1g68200) with the TZF motif (C-x8-C-x5-C-x3-H-x18-C-x8-C-x5-C-x3-H) spaced identically to TTP. Their lead-in sequence (MM/TKTEL or RYKTEV) before each finger is also similar to the conserved pattern R/KYKTEL. As these two proteins are also localized in PB/SG-like cytoplasmic foci (Fig. 3B), it will be informative to determine whether or not they are authentic ARE-binding proteins. Another critical area of study is to identify the cis-regulatory elements in the promoter, 5′ UTR, or 3′ UTR that allow AtTZF1 to bind and hence may affect transcription, RNA stability, and/or translation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Unless specified otherwise, Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) plants for microscopy were grown on Murashige and Skoog (MS) plates (MS salts, B5 vitamins, pH 5.7, 0.7% Phytagar, and 2% Suc) at 24°C in either continuous white light (100 μmol m−2 s−1) or dark in a tissue culture room.
Molecular Cloning, RNA/Protein Isolation, and Gel-Blot Analyses
Unless specified otherwise, all cloning procedures were carried out using Gateway (Invitrogen) technologies. Genes were first inserted into pENTR vectors and then transferred to various destination vectors via recombination reactions. Binary vectors used for promoter fusions and translational fusions can be found at http://www.psb.ugent.be/gateway/index.php?_app=vector&_act=construct_list_plant&. cDNAs were amplified from either full-length cDNA clones from Arabidopsis Biological Resource Center SSP pUNI clones (http://www.arabidopsis.org/abrc/catalog/cdna_clone_1.html) or the pFL61 cDNA library (Minet et al., 1992) via PCR. hTTP was cloned from hTTP/pEGFP plasmid consisting of hTTP coding sequence (NM_003407; bp 54–1,040) inserted into multicloning sites of vector pEGFP-N1 (GenBank accession no. 55762). For protoplast transient expression, a recombination cassette derived from pK7FWG2 was transferred to multicloning sites of pBluescript KS+ to serve as destination vector for high-yield plasmid production in Escherichia coli and enhanced expression in protoplasts. Primers used for cloning coding sequences are as follows: AtTZF1 (At2g25900; 5′-ATGATGATCGGCGAAAATAAAAA-3′ and 5′-ACCGAGTGAGTTCTCTCTAC-3′), OsDOS (Os01g09620; 5′-ATGATGATGGGGGAAGGAGTC-3′ and 5′-ATGTTGATGAGGTCGGACACCCAATC-3′), hTTP (P26651; 5′-ATGGATCTGACTGCCATCTA-3′ and 5′-ATCTCAGAAACAGAGATGCGAT-3′), bZIP10 (At4g02640; 5′-ATGAACAGTATCTTCTCC-3′ and 5′-GTCCACGCATTTTTTCG-3′), HUA1 (At3g12680; 5′-ATGGCACATCGTCAATTG-3′ and 5′-CATTGAGTAGTGTCGGTG-3′), ZFN1 (At3g02830; 5′-ATGGATTTTAATGCCGGAG-3′ and 5′-CACTGCTGTGTATTATCA-3′), AGO1 (At1g48410; 5′-ATGGTGAGAAAGAGAAGAAC-3′ and 5′-AAGCAGTAGAACATGACACGC-3′), DCP2 (At5g13570; 5′-ATGTCGGGCCTCCATCGA-3′ and 5′-AAAGCTGAATTACCAGATTCCA-3′), XRN4 (At1g54490; 5′-ATGGGAGTACCGGCGTTC-3′ and 5′-AACAAGTTTGCACCTCGATGA-3′), and PABP8 (At1g49760; 5′-ATGGCTCAGATTCAGCATCA-3′ and 5′-AAAGGTACGATGTTGTCTCCAA-3′). Probes used for RNA gel-blot analyses were amplified from full-length cDNA of AtTZF1. Probes used for gel-shift assays were cloned into pBluescript KS+ using the following primers: TNF-α (5′-TTATTTATTTATTATTTATTTTTT-3′ and 5′-AAAAAATAAATAATAAATAAATAA-3′), mutG (5′-TTGTTTGTTTGTTGTTTGTTTTTT-3′ and 5′-AAAAAACAAACAACAAACAAACAA-3′), and GASA6 3′ UTR (5′-ACATTGAGAGAGAAACCCCA-3′ and 5′-ATACGAAAGATTATCTAATTAATAA-3′). RNA isolation and RNA gel-blot analyses were conducted as described (Price et al., 2004). Protein isolation and gel-blot analysis were conducted as described (Jang et al., 2000).
Transient and Stable Plant Transformation
Protoplast isolation and transformation were conducted essentially as described (Jang and Sheen, 1994; Chen et al., 2006; Yoo et al., 2007), except for a few modifications. In the Arabidopsis protocol, leaf slices were vacuum infiltrated with digestion solution without enzymes (0.5 m mannitol, 20 mm KCl, 20 mm MES, pH 5.7, 10 mm CaCl2, 5 mm β-mercaptoethanol, and 0.1% BSA) for 10 min and then replaced with fresh enzyme solution containing 3% cellulase RS and 0.8% macerozyme R10. The polyethylene glycol transformation time was 5 min. The recipes of three solutions were modified as follows: WI solution (0.5 m mannitol, 20 mm MES, and 20 mm KCl, pH 5.7), W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 5 mm MES, pH 5.7), MMg solution (0.5 m mannitol, 20 mm MgCl2, and 5 mm MES, pH 5.7). Transient expression in lima bean (Phaseolus lunatus) cotyledons using particle bombardment was conducted as described (Chiera et al., 2007). For stable transformation, floral dip or vacuum infiltration was used as described (Bechtold and Pelletier, 1998; Clough and Bent, 1998).
Microscopy
A Nikon Eclipse E600 fluorescence microscope with filter sets was used for microscopy. GFP fluorescence was visualized with a filter set consisting of an excitation filter of 450 to 490 nm, a dichroic mirror of 510 nm, and a barrier filter of 520 to 560 nm. Images were captured by a SPOT RT Slider multimode camera and Advanced SPOT Software (Diagnostics Instruments). For protoplasts, 10-μL samples were loaded onto a hemacytometer for observation. Plate-grown seedlings were placed on glass slides and immersed in an MS salt solution for observation.
In Vitro Bead Binding and REMSA Assays
Ribohomopolymer binding assays (Yang et al., 1998) were conducted by incubating 1 μg of AtTZF1 protein with 20 μL of ribohomopolymer beads (Sigma) in RHPA buffer (10 mm Tris, pH 7.4, 2.5 mm MgCl2, 5% Triton X-100, and 1 mg mL−1 heparin) with NaCl at various concentrations. Reactions were placed on a Nutator plate at room temperature for 10 min and then washed four times for 5 min each. Beads were then boiled in 5× SDS buffer for 3 min and run on 12.5% SDS-PAGE for western-blot analysis. Low-melting-point agarose (NuSieve; Cambrex) was used as a control for nonspecific protein binding. REMSAs were conducted as described (Brewer et al., 2004). RNA probes were synthesized by cloning probe sequence into pBluescript KS+ containing an upstream T7 promoter. Plasmid vectors were linearized using KpnI restriction enzyme and then used for RNA synthesis labeled with [32P]α-UTP using the Promega T7 Riboprobe Kit. Binding reactions consisted of each protein incubated with 2 × 106 cpm of various radiolabeled RNA probes. The reaction was assembled in a final volume of 10 μL containing 10 mm Tris-HCl (pH 8.0), 40 mm KCl, 2 mm dithiothreitol, 3 mm MgCl2, 10 mm HEPES, 10% glycerol, and heparin (to 25 μg μL−1). Binding reactions were incubated at room temperature for 15 min and then separated using electrophoresis through a 6% (40:1 acrylamide:bis-acrylamide) native gel at 4°C. After electrophoresis, gels were dried and exposed to x-ray film.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtTZF1 (At2g25900), OsDOS (Os01g09620), hTTP (P26651), bZIP10 (At4g02640), HUA1 (At3g12680), ZFN1 (At3g02830), AGO1 (At1g48410), DCP2 (At5g13570), XRN4 (At1g54490), and PABP8 (At1g49760).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The CaMV 35S:AtTZF1-GFP is not expressed in the maize mesophyll protoplasts.
Supplemental Figure S2. Cytoplasmic foci formation in relation to the levels of gene expression.
Supplementary Material
Acknowledgments
We thank Li Zhang for rice protoplast transformation, the Arabidopsis Biological Resource Center for providing DNA clones and seeds, Dr. Biao Ding for use of the microscopy facility, Dr. Daniel Schoenberg for advice on RNA EMSA, Dr. Fred Sack for advice on the interpretation of the AtTZF1-GFP expression pattern in planta, Dr. Guo-Liang Wang for advice on rice protoplast transformation, Drs. Esther van der Knaap and Randy Scholl for reading the manuscript, Jie Qu for the GASA6-mCherry construct, and Joe Takayama for greenhouse support.
This work was supported by the National Science Foundation (grant no. IOB–0543751 to J.-C.J.).
This article is dedicated to the memory of Pei-Chi Lin (1978–2007), who was a great coworker and made an important contribution to this work.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jyan-Chyun Jang (jang.40@osu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2008) Stress granules: the tao of RNA triage. Trends Biochem Sci 33 141–150 [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R (2009) Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 21 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al (2006) Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Dutertre S, Paillard L, Osborne HB (2006) Liposome-mediated RNA transfection should be used with caution. RNA 12 1790–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB (2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82 259–266 [DOI] [PubMed] [Google Scholar]
- Blackshear PJ, Phillips RS, Lai WS (2005) Tandem CCCH zinc finger proteins in mRNA binding. In S Iuchi, N Kuldell, eds, Zinc Finger Proteins: from Atomic Contact to Cellular Function. Kluwer Academic/Plenum Publishers, London, pp 80–90
- Brengues M, Teixeira D, Parker R (2005) Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BY, Malicka J, Blackshear PJ, Wilson GM (2004) RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of AU-rich MRNA-destabilizing motifs. J Biol Chem 279 27870–27877 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brogna S, Ramanathan P, Wen J (2008) UPF1 P-body localization. Biochem Soc Trans 36 698–700 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R (2008) P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ (1998) Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281 1001–1005 [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ (2000) Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95 1891–1899 [PubMed] [Google Scholar]
- Carrick DM, Lai WS, Blackshear PJ (2004) The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res Ther 6 248–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MT, Yu SM (1998) The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc Natl Acad Sci USA 95 6543–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defense gene expression and protein-protein interactions in rice. Mol Plant Pathol 7 417–427 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1999) Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc Natl Acad Sci USA 96 10512–10517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X (2003) Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell 4 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiera JM, Bouchard RA, Dorsey SL, Park E, Buenrostro-Nava MT, Ling PP, Finer JJ (2007) Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26 1501–1509 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R (2005) General translational repression by activators of mRNA decapping. Cell 122 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R (2007) RNA quality control in eukaryotes. Cell 131 660–668 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8 9–22 [DOI] [PubMed] [Google Scholar]
- Farley BM, Pagano JM, Ryder SP (2008) RNA target specificity of the embryonic cell fate determinant POS-1. RNA 14: 2685–2697 [DOI] [PMC free article] [PubMed]
- Farmer EE, Ryan CA (1990) Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA 87 7713–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J (2005) Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20 905–915 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446 [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J (2007) TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J (2008) The control of mRNA decapping and P-body formation. Mol Cell 32 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8 113–126 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R (2004) A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048 [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YH, Yu YP, Wang D, Wu CA, Yang GD, Huang JG, Zheng CC (2009) GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol 183 62–75 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35 D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11 257–264 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Takeda A, Motose H, Watanabe Y (2007) Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett 581 2455–2459 [DOI] [PubMed] [Google Scholar]
- Jadhav S, Rana M, Subramaniam K (2008) Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J (2000) A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev 14 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Sheen J (1994) Sugar sensing in higher plants. Plant Cell 6 1665–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P (2007) Mammalian stress granules and processing bodies. Methods Enzymol 431 61–81 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 96 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Kennington EA, Blackshear PJ (2002) Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem 277 9606–9613 [DOI] [PubMed] [Google Scholar]
- Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ (2006) Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26 9196–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13 2269–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian XJ, Gallouzi IE (2009) Oxidative stress increases the number of stress granules in senescent cells and triggers a rapid decrease in p21waf1/cip1 translation. J Biol Chem 284 8877–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R (2005) MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E (2005) Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR (1996) The PIE-1 protein and germline specification in C. elegans embryos. Nature 382 710–712 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–422 [DOI] [PubMed] [Google Scholar]
- Munemasa S, Oda K, Watanabe-Sugimoto M, Nakamura Y, Shimoishi Y, Murata Y (2007) The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells: specific impairment of ion channel activation and second messenger production. Plant Physiol 143 1398–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury SF, Muhlemann O, Stoecklin G (2006) Turnover in the Alps: an mRNA perspective. Workshops on mechanisms and regulation of mRNA turnover. EMBO Rep 7 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai M, Roncato MA, Canoy AS, Rouquie D, Sarda X, Freyssinet G, Robaglia C (2006) Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol 141 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D (1983) Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol 3 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D (1989) Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 9 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Taylor CB, Newman TC, Green PJ (1993) The effect of sequences with high AU content on mRNA stability in tobacco. Proc Natl Acad Sci USA 90 11811–11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, McCoig LM, Ryder SP (2007) Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem 282 8883–8894 [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25 635–646 [DOI] [PubMed] [Google Scholar]
- Phillips RS, Ramos SB, Blackshear PJ (2002) Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J Biol Chem 277 11606–11613 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al (2003) ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res 31 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G (2000) Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell 6 445–455 [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N, Munemasa S, Nakamura Y, Shimoishi Y, Mori IC, Murata Y (2008) Roles of RCN1, regulatory A subunit of protein phosphatase 2A, in methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid. Plant Cell Physiol 49 1396–1401 [DOI] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2006) Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Yokosawa H, Kawahara H (2006) OMA-1 is a P granules-associated protein that is required for germline specification in Caenorhabditis elegans embryos. Genes Cells 11 383–396 [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A (2008) Messenger RNA regulation: to translate or to degrade. EMBO J 27 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev 13 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15 173–183 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Anderson P (2007) In a tight spot: ARE-mRNAs at processing bodies. Genes Dev 21 627–631 [DOI] [PubMed] [Google Scholar]
- Strome S (2005) Specification of the germ line. WormBook 28 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Lehmann R (2007) Germ versus soma decisions: lessons from flies and worms. Science 316 392–393 [DOI] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A (2004) Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol 134 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48 1148–1158 [DOI] [PubMed] [Google Scholar]
- Taylor GA, Thompson MJ, Lai WS, Blackshear PJ (1996) Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol Endocrinol 10 140–146 [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Subramaniam K, Dunn MA, Seydoux G (2001) PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev 15 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20 515–524 [DOI] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed]
- Xu J, Yang JY, Niu QW, Chua NH (2006) Arabidopsis DCP2, DCP1, VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB (1998) The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci USA 95 13254–13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K (2006) The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J 47 49–62 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.