Abstract
Stem cell number in shoot and floral meristems of Arabidopsis (Arabidopsis thaliana) is regulated by the CLAVATA3 (CLV3) signaling pathway. Perception of the CLV3 peptide requires the receptor kinase CLV1, the receptor-like protein CLV2, and the kinase CORYNE (CRN). Genetic analysis suggested that CLV2 and CRN act together and in parallel with CLV1. We studied the intracellular localization of receptor fusions with fluorescent protein tags and their capacities for interaction via efficiency of fluorescence resonance energy transfer. We found that CLV2 and CRN require each other for export from the endoplasmic reticulum and localization to the plasma membrane (PM). CRN readily forms homomers and interacts with CLV2 through the transmembrane domain and adjacent juxtamembrane sequences. CLV1 forms homomers independently of CLV2 and CRN at the PM. We propose that the CLV3 signal is perceived by a tetrameric CLV2/CRN complex and a CLV1 homodimer that localize to the PM and can interact via CRN.
In Arabidopsis (Arabidopsis thaliana), the stem cell number in the shoot apical meristem is regulated by negative feedback regulation. Stem cell induction and maintenance are controlled by the homeodomain protein WUSCHEL (WUS), and WUS expression is in turn repressed by CLAVATA3 (CLV3; Brand et al., 2000; Schoof et al., 2000), which encodes a 13-amino acid arabinosylated glycopeptide that is secreted from stem cells (Ohyama et al., 2009). Three genes have been identified that encode receptors for CLV3 signaling. Mutations in CLV1 (Clark et al., 1997), encoding a leucine-rich repeat (LRR) receptor kinase, CLV2, encoding a LRR receptor-like protein (Jeong et al., 1999), and CORYNE (CRN), encoding a receptor-like kinase, disrupt CLV3 signaling and allow the stem cell domain to expand (Sablowski, 2007; Müller et al., 2008). Binding of CLV3 to the LRR domains of CLV1 was recently shown (Ogawa et al., 2008).
A simple readout for CLV3 signaling is carpel number. Stem cells of floral meristems are normally consumed with the production of two central carpels. Any reduction in CLV3 signaling, which results in increased WUS expression and production of more stem cells, causes an increase in carpel number. Mutations in CLV1, CLV2, or CRN showed an intermediate carpel number phenotype and reduced CLV3 signaling (Müller et al., 2008). Double mutants of clv2 with crn were epistatic, but double mutants of clv1 with clv2 or crn were synergistic and abolished CLV3 signaling. This indicated that CLV1 acts independently from, and in parallel with, CLV2 and CRN to transmit the CLV3 signal. Furthermore, clv2 and crn mutants showed additional phenotypes, such as elongated pedicels and defects in stamen development, suggesting that CLV2 and CRN act in a common pathway (Müller et al., 2008). Both CRN and CLV2 were proposed to be membrane localized and may physically interact via their transmembrane domains or immediately adjacent sequences (Fig. 1A). A loss-of-function mutation of CRN, crn-1, is caused by an amino acid exchange within the predicted transmembrane domain, suggesting that membrane localization, interaction with a partner protein, or both is essential for CRN function.
Figure 1.
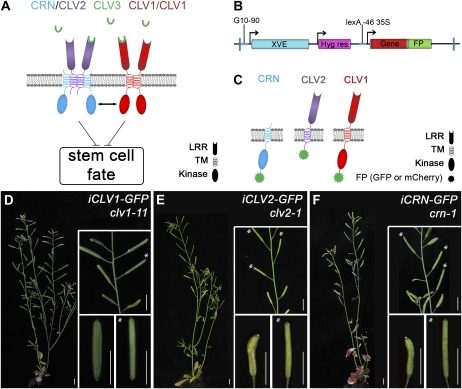
Inducible transgene expression rescues the corresponding mutants. A, Speculative model for interactions of CLV3 with receptor complexes. CLV3 peptide is proposed to bind to two separate receptor systems, consisting of CLV1 and CLV2 together with CRN. CLV2 homodimers could interact with CRN via their transmembrane domains. Receptor activity restricts stem cell fate, and the separate receptor complexes may interact through their kinase domains. B, T-DNA for inducible expression of translational fusions with the FPs GFP, mCherry, or both. G10-90, Constitutive promoter; XVE, chimeric transcription factor that activates transcription from the lexA-46 35S promoter upon estradiol induction. FPs are GFP in pABindGFP, mCherry in pABindmCherry, and GFP-mCherry in pABindFRET. C, Schematic representation of CLV1-FP, CLV2-FP, and CRN-FP. D to F, Examples of partial phenotypic restoration in clv1-11, clv2-1, and crn-1 mutants upon induced expression of the corresponding FP fusion protein. Left panels show whole plants; top insets show higher magnification of the primary shoot; bottom insets show mutant siliques with four carpels and rescued siliques (asterisks) with two carpels. D, clv1-11 carrying the iCLV1-GFP transgene. Inducing iCLV1-GFP expression led to the formation of a single silique with two carpels, whereas older and younger siliques form four carpels. E, clv2-1 carrying iCLV2-GFP. iCLV2-GFP induction restored carpel number to two in four siliques. F, crn-1 carrying iCRN-GFP. iCRN-GFP induction led to the formation of three siliques with only two carpels; all older and younger siliques consisted of four carpels. Bars = 1 cm.
We have here investigated the intracellular localization of CLV1, CLV2, and CRN in plant cells and their tendencies for protein-protein interactions. Using fluorescent protein (FP) tags, we show that CLV1 resides at the plasma membrane (PM). We found that CLV2 and CRN require each other for transport from the endoplasmic reticulum (ER) to the PM. Via fluorescence resonance energy transfer (FRET), we show that CLV2 and CRN form complexes at the ER that then relocalize to the PM, and we identified the protein domains required for this interaction. Furthermore, we found that CLV1 homomerizes but can also interact with CRN and CLV2, suggesting a mechanism for cross talk between the two receptor complexes for CLV3 signaling.
RESULTS
Mutant Complementation with FP-Tagged Receptors
For FRET-based analyses of protein-protein interactions, the proteins CLV1, CLV2, and CRN were expressed as fusions with FPs. We found that constitutive expression of most receptor-GFP fusions from the cauliflower mosaic virus (CaMV) 35S promoter failed to rescue the corresponding mutants in transgenic Arabidopsis. Therefore, we decided to use an inducible gene expression system, which allowed us to study protein localization and interaction at variable protein concentrations.
A set of transgenic Arabidopsis plants was generated that expressed fusions of CLV1, CLV2, or CRN with either GFP or mCherry at their C terminus (CLV1-GFP, CLV1-mCherry, etc.) from an estradiol-inducible promoter (Zuo et al., 2002; Fig. 1, B and C). We studied the functionality of the different receptor-FPs by analyzing their capability to complement the corresponding loss-of-function mutant plants, clv1-11, clv2-1, or crn-1 (Fig. 1, D–F). Transgenic plants were first cultivated for 28 d on soil, before transgene expression was induced with 20 μm β-estradiol on days 28, 30, and 32. At 8 weeks after germination, we assayed for restoration of CLV3 signaling by counting the carpel number of 10 siliques on each plant. The nontransgenic control plants as well as mock-induced transgenic plants produced on average 3.9 carpels per silique. Importantly, siliques with less than three carpels were never observed under our growth conditions. Induced transgenic plants showing at least two consecutively formed siliques with wild-type carpel number were regarded as complemented, and those carrying a single rescued silique were regarded as partially complemented. We found that all our inducible transgenes were functional in vivo and complemented the respective mutations, albeit with different frequencies (Table I). Although up to two flower primordia are normally initiated per day (Smyth et al., 1990) and transgene expression was induced for at least 5 d, we found that many transgenic plants carried only two siliques with two carpels. Transient restoration of CLV3 signaling could indicate delayed floral primordium initiation, rapid loss of transgene activity due to cosuppression, or sensitivity of the system to receptor concentrations.
Table I.
Rescue of clv signaling mutants by inducible expression of receptor-FP fusions
Transgenic T1 plants of the indicated genotypes were estradiol induced at 4 weeks after germination. clv1-11, clv2-1, and crn-1 mutants carry siliques with on average 3.9 ± 0.1 carpels (Müller et al., 2008). Carpel number of 10 siliques was analyzed per plant. N, Number of T1 lines; C, number of complementing T1 plants, with percentage of total, carrying more than one silique with only two carpels (wild type); PC, partially complemented, carrying one silique with two carpels; NC, noncomplemented, producing only mutant carpels.
| Fusion | N | C | PC | NC |
|---|---|---|---|---|
| iCLV1-FP clv1-11 | 48 | 16 (33%) | 4 (8%) | 28 (59%) |
| iCLV2-FP clv2-1 | 23 | 21 (91%) | 2 (9%) | 0 (0%) |
| iCRN-FP crn-1 | 20 | 11 (55%) | 4 (20%) | 5 (25%) |
|
crn-1 |
10 |
0 (0%) |
0 (0%) |
10 (100%) |
Intracellular Localization of CLV1, CLV2, and CRN
In Arabidopsis, we detected induced CLV1-GFP fusion proteins in hypocotyl and root cells and very faint CRN-GFP signals when expressed from the CRN promoter, but we failed to visualize CLV2-GFP. However, we were unable to reproducibly detect the fusion proteins in shoot or floral meristems (Supplemental Fig. S1).
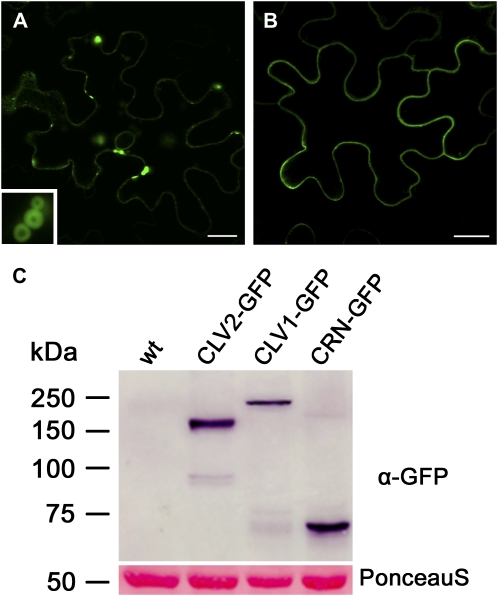
Because localization studies with stably transformed Arabidopsis plants proved not feasible, we used a transient expression system in Nicotiana benthamiana leaf epidermis cells for further experiments. Constitutively expressed CLV1-GFP localized to the PM, but we also noted the formation of larger, fluorescing aggregates (Supplemental Fig. S2). Similar aggregates were observed for CaMV35S::CLV2-GFP and CaMV35S::CRN-GFP, which could be caused by protein overexpression. To control protein expression levels, we then used the estradiol-inducible system. Vectors were transformed into intact leaves via Agrobacterium tumefaciens infiltration, and expression was induced by spraying leaves with β-estradiol. Signals from FPs were first detectable 3 h after induction. Upon extended induction (>12 h), most cells carried large FP aggregates, similar to those observed in our constitutive expression experiments (Fig. 2A; Supplemental Figs. S1 and S2). Such an aggregation of receptor-FP fusions upon extended induction could be responsible for the only transient complementation of the Arabidopsis mutants. Therefore, all measurements were performed with cells that did not show this overexpression phenotype (Fig. 2B). Integrity of the fusion proteins was confirmed by western blotting using an anti-GFP antibody (Fig. 2C). The anti-GFP antibody identified a single specific band each for CLV1-GFP (138 kD), CLV2-GFP (110 kD), and CRN-GFP (75 kD). The sizes of the detected bands for CLV1-GFP and CLV2-GFP were slightly increased compared with the calculated fusion protein sizes. Similar discrepancies have been noted for other proteins carrying LRR domains and are likely due to posttranslational modifications, such as glycosylation (van der Hoorn et al., 2005).
Figure 2.
Analysis of receptor-GFP fusion protein expression. A and B, Transient expression of iCLV1-GFP in leaf epidermis cells of N. benthamiana. Bars = 20 μm. A, Long induction (>12 h) of iCLV1-GFP causes formation of fluorescent aggregates (inset shows close-up). B, At 4 h after induction, CLV1-GFP localizes predominantly to the PM. C, Western-blot analysis of protein extracts from N. benthamiana leaf cells transiently expressing CLV1-GFP, CLV2-GFP, or CRN-GFP. An anti-GFP antibody was used for detection; sizes of protein markers are given in kD. The Ponceau S-stained protein bands of Rubisco are shown as a loading control. wt, Wild type.
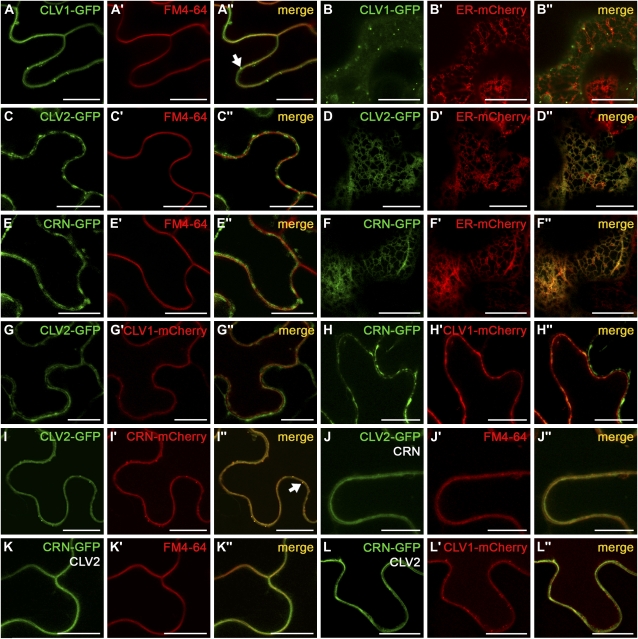
CLV1-GFP predominantly localizes to the PM, which was confirmed by colocalization with the lipophilic fluorescent dye FM4-64 (Fig. 3A). Staining was also observed in vesicles, but only faintly in the ER, which reflects transport to the PM and the site of synthesis (Fig. 3B). In contrast, cells expressing CLV2-GFP or CRN-GFP revealed a spotty GFP pattern near the PM, which did not colocalize with the PM marker FM4-64 (Fig. 3, C and E). Transformation of CLV2-GFP or CRN-GFP together with an ER-tagged mCherry reporter (Nelson et al., 2007) revealed complete colocalization in the ER network (Fig. 3, D and F). Because GFP-positive transport vesicles were never observed, we conclude that CLV2 and CRN are predominantly ER localized. This contrasts with our expectation of CLV3 perception and signaling at the PM.
Figure 3.
Intracellular localization of CLV1, CLV2, and CRN. Transient expression of FP-tagged receptor chimeras in epidermis cells of N. benthamiana. Confocal sections of epidermis cells either through the middle of a cell (A–A″, C–C″, E–E″, and G–L″) or beneath the outer cell wall (B–B″, D–D″, and F–F″) are shown. A to A″, C to C″, E to E″, J to J″, and K to K″, Receptor-GFP colocalization with FM4-64. B to B″, D to D″, and F to F″, Receptor-GFP colocalization with an mCherry ER-reporter. A to A″, CLV1-GFP colocalizes with the FM4-64 dye at the PM and in a few transport vesicles (arrow). B to B″, Weak CLV1-GFP expression in the ER. C to C″, CLV2-GFP is found next to the PM in the ER and does not colocalize with FM4-64. D to D″, Colocalization of CLV2-GFP with the ER reporter mCherry. E to E″, Like CLV2, CRN-GFP is not found at the PM. F to F″, CRN-GFP colocalization with the ER-mCherry protein. G to G″, Coexpression of CLV1-mCherry with CLV2-GFP does not affect their localization in the cell. H to H″, Coexpression of CLV1-mCherry with CRN-GFP does not affect their localization in the cell. I to I″, Coexpression of CLV2-GFP with CRN-mCherry leads to their relocation to the PM and the formation of transport vesicles (arrow). J to J″, CLV2-GFP colocalizes with FM4-64 at the PM if CRN is coexpressed. K to K″, CRN-GFP colocalizes at the PM with FM4-64 in the presence of CLV2. L to L″, CRN-GFP colocalized with CLV1-mCherry at the PM in the presence of CLV2. Bars = 20 μm.
Protein localization may depend on the establishment of functional complexes; therefore, we also tested our fusion proteins in coexpression experiments. Coexpression of CLV2-GFP with CLV1-mCherry (Fig. 3G) or CRN-GFP with CLV1-mCherry (Fig. 3H) did not affect their localization at the ER and PM, respectively. However, coexpression of CLV2-GFP and CRN-mCherry caused a relocation of both proteins from the ER to the PM (Fig. 3I) and the formation of transport vesicles. This relocation was confirmed by colocalization of CLV2-GFP with FM4-64 in the presence of CRN (Fig. 3J) and of CRN-GFP with FM4-64 in the presence of CLV2 (Fig. 3K). Cotransformed cells were easily detectable by their decreased ER fluorescence, the production of GFP-positive vesicles, and PM staining. When CLV1, CLV2, and CRN were coexpressed, the tagged proteins were always located at the PM (Fig. 3L).
PM Localization Requires Transmembrane and Extracellular Domains of CRN
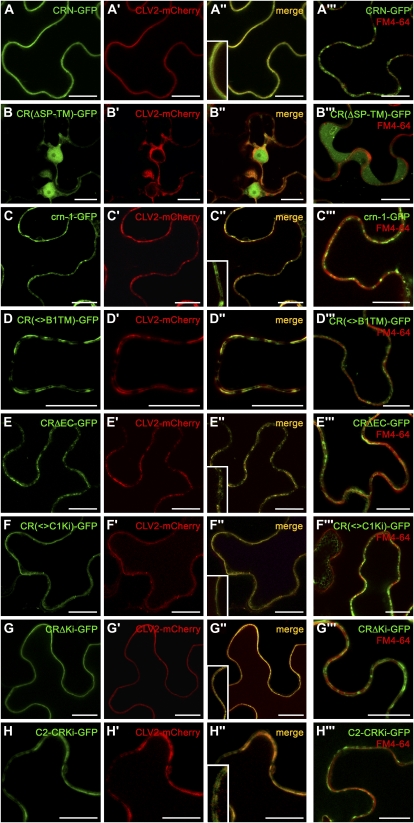
In silico studies indicated that CRN consists of a signal peptide (SP; amino acids 1–33), a short extracellular domain (EC; amino acids 34–61), the transmembrane domain (TM; amino acids 62–84), and the protein kinase domain (amino acids 118–393; Müller et al., 2008). To identify the protein domains required for interaction with CLV2 and PM localization, we designed CRN derivatives and coexpressed them with CLV2. Deleting the SP, EC, and TM of CRN [CR(ΔSP-TM)] resulted in a fusion protein of 56 kD that was detected in the cytoplasm and the nucleus and failed to relocate CLV2 to the PM upon coexpression (Fig. 4, A and B). crn-1 encodes a mutant protein that carries an amino acid exchange in the TM, which shortens the predicted TM from 23 to only 19 amino acids (Müller et al., 2008). The mutant phenotype was explained by CRN protein mislocalization or instability. We found that crn-1-GFP was stably expressed and ER localized (Fig. 4C). Coexpression of crn-1-GFP with CLV2-mCherry induced only a partial relocalization of both proteins from the ER to the PM, and very few transport vesicles were detectable. We then swapped the TM of CRN with the TM of BRI1-ASSOCIATED RECEPTOR KINASE (BAK1; Russinova et al., 2004; Chinchilla et al., 2007), a PM-localized LRR receptor protein kinase acting in the brassinosteroid and flagellin signaling pathways [CR(<>B1TM)]. Even when coexpressed with CLV2, CR(<>B1TM) was mostly retained in the ER, indicating that the specific TM of CRN is essential for CLV2-dependent PM localization (Fig. 4D). Similarly, deletion of the EC of CRN (CRΔEC) caused a complete retention in the ER, even when CLV2 was coexpressed (Fig. 4E). The extracellular juxtamembrane domains (28 amino acids) of CRN and CLV2 are oppositely charged, with pI 11.7 for the EC of CRN and pI 3.7 for the EC of CLV2. Close apposition and neutralization of these charges may be required to permit the relocalization from the ER to the PM. Next, we addressed the role of the kinase domain of CRN. An exchange against the kinase domain of CLV1 [CR(<>C1Ki)] caused increased retention of CLV2 and CR(<>C1Ki) at the ER when coexpressed, suggesting that the CRN kinase domain contributes to PM localization (Fig. 4F). However, deletion of the entire kinase domain (CRΔKi) still allowed PM expression together with CLV2 (Fig. 4G), which shows that the localization of CRN is independent of its kinase domain or function and depends only on sequences that allow the interaction with CLV2. The presence of the CLV1 kinase domain in CR(<>C1Ki) may destabilize the fusion protein or interfere with CLV2 interaction. A fusion of CLV2 with the intracellular domain of CRN (C2-CRKi) was retained in the ER, even in the presence of coexpressed CLV2 (Fig. 4H). We conclude that interaction of the TM and EC of CRN with CLV2 is essential to direct both proteins to the PM. Integrity of the CRN derivatives was confirmed by western blotting using an anti-GFP antibody (Supplemental Fig. S3).
Figure 4.
CRN localization at the PM requires the TM domain and adjacent sequences. Transient expression of receptor-FP fusions in N. benthamiana. Confocal sections through the middle of an epidermis cell are shown. First to third columns show coexpression of CRN-GFP or derivatives with CLV2-mCherry. Fourth column shows localization of CRN-GFP or derivatives in the absence of CLV2 and PM labeling with FM4-64 (red). A to A″′, CRN and CLV2 localize to the PM. B to B″′, CR(ΔSP-TM)-GFP is found in the cytoplasm and nucleus, and CLV2 is now found in the ER. C to C″′, The point mutation in crn-1 reduces PM localization. D to D″′, The TM of BAK1 is insufficient to replace the TM of CRN. E to E″′, Deleting the EC of CRN abolishes PM localization of CRN and CLV2. F to F″′, Exchanging the CRN kinase domain against the CLV1 kinase domain weakly interferes with PM localization. G to G″′, Deleting the CRN kinase domain does not affect PM localization. H to H″′, Fusion of CLV2 to the CRN kinase domain abolishes PM localization. Insets show close-ups. Scale bars = 20 μm.
CLV1 and CRN Homomerization
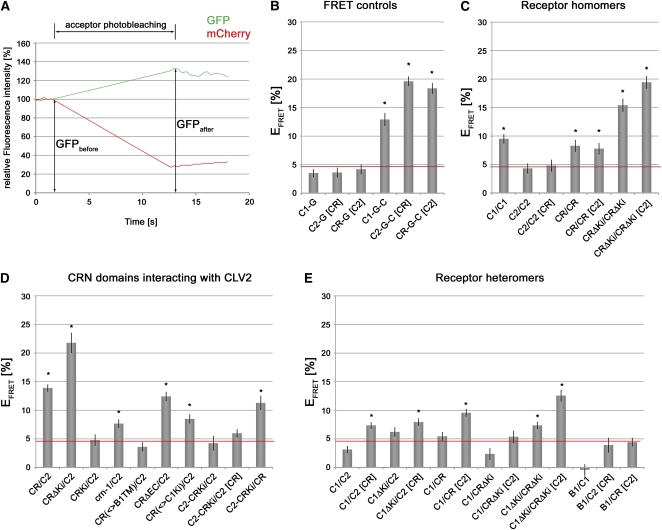
We measured FRET efficiency (EFRET) between FP-labeled receptor proteins for more detailed interaction studies. EFRET has to be considered as “apparent,” because the signal was not corrected for background contributions, spectrum-dependent cross talk, and sensitivity (Raicu, 2007). As FRET pair we used the combination of GFP with mCherry, which were previously described to be sufficiently stable to enable EFRET measurements (Albertazzi et al., 2009). Emission spectra of these proteins were fully separable from autofluorescence of plant cells. We then quantified donor (GFP) fluorescence after photobleaching of the acceptor (mCherry) and calculated EFRET as the resulting percentage GFP expression change (Fig. 5A). Unless noted otherwise, all measurements were performed at the PM, which required coexpression of untagged CRN for CLV2-FPs and of untagged CLV2 for CRN-FPs. EFRET depends on the orientation of chromophores to each other and on their distance. As a control for minimum distance, we fused GFP with mCherry to the C termini of CLV1 (C1-G-C), CLV2 (C2-G-C), and CRN (CR-G-C; Fig. 5B). Intramolecular EFRET ranged from 13% to 19%. Even in the absence of mCherry, GFP fluorescence fluctuated by 3% to 4% during the course of an acceptor photobleaching experiment. This fluctuation could result from GFP chromophore reconstitution during the bleaching period (Fig. 5B). A similar level of EFRET “background” was previously observed by others for the cyan fluorescent protein/yellow fluorescent protein pair (Karpova et al., 2003). In all further experiments, only EFRET significantly higher than 4% was regarded to indicate close proximity or physical interaction between proteins (Supplemental Table S1). These significance levels were confirmed by independent fluorescence measurement of donor lifetimes (Weidtkamp-Peters et al., 2009).
Figure 5.
Receptor interaction revealed by EFRET. A, Principle of EFRET measurement by acceptor photobleaching. Fluorescence intensities of GFP and mCherry are recorded. EFRET is calculated as relative increase of GFP fluorescence intensity (%) after photobleaching of the mCherry FRET acceptor. B, EFRET control measurements. GFP background fluctuations of C1-G, C2-G, and CR-G at the PM and measurement of intramolecular FRET are shown. C, Receptor homomers. D, Capacity of CRN deletion derivatives and domain swaps to interact with CLV2. E, Formation of receptor heteromers. Gray bars show mean values, with se indicated. Asterisks mark EFRET levels significantly different from GFP fluorescence fluctuation. Red lines at 4% indicate background fluctuation level of GFP. B1, BAK1; C, mCherry; C1, CLV1; C2, CLV2; CR, CRN; G, GFP; Ki, kinase domain; square brackets, coexpression of unlabeled protein; <>, domain exchange.
To analyze the formation of receptor homomers (Fig. 5C), we coexpressed CLV1, CLV2, and CRN fused to GFP and mCherry. Importantly, only homomers formed between receptors labeled with different fluorophores can be quantified by these measurements, and interactions such as CLV1-GFP/CLV1-GFP and CLV1-mCherry/CLV1-mCherry are undetectable. CLV1 clearly formed homomers at the PM (9.5% ± 0.8%). Importantly, homomers were formed in the absence of exogenous CLV3, suggesting that CLV1 homointeraction is ligand independent. EFRET values for CLV2 were close to background (4.3% ± 0.8%), indicating no dimerization. Furthermore, CLV2 homointeraction was not stimulated by coexpression of CRN (4.8% ± 1.0%). However, CRN showed significant self-interaction already in the ER (8.3% ± 1.0%). Deleting the kinase domain (CRΔKi) increased EFRET (15.4% ± 1.1%), possibly due to decreased distance between the chromophores. Alternatively, the kinase domain could promote the disassembly of the protein complex. CRN homomerization must be mediated by the TM and immediately flanking sequences. Coexpression of CLV2 relocalized CRΔKi to the PM and resulted in even higher EFRET values (19.4% ± 1.1%), suggesting the formation of a CRN/CLV2 complex.
We conclude that both CLV1 and CRN self-interact and that the kinase domain is not required for CRN homodimers. CRN homomeric complexes are already formed in the ER and are translocated in the presence of CLV2 to the PM. For CLV2, no homomerization was detected.
CRN Interacts with CLV2 in the ER and at the PM
The interdependence between CRN and CLV2 for PM localization suggested that both proteins may bind to each other (Fig. 5D). We found the CRN/CLV2 interaction to be the strongest we observed in all our EFRET measurements, giving 13.9% ± 0.6% for CRN/CLV2 in both GFP/mCherry combinations and 21.8% ± 1.7% for the CRΔKi/CLV2 interaction. This suggested that the TM and juxtamembrane domains are not only essential for PM localization but also for the interaction of CRN with CLV2. Consistent with this, CR(ΔSP-TM)-GFP and CLV2-mCherry did not interact (4.8% ± 0.9%). Furthermore, the mutant crn-1 protein, which showed reduced PM localization, interacted only weakly with CLV2 (7.7% ± 0.7%). Replacing the CRN TM with that of BAK1 abolished CLV2 interaction (3.6% ± 0.8%). Interestingly, deleting the EC of CRN, which caused complete ER retention, did not affect the interaction with CLV2 (12.4% ± 0.7%). We conclude that CRN and CLV2 form complexes via their TM domains and that the EC of CRN is required for ER exit and localization of the complex to the PM.
CRN Mediates the Binding of CLV2 to CLV1
We did not detect interaction between CLV1 and CLV2 (3.2% ± 0.5%; Fig. 5E). This is not unexpected, because CLV2 is localized to the ER (in the absence of CRN) and only a subpopulation of CLV1 is found in the ER before transfer to the PM. Deleting the CLV1 kinase should shorten the distance between the FP tags that are located at the C termini of CLV1 and CLV2; however, no significant EFRET was observed (6.2% ± 0.7%). Importantly, when we coexpressed untagged CRN, significant EFRET between CLV1 and CLV2 was recorded (7.4% ± 0.5%). We then tested whether CLV1 can directly interact with CRN. In the absence of CLV2, we observed no significant interaction of CRN with CLV1 (5.5% ± 0.7%). Deleting the kinase domains (CRΔKi and C1ΔKi) should increase the sensitivity of the FRET assay. For these deletion constructs, we found significant interaction between CRΔKi and C1ΔKi in the ER (7.4% ± 0.6%). Upon coexpression with untagged CLV2, CLV1/CRN heteromers were now found at the PM and EFRET values increased further (9.6% ± 0.6% for CRN and CLV1, 12.6% ± 0.9% for CRΔKi and C1ΔKi).
To test for the specificity of the observed interactions, we analyzed the behavior of BAK1, a LRR receptor kinase that was previously shown to heterodimerize with BRI1 (Russinova et al., 2004), FLS2 (Chinchilla et al., 2007), and possibly other LRR-RLKs as coreceptor (Kemmerling et al., 2007). BAK1-GFP localized to the PM as expected, but we did not observe significant EFRET with mCherry-tagged CLV1, CLV2, CRN, or combinations of them (Fig. 5E; Supplemental Table S1).
Our experiments revealed that CRN binds to CLV2 via its TM. The CRN/CLV2 heteromer can interact with CLV1 at the PM. Translocation to the PM requires the EC domain of CRN. CLV2 interacts with CLV1 only indirectly, probably as part of the CRN/CLV2 heteromer. Thus, CRN mediates both the localization of the CRN/CLV2 heteromer to the PM and also its interaction with CLV1 monomers or homomers.
DISCUSSION
CLV3 controls stem cell proliferation in shoot and floral meristems and requires CLV1, CLV2, and CRN for its activity. Genetic studies suggested that the CLV3 signal is transmitted by CLV1 and CLV2 with CRN in two separate receptor pathways that can cross talk (Müller et al., 2008). It is always desirable to analyze interaction and signaling capacity of receptor proteins in their native context (i.e. in those cells where they are normally expressed). Attempts to express components of the CLV3 signaling pathways in Arabidopsis suffered from low expression signals when the endogenous promoters were used or from protein aggregation and degradation when strong constitutive promoters were employed. Inducible expression of receptor-FP fusions allowed complementation of the respective mutants, but in most cases only in a limited number of floral meristems, indicating integrity of the receptor fusions and that their dosage can be critical for in vivo function. To analyze receptor localization and interaction, we switched to the N. benthamiana leaf system, which allowed us to precisely control and monitor expression levels and to avoid any overexpression artifacts. Our transient expression studies of translational fusions with GFP and mCherry now showed that all three receptor proteins can localize to the PM and have the capacity to undergo multiple interactions.
First, we found that CLV1 forms homomers at the PM of N. benthamiana cells. Because this interaction was revealed by FRET between differentially labeled CLV1 fusion proteins, we have no information on the exact composition of this complex. The native ligand for CLV1 and CLV3 is normally expressed only from stem cells and not in epidermal cells, indicating that receptor oligomerization is not induced by ligand binding and that CLV1 can preassemble into binding-competent homotypic complexes. We also noted that the addition of a 12-amino acid CLV3 peptide, which suffices to activate CLV signaling (Kondo et al., 2006), did not further stimulate heterotypic interactions (Supplemental Fig. S4). This indicates that the signaling mode for CLV1 differs from that of other peptide-binding LRR-RLKs, such as the flagellin receptor FLS2. Here, interaction with BAK1 together with mobility reduction was induced by flagellin, indicating that ligand binding is required to trigger heteromerization (Ali et al., 2007; Chinchilla et al., 2007). A capacity for homotypic interaction was also found for CRN but not for CLV2. Notably, both CRN and CLV1 homomeric complexes may assemble already in the ER.
Second, CRN and CLV2 interact in the ER and require each other for localization to the PM. Deletion mapping and domain swaps showed that the TM of CRN specifically mediates binding to CLV2. The mutation in crn-1 introduces a charged amino acid into the TM, which decreases interaction with CLV2 and also PM localization. A similar mutation in the GABAA receptor caused ER retention but was also associated with protein degradation via the ERAD pathway (Gallagher et al., 2005), whereas crn-1-GFP fusions remained expressed at levels comparable to the wild-type protein.
The CLV2/CRN interaction is not sufficient for PM localization but requires also the EC domain of CRN, which may serve to neutralize charged residues in the juxtamembrane domain of CLV2. Because CLV2 also remains in the ER if CRN is absent, we propose that the formation of a CLV2/CRN complex has to shield ER retention signals that are found in both proteins. Such quality control via a trafficking checkpoint was previously described for the GABA receptor. Here, a possible ER retention signal RXR at the C-terminal domain of the GB1 receptor must be shielded by GB2 for PM expression of only functionally assembled receptor complexes (Margeta-Mitrovic et al., 2000). Although a related sequence motif is found at the cytoplasmic tail of CLV2, it cannot be solely responsible for ER retention, because complexes between CLV2 and CRN lacking the EC domain still fail to localize to the PM. The cytoplasmic juxtamembrane domain of CRN further contributes to complex assembly in the absence of CLV3, and together with the TM and EC sequences it forms the pre-ligand-binding assembly domains of CRN. Such receptor preassembly in the ER has been proposed to assist in signaling specificity and rapidity (Chan et al., 2000). Furthermore, it represents a mechanism to avoid surface expression of unassembled receptors. This could be especially important to safeguard against interactions at the PM between CRN and CLV2-related RLPs that respond to danger signals and trigger plant immune responses (Boller and Felix, 2009).
It is possible that both CLV2/CRN heteromers and CLV1 homomers assemble already in the ER, which could be investigated using fluorescence-based techniques such as fluorescence lifetime imaging microscopy with high local resolution (Weidtkamp-Peters et al., 2009). Given that the expression patterns of CRN, CLV2, and CLV3 overlap in meristem cells of the L3 layer, CLV3 could act there as an intracrine signal, which may serve to sharpen the boundary between stem cells and the organizing center.
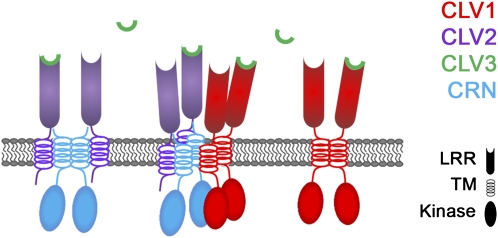
Based on our results, we propose here that CLV1 homodimers and CLV2/CRN heterotetramers coexist at the PM of shoot meristem cells (Fig. 6). Both receptor complexes should independently bind the processed CLV3 peptide and activate intracellular signal transduction. Formation of a larger complex, comprising CLV1, CLV2, and CRN, could allow for cross talk between the receptors and may be necessary to fine-tune the response to CLV3 signaling. In addition to its role in CLV3 signaling, the CLV2/CRN complex would contribute to the regulation of other developmental pathways, including the regulation of pedicel length, anther development, and root meristem growth (Müller et al., 2008).
Figure 6.
Model of CLV receptor complexes. CRN dimer interacts with two CLV2 receptors via the TM domain. Juxtamembrane sequences are required to secure interaction and for localization of the complex at the PM (left). CLV1 forms homodimers (right), which can also bind the tetrameric CLV2/CRN complex (middle). This interaction is mediated by CRN. Binding of CLV3 to the three complexes may trigger different signal transduction cascades.
The secreted bone morphogenetic proteins (BMPs) control many aspects of vertebrate development and are perceived by homomeric and heteromeric receptor complexes, consisting of different receptor types. Importantly, separate signaling pathways are activated when BMP-2 binds to preassembled complexes or when receptors are first recruited by BMP-2 (Gilboa et al., 2000; Nohe et al., 2002). We now need detailed studies on downstream events to elucidate the consequences of CLV3 signaling via different receptor complexes that comprise CLV1, CLV2, and CRN.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Nicotiana benthamiana plants were grown for 4 weeks in a greenhouse under controlled conditions. Arabidopsis (Arabidopsis thaliana) mutant lines were obtained from the Nottingham Arabidopsis Stock Centre. clv1-11 (Dievart et al., 2003), clv2-1 (Jeong et al., 1999), and crn-1 (Müller et al., 2008) mutations are in a Landsberg erecta background and were described previously.
Construction of Inducible Receptor Fusions
The destination plasmids pABindGFP, pABindmCherry, and pABindFRET were designed by inserting coding regions of GFP(S65T) (pABindGFP), mCherry (pABindmCherry), and GFP(S65T) + mCherry (pABindFRET) 3′ to the Gateway cassette of pMDC7. To create receptor fusions, attB sites were added via PCR-mediated ligation to coding regions of CLV1 (AT1G75820.1), CLV2 (AT1G65380.1), BAK1 (AT4G33430.1), or CRN (AT5G13290.2) and recombined into pDONR201 according to the manufacturer's instructions (Gateway manual; Invitrogen). The following derivatives were created via PCR-mediated mutagenesis: (1) CRΔEC, deletion of amino acids 36 to 61; (2) CR(<>B1TM), exchange of amino acids 62 to 84 from CRN against amino acids 225 to 247 of BAK1; (3) CRΔKi, deletion of amino acids 88 to 401; (4) CR(<>C1Ki), exchange of amino acids 91 to 401 against amino acids 665 to 980 of CLV1; (5) C2-CRKi, fusion of amino acids 1 to 705 of CLV2 with amino acids 86 to 401 of CRN; and (6) CR(ΔSP-TM), deletion of amino acids 1 to 92.
Construction of Transgenic Arabidopsis
The transgenes were transformed into Arabidopsis plants via the floral dip method and selected as described previously (Stahl et al., 2009). Transgene expression was induced 4 weeks after germination by spraying with 20 μm β-estradiol and 0.1% Tween (three times every 48 h).
Phenotypical Analysis
Carpel number analysis was used to determine CLV signaling pathway activity. Mature siliques were individually observed with a dissection microscope. The siliques were examined to determine the carpel number, and a partially formed carpel was counted as one. Photographs were taken with a Canon Powershot G2 digital camera or with a AxioCam ICc3 mounted onto a Zeiss dissecting microscope. Digital photographs were collated with Adobe Photoshop.
Transient Gene Expression in N. benthamiana Leaves
The Agrobacterium tumefaciens strain GV3101 pMP90 was transformed with expression clones and cultured on double yeast tryptone medium supplemented with rifampicin (50 μg mL−1) gentamycin (50 μg mL−1), and spectinomycin (100 μg mL−1). Bacterial cultures were grown, precipitated, and dissolved in 5% (w/v) Suc, 150 μm acetosyringone, and 0.01% (v/v) Silwet. To reduce gene silencing in planta, cultures were mixed with an Agrobacterium culture that allows expression of the silencing suppressor p19 (Voinnet et al., 2003), giving an optical density at 600 nm of 0.3 for each strain. Leaves of 4-week-old N. benthamiana plants were infiltrated with the culture. Transgene expression was induced 48 to 96 h after infiltration with 20 μm β-estradiol + 0.1% Tween 20 and analyzed within 4 to 24 h.
Confocal Microscopy
Epidermis cells were examined with a 40× 1.3 numerical aperture Zeiss oil-immersion objective using a Zeiss LSM 510 Meta confocal microscopy system. GFP was excited with a 488-nm argon laser with emission detection through the meta-channel at 497 to 550 nm. mCherry was excited at 561 nm using a diode, and emission was detected at 572 to 636 nm via the meta-channel. EFRET was measured via GFP fluorescence intensity increase after photobleaching of the acceptor mCherry. Frame size was kept constant at 256 × 256 pixels, with a pixel time of 2.55 μs per pixel. A region of interest around the PM was bleached after five detection frames with 100% laser intensity of the 561-nm diode and 120 iterations. Fifteen frames were recorded after photobleaching. The GFP fluorescence intensity change was analyzed around the PM in the region of interest. Only measurements with less than 10% GFP intensity fluctuations before acceptor bleaching were further analyzed. The percentage change of the GFP intensity directly before and after bleaching was analyzed as EFRET = (GFPafter − GFPbefore)/GFPafter × 100. FM4-64 (Invitrogen) staining was performed at a final concentration of 20 μm for 5 to 20 min. A minimum of 15 measurements were performed for each experiment. Significance was analyzed using Student's t test.
Western-Blot Analyses
Approximately 0.1 g of N. benthamiana leaf tissue was homogenized with a Precellys Homogenisator (PeqLab) for 20 seconds in 740 μL of extraction buffer (0.1 m Tris-HCl, pH 8.3, 5 mm dithiothreitol, 5 mm EDTA, and 5 μL of protease inhibitor cocktail [Sigma P9599]). After 1 h incubation at 4°C, 90 μL of 10× SDS loading buffer (0.25 m Tris, 1.92 m glycine, and 1% [w/v] SDS) was added, heated for 10 min at 95°C, and separated by 8% (v/v) SDS-PAGE. The separated proteins were transferred to a polyvinylidene difluoride membrane and probed with the primary anti-GFP antibody (Roche) and a secondary anti-mouse alkaline phosphatase-conjugated antibody (Dianova). Nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate was used as detection substrate.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CLV1 (NM_106232), CLV2 (NM_105212), BAK1 (NM_119497), and CRN (NM_180481).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein expression analysis in Arabidopsis.
Supplemental Figure S2. Localization of constitutively and inducibly expressed receptor-GFP fusion proteins.
Supplemental Figure S3. Western-blot analysis of protein expression in N. benthamiana.
Supplemental Figure S4. Receptor interaction independency on CLV3 treatment.
Supplemental Table S1. EFRET measurements via acceptor photobleaching.
Supplementary Material
Acknowledgments
We are grateful to Carin Theres, Cornelia Gieseler, and Silke Winters for technical support and to Marcel Lafos, Nicole Schatlowski, and members of the R.S. laboratory for critical comments.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SFB590 to R.S. and C.A.M.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rüdiger Simon (ruediger.simon@uni-duesseldorf.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albertazzi L, Arosio D, Marchetti L, Ricci F, Beltram F (2009) Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem Photobiol 85 287–297 [DOI] [PubMed] [Google Scholar]
- Ali GS, Prasad KV, Day I, Reddy AS (2007) Ligand-dependent reduction in the membrane mobility of FLAGELLIN SENSITIVE2, an Arabidopsis receptor-like kinase. Plant Cell Physiol 48 1601–1611 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60 379–406 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619 [DOI] [PubMed] [Google Scholar]
- Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ (2000) A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288 2351–2354 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448 497–500 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585 [DOI] [PubMed] [Google Scholar]
- Dievart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, Clark SE (2003) CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher MJ, Shen W, Song L, Macdonald RL (2005) Endoplasmic reticulum retention and associated degradation of a GABAA receptor epilepsy mutation that inserts an aspartate in the M3 transmembrane segment of the alpha1 subunit. J Biol Chem 280 37995–38004 [DOI] [PubMed] [Google Scholar]
- Gilboa L, Nohe A, Geissendorfer T, Sebald W, Henis YI, Knaus P (2000) Bone morphogenetic protein receptor complexes on the surface of live cells: a new oligomerization mode for serine/threonine kinase receptors. Mol Biol Cell 11 1023–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG (2003) Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J Microsc 209 56–70 [DOI] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Mussig C, et al (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17 1116–1122 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 845–848 [DOI] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY (2000) A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27 97–106 [DOI] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, Knaus P (2002) The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem 277 5330–5338 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5 878–880 [DOI] [PubMed] [Google Scholar]
- Raicu V (2007) Efficiency of resonance energy transfer in homo-oligomeric complexes of proteins. J Biol Phys 33 109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R (2007) The dynamic plant stem cell niches. Curr Opin Plant Biol 10 639–644 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed]
- van der Hoorn RA, Wulff BB, Rivas S, Durrant MC, van der Ploeg A, de Wit PJ, Jones JD (2005) Structure-function analysis of Cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17 1000–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Felekyan S, Bleckmann A, Simon R, Becker W, Kuhnemuth R, Seidel CA (2009) Multiparameter fluorescence image spectroscopy to study molecular interactions. Photochem Photobiol Sci 8 470–480 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30 349–359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.