Abstract
Many plants flower in response to seasonal changes in daylength. This response often varies between accessions of a single species. We studied the variation in photoperiod response found in the model species Arabidopsis (Arabidopsis thaliana). Seventy-two accessions were grown under six daylengths varying in 2-h intervals from 6 to 16 h. The typical response was sigmoidal, so that plants flowered early under days longer than 14 h, late under days shorter than 10 h, and at intermediate times under 12-h days. However, many accessions diverged from this pattern and were clustered into groups showing related phenotypes. Thirty-one mutants and transgenic lines were also scored under the same conditions. Statistical comparisons demonstrated that some accessions show stronger responses to different daylengths than are found among the mutants. Genetic analysis of two such accessions demonstrated that different quantitative trait loci conferred an enhanced response to shortening the daylength from 16 to 14 h. Our data illustrate the spectrum of daylength response phenotypes present in accessions of Arabidopsis and demonstrate that similar phenotypic variation in photoperiodic response can be conferred by different combinations of loci.
Growth and reproduction of many plant species are regulated by seasonal changes in daylength. Specific traits controlled by daylength include flowering, bud dormancy in trees, and tuberization of potato (Solanum tuberosum; Thomas and Vince-Prue, 1997). Within a species, there is often quantitative variation for the precise length of day that induces a response, and the distribution of accessions that respond to different daylengths suggests that this trait is associated with adaptation to growth at particular latitudes. Examples of such distributions include induction of flowering by daylength in cultivated populations of soybean (Glycine max; Borthwick and Parker, 1939) and natural populations of Xanthium strumarium (Ray and Alexander, 1966) or repression of bud growth in poplar (Populus spp.; Bohlenius et al., 2006). The mechanisms controlling photoperiodic flowering are best understood in Arabidopsis (Arabidopsis thaliana; Kobayashi and Weigel, 2007; Turck et al., 2008), but no comprehensive analysis of quantitative variation in photoperiod response within this species has been reported.
Arabidopsis is a quantitative long-day plant that flowers earlier under long days (LDs) of spring and early summer than during short days (SD) of winter. Commonly used laboratory accessions such as Columbia (Col) and Landsberg erecta (Ler) show a marked flowering response to daylength and were used to screen for mutations that impair photoperiodic flowering (Redei, 1962; Koornneef et al., 1991). The genes identified by these mutations defined a pathway that promotes flowering in response to LDs. GIGANTEA, CONSTANS (CO), and FLOWERING LOCUS T (FT) are central to this pathway (Putterill et al., 1995; Fowler et al., 1999; Kardailsky et al., 1999; Kobayashi et al., 1999; Park et al., 1999). Transcription of each of these genes is regulated by the circadian clock (Fowler et al., 1999; Park et al., 1999; Suarez-Lopez et al., 2001), while CO activity is promoted by exposure to light both at the transcriptional and posttranscriptional levels (Valverde et al., 2004; Imaizumi et al., 2005). This complex regulation ensures that CO activates FT transcription only under LDs.
In addition to the mutational analysis performed in Arabidopsis accessions commonly used in the laboratory, natural genetic variation has been studied by analyzing genetic differences between a wider range of accessions. The most dramatic variation in flowering is between accessions that show a strong requirement for vernalization (extended exposure to low temperatures) to induce flowering and those that flower rapidly without vernalization. These distinct types are often referred to as winter annuals and summer annuals, respectively. Detailed genetic analysis based on crossing both types identified the semidominant locus FLOWERING LOCUS C (FLC) and the dominant locus FRIGIDA (FRI), which are present in winter annuals and are required for the vernalization response (Burn et al., 1993; Clarke and Dean, 1994; Koornneef et al., 1994; Lee et al., 1994). FLC encodes a MADS box transcription factor that represses flowering, and FRI promotes FLC transcription prior to vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999). Exposure of plants to vernalization for several weeks causes FLC transcript levels to fall due to changes in chromatin structure at the FLC locus (Bastow et al., 2004; Sung and Amasino, 2004). Summer annuals carry alleles of FLC or FRI that reduce the activity of one or both genes. Mutations at FRI and FLC appear to have occurred independently many times, conferring the summer annual habit (Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Michaels et al., 2003; Lempe et al., 2005; Shindo et al., 2005).
Although genetic differences at loci involved in vernalization are responsible for much of the variation in flowering time among Arabidopsis accessions, allelic differences at genes contributing to the photoperiodic response can also have important effects on flowering time. This variation was mainly characterized by comparing flowering time under extreme LDs of 16 h and SDs of 8 or 10 h (Alonso-Blanco et al., 1998; Lempe et al., 2005; Werner et al., 2005b). However, in one set of experiments, daylength and temperature were varied continuously through the experiment, recreating the effect of the changing seasons, which allowed identification of quantitative trait loci (QTLs) that regulate flowering time and interact with seasonal changes in environmental parameters (Li et al., 2006). Also, the flowering times of the Ler and Wassilewskija (Ws) accessions were recently characterized under a wide range of photoperiods, describing a quantitative response to photoperiod that exhibited a sigmoidal shape (Pouteau et al., 2008; Wilczek et al., 2009). Most of the described natural genetic variation in photoperiod response causes earlier flowering under SDs. For example, recessive alleles at PHYTOCHROME C (PHYC) and FLOWERING LOCUS M (FLM) were identified as causing earlier flowering under SDs and therefore reducing the difference in flowering time between LDs and SDs (Werner et al., 2005b; Balasubramanian et al., 2006; Li et al., 2006). Similarly, a single amino acid change in the CRYPTOCHROME2 (CRY2) photoreceptor was shown to be present in the Cape Verde Islands (Cvi) accession and to promote early flowering under SDs (El-Assal et al., 2001). Sequence variation at PHYC was proposed to be significant in natural populations, because both active and inactive alleles are common and these formed a latitudinal cline with the inactive allele more frequent at lower latitude while the active allele was more prevalent at higher latitude (Balasubramanian et al., 2006). Such a distribution may be consistent with the notion that a strong response to daylength is more significant at higher latitudes where more extreme seasonal variation in daylength occurs. Similarly, the dominant CRY2 allele that reduces daylength sensitivity was identified in an accession from low latitude (El-Assal et al., 2001).
Across the latitudinal range of Arabidopsis, daylength varies continuously and the LDs or SDs most commonly used in laboratory experiments are experienced only rarely. To more thoroughly analyze the daylength response of Arabidopsis, we tested over 70 accessions under six different daylengths, and the results were compared with those for many characterized mutants and transgenic lines. Accessions that show a greater delay in flowering time than Ler as daylength is reduced from 16 to 14 h were selected, and the genetic basis of this response was examined. Our results provide an extensive analysis of variation in photoperiodic flowering responses in Arabidopsis and indicate that the enhanced responses to daylength shown by two accessions are caused by different combinations of QTLs.
RESULTS
Variation in Flowering Time of Arabidopsis Accessions under a Wide Range of Daylengths
Quantitative responses to photoperiod were measured in Arabidopsis by scoring flowering time of 72 genetically divergent accessions under six different daylengths. The daylengths used varied in 2-h intervals from 6 to 16 h. The accessions included Ler, Col, and Ws to act as points of reference with previous analyses of photoperiod response and with studies on mutants recovered in these backgrounds. Accessions showing extreme late flowering under standard 16-h daylengths were excluded from the analysis, because most of these exhibit a strong vernalization requirement. The accessions used are listed in Supplemental Table S1, and their genetic relatedness is illustrated in Supplemental Figure S1 (see “Materials and Methods”).
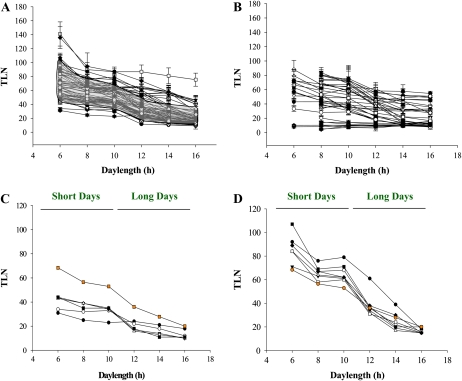
Flowering time was measured by counting total leaf number for each accession under all daylengths (Fig. 1; Supplemental Table S1; see “Materials and Methods”). In most cases, the photoperiod response curve was sigmoidal (Fig. 1A), as described previously for Ws and Ler (Pouteau et al., 2008; Wilczek et al., 2009). Typically, flowering time of these accessions occurred only slightly earlier when daylength was lengthened from 6 to 8 h, was broadly the same between daylengths of 8 and 10 h, accelerated markedly as daylength was lengthened from 10 to 12 h, and was little affected as daylength was extended from 12 to 16 h.
Figure 1.
Characterization of flowering responses of Arabidopsis accessions, mutants, and transgenic lines under a range of daylengths. A, Flowering times of 72 accessions under six daylengths. Daylength is plotted in hours along the horizontal axis. Flowering time, measured as total leaf number (TLN), is plotted on the vertical axis. The flowering time data are also presented in Supplemental Table S1. The photoperiodic response curve of each accession is represented. The gray region highlights those accessions showing a sigmoidal photoperiodic response curve, as determined by the correlation coefficients between the theoretical sigmoidal pattern and the actual data for the accessions. B, Flowering times of 31 mutants and transgenic lines under six different daylengths. Daylength is plotted in hours along the horizontal axis. Flowering time, measured as total leaf number, is plotted on the vertical axis. The flowering time data are also presented in Supplemental Table S1. C, Flowering times of a set of early-flowering accessions under all daylengths. Black circles, Cvi; white circles, Sha; black triangles, Dra-0; white triangles, Ws-0; black squares, Ler; orange squares, average response. D, Flowering times of a set of accessions showing a greater than average difference in flowering time between 6- and 16-h days and similar or earlier flowering than average at 16 h. Black circles, Cen-0; white circles, Col-2; black triangles, Col-0; white triangles, Col-3; black squares, Fr-4; white squares, Enkheim-T; black diamonds, Bs-1; orange circles, average response. [See online article for color version of this figure.]
Although the sigmoidal pattern observed in Figure 1A described the behavior of most accessions, others deviated from the standard response. For example, one group showed earlier flowering than the mean under all daylengths (e.g. Dra-0, Ler, and Ws; Fig. 1C), and this group included Cvi, which exhibited a strongly diminished response to photoperiod. A second set of accessions showed an enhanced response to daylength, flowering earlier than or similar to the mean under 16-h LDs but later than the mean under shorter days (e.g. Bs-1 and Cen-0; Fig. 1D).
Flowering Times of Arabidopsis Mutants and Transgenic Plants under a Range of Daylengths
To facilitate comparisons between accessions and mutants that impair photoperiodic responses, the flowering times of 31 previously described mutants and transgenic lines were scored under the same range of daylengths (Fig. 1B; Supplemental Table S2). These lines showed a wide range of responses. Early- and late-flowering lines showing little response to changing daylength were analyzed, and neither showed the sigmoidal pattern characteristic of wild-type plants (Supplemental Fig. S2, A and B; Supplemental Table S2).
Mutants impaired in circadian clock function were also tested (Supplemental Fig. S2C). The photoperiodic responses of late elongated hypocotyl-11 (lhy-11) circadian clock associated1-1 (cca1-1) double mutants and transgenic lines overexpressing both LHY and CCA1 (called 35S:CCA1 lhy-1) were compared. Both of these genotypes are severely impaired in circadian clock function (Schaffer et al., 1998; Wang and Tobin, 1998; Mizoguchi et al., 2002), but 35S:CCA1 lhy-1 is late flowering, whereas lhy-11 cca1-1 is early flowering. The 35S:CCA1 lhy-1 plants were almost entirely insensitive to daylength. The lhy-11 cca1-1 plants were similarly insensitive to changes in daylength from 16 to 8 h, so that in this interval flowering time only varied from 7 to 15 leaves. However, when daylength was shortened to 6 h, flowering was strongly delayed to 42 leaves. These data indicate that photoperiodic responses are almost completely abolished across a wide range of daylengths by strong impairment of the circadian clock and that lhy-11 cca1-1 plants show a very different response, being strongly delayed in flowering only when daylength falls below 8 h.
Photoperiodic responses also require active photoreceptors; therefore, the photoperiodic response curves of phyA-201 and phyB-1 mutants were compared with those of wild-type plants (Supplemental Fig. S2D). The curves describing the flowering time behavior of these mutants were similar in shape to those of wild-type plants. However, phyB-1 mutants were generally earlier flowering than wild-type plants, especially under SDs (10, 8, and 6 h). In contrast, the flowering time of phyA-201 mutants tended to be slightly later than that of wild-type plants, particularly under 12- and 8-h days.
Taken together, these data define the photoperiodic responses of a wide range of flowering time mutants and provide a basis for comparison with the responses of the accessions.
Comparisons of Daylength Responses of Mutants, Transgenic Plants, and Accessions
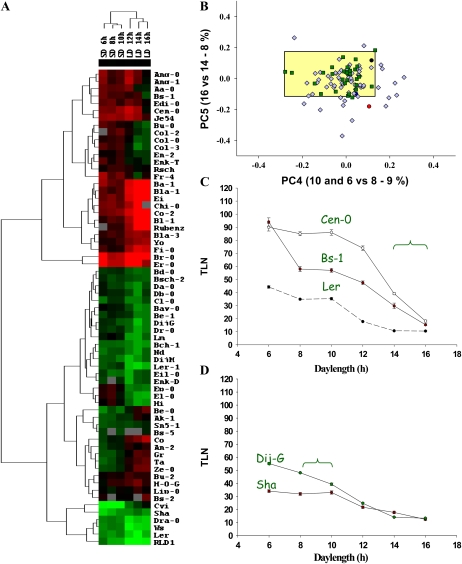
To classify the accessions according to their photoperiodic responses, the flowering time data were analyzed using the Cluster program (Fig. 2A; see “Materials and Methods”). Accessions with a generally later flowering phenotype formed clusters located at the top of the figure, while mostly earlier flowering accessions are located toward the bottom. This analysis identified several groups, each of which contained accessions showing broadly similar photoperiodic responses, and the photoperiod response curves of these accessions were compared.
Figure 2.
Characterization of flowering responses of Arabidopsis accessions under a range of daylengths. A, Hierarchical clustering analysis of daylength responses of 72 accessions. The flowering time responses of each genotype under six different daylengths were analyzed using Cluster. The colors are as follows: green, earlier flowering than the mean; red, later flowering than the mean; gray, missing values; black, equal to the mean. B, PC analysis comparing photoperiod discrimination of mutants and accessions. The y axis is PC5, which is flowering time under 16 h versus flowering time under 14 h. The x axis is PC4, which is flowering time under 10 and 6 h versus flowering time under 8 h. PC5 explains 8% of the phenotypic variation, while PC4 explains 9% of the variation. The large rectangle represents the space defined by the photoperiodic responses of mutants. The green squares represent mutants, and the light blue diamonds indicate accessions. Black circles, Ler; dark blue circles, Bs-1; red circles, Cen-0. C, Flowering times of accessions Cen-0 and Bs-1 compared with Ler. The bracket illustrates that flowering of Bs-1 and Cen-0 is delayed by shortening daylength from 16 to 14 h but flowering of Ler is not. D, Flowering times of accessions Dijon-G and Sha that show different responses to decreasing daylength below 10 h. The bracket illustrates the difference between 10- and 8-h days, which delays flowering in Dijon-G but not in Sha. TLN, Total leaf number.
Principal components (PC) analysis was used to compare the responses of individual accessions and to contrast the accessions with the mutants (Fig. 2B; Supplemental Figs. S3 and S4). Components that most effectively separated the responses of accessions and mutants were identified (Fig. 2B). These were PC5, which compared flowering time under 16 h with flowering time under 14 h, and PC4, which compared flowering time under 10 and 6 h with flowering time under 8 h (for component loading, see Supplemental Fig. S4). These components clearly identified accessions that showed stronger photoperiodic flowering responses than those present in the mutants and transgenic lines.
Based on the cluster and PC analyses, four accessions that showed interesting differences in their photoperiodic responses compared with the laboratory accession Ler were selected for further analysis. The photoperiod response curves of these accessions also confirm the differences inferred from the PC analyses. Cen-0 and Bs-1 both flowered markedly later under 14-h days than under 16-h days, whereas Ler flowered at the same time under both daylengths (Fig. 2C). Therefore, Bs-1 and Cen-0 show increased photoperiod discrimination compared with Ler, distinguishing between 14- and 16-h daylengths, whereas Ler does not. A few other accessions, such as Ang-0, showed similar responses to Bs-1 and Cen-0 (Fig. 2A; Supplemental Table S1). However, not all accessions that were later flowering than Ler at 14 h showed such a steep acceleration in flowering between 14- and 16-h days (Supplemental Fig. S2E). Similarly, the photoperiod response curves of Dijon-G and Shakdara (Sha) differed under SDs (Fig. 2D). Dijon-G distinguished between 10- and 8-h photoperiods, flowering later under 8-h days than under 10-h days. In contrast, Sha flowered at similar times under both daylengths. Thus, the statistical analysis of the photoperiodic responses of the accessions defined genotypes exhibiting increased discrimination of photoperiod under LD (16 versus 14 h) or SD (10 versus 8 h) and provided a basis for subsequent genetic analysis.
Overall, the analysis of flowering time under six different daylengths described considerable quantitative variation for photoperiodic response among the 72 accessions that were tested, placed these accessions in response groups, and identified accessions exhibiting phenotypes distinct from those found among the tested mutants and transgenic lines.
Effect of Vernalization on Accessions Showing Enhanced Response to Changing Daylength
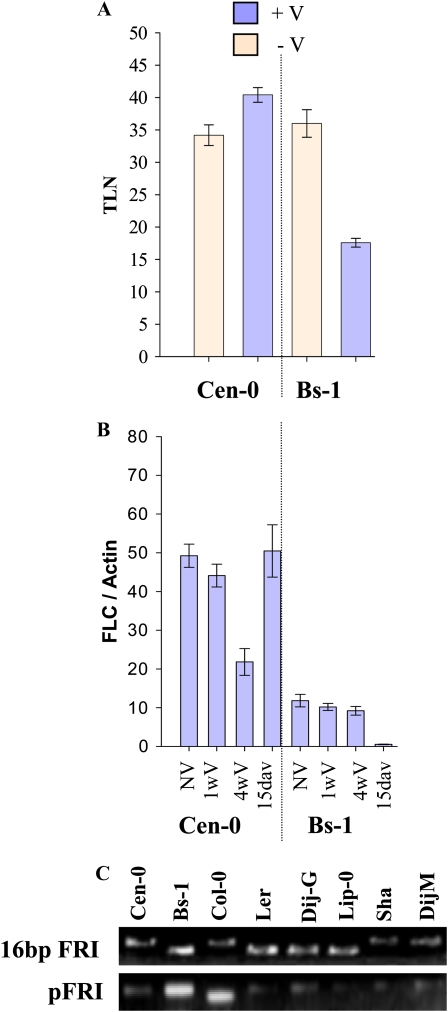
In many late-flowering Arabidopsis accessions, the delay in flowering observed under inductive photoperiods can be overcome by exposure to low temperatures (vernalization). Therefore, whether vernalization suppresses the late flowering of Bs-1 and Cen-0 under 14-h days was tested. The accessions were exposed to 4°C for 4 weeks under 8-h days and then returned to 14-h days at normal growth temperatures (Fig. 3; see “Materials and Methods”). Cen-0 plants exposed to vernalization flowered at a similar time to those that were not vernalized, indicating that the delay in flowering of this accession under 14-h days was not caused by a requirement for vernalization (Fig. 3A). In contrast, Bs-1 flowered approximately 20 leaves earlier after vernalization, indicating that the later flowering of this accession under 14-h days was at least partially suppressed by vernalization (Fig. 3A). The insensitivity of Cen-0 to vernalization was consistent with previous reports (Shindo et al., 2005); however, Bs-1 was previously described as almost insensitive to vernalization (Shindo et al., 2005). The stronger response to vernalization that we observed may be due to the plants being returned after vernalization into 14-h days rather than longer days of 16 h. Flowering of nonvernalized plants is delayed under 14 h; therefore, the difference between the flowering times of vernalized and nonvernalized plants may be more pronounced.
Figure 3.
Vernalization response of Bs-1 and Cen-0 accessions under 14-h days and its relationship to FLC expression levels. A, Flowering times of Bs-1 and Cen-0 accessions with or without vernalization. Flowering time is plotted as total leaf number (TLN) on the y axis. The accessions with or without vernalization are arranged on the x axis. B, FLC mRNA abundance before, during, and after vernalization in Bs-1 and Cen-0 accessions. FLC mRNA level relative to actin mRNA level is plotted on the y axis. Each accession without vernalization (NV), exposed to vernalization for 1 or 4 weeks (1wV and 4wV, respectively), or exposed to vernalization for 4 weeks and then returned to normal growth temperatures for 15 d (15dav), are arranged on the x axis. C, Analysis of FRI alleles present in selected accessions. Polymorphisms associated with a 16-bp deletion in the coding sequence (top) or a deletion in the promoter (pFRI; bottom) were tested. Ler and Col-0 act as controls. Ler carries the 16-bp deletion in the FRI coding sequence, whereas Col-0 carries the deletion in the FRI promoter. [See online article for color version of this figure.]
The vernalization requirement in Arabidopsis is mainly conferred by the floral repressor FLC, whose transcription is in turn repressed by vernalization. The abundance of FLC mRNA in Cen-0 and Bs-1 was measured before and after vernalization and compared with other accessions. Prior to vernalization, FLC mRNA accumulated to relatively high levels in the Cen-0 accession but to low levels in Bs-1 (Fig. 3B). These results are broadly in agreement with previous data (Shindo et al., 2005). In Bs-1, the abundance of FLC mRNA was further reduced by vernalization, and the difference was mainly observed after return to normal growth temperatures. This reduction was consistent with the earlier flowering induced by the treatment. In contrast, the high level of FLC mRNA in Cen-0 was not stably reduced by vernalization. However, previous data indicated that Cen-0 FLC mRNA is improperly spliced (Lempe et al., 2005), suggesting that Cen-0 FLC may contribute little to flowering time of this accession despite the relatively high level of its mRNA.
Sequence variation at the FRI locus plays an important role in determining the level of FLC expression. To further study the contribution of the FRI/FLC system to the flowering time of the Bs-1 and Cen-0 accessions, the FRI allele of each genotype was tested for polymorphisms characterized in other accessions (Le Corre et al., 2002; Caicedo et al., 2004; Lempe et al., 2005; Shindo et al., 2005). The inactive FRI allele present in Ler carries a 16-bp deletion within the protein-coding sequence, and a similar deletion is present in the Bs-1 allele (Fig. 3C). Therefore, the low level of FLC mRNA in Bs-1 is consistent with the presence of an inactive FRI allele (Lempe et al., 2005). In contrast, the FRI allele of Cen-0 does not carry the deletions present in Col or Ler and was previously characterized as an active allele. This conclusion is consistent with the higher level of FLC mRNA detected in Cen-0.
Taken together, these results suggest that the relatively low level of FLC mRNA in Bs-1 may nevertheless contribute to its late-flowering phenotype under 14-h days, creating the observed vernalization response. In contrast, Cen-0 exhibits high levels of FLC mRNA that are improperly spliced, consistent with the observation that it did not show a response to vernalization.
Genetic Basis of Enhanced Response to Changing Daylength Differs between Accessions
QTL mapping was carried out to identify loci contributing to the enhanced response to photoperiod of the Bs-1 and Cen-0 accessions. Each of these accessions was crossed to Ler. The flowering times of F2 populations were scored under 14- and 16-h days (Supplemental Fig. S5). Under 14-h days, approximately 17% of F2 plants derived from the cross of Bs-1 to Ler flowered later than any of the corresponding F2 plants grown under 16-h days (Supplemental Fig. S5, A and B). These data demonstrate that a proportion of F2 plants derived from this cross can be identified as late flowering under 14-h days compared with 16-h days. Analysis of late-flowering F2 plants under 14-h days allows mapping of Bs-1 alleles that delay flowering under these conditions. However, loci identified by this approach might also cause late flowering under 16-h days and, therefore, might not be responsible for generating a difference in flowering time between 14- and 16-h days (see below). The F2 population derived from the Cen-0 cross to Ler was also scored under the same conditions, and approximately 40% of plants appeared to flower later under 14-h days than under 16-h days, although only around 5% were outside the range of flowering times observed under 16-h days (Supplemental Fig. S5, E and F).
The effect of vernalization on flowering time of the segregating F2 populations was also tested (Supplemental Fig. S5, C and D). As observed in the parental Bs-1 line, the number of late-flowering plants under 14 h in the F2 population derived from the Bs-1 cross to Ler was greatly reduced by vernalization. In contrast, vernalization had little effect on flowering time of the F2 population derived from the cross of Cen-0 to Ler (Supplemental Fig. S5, G and H).
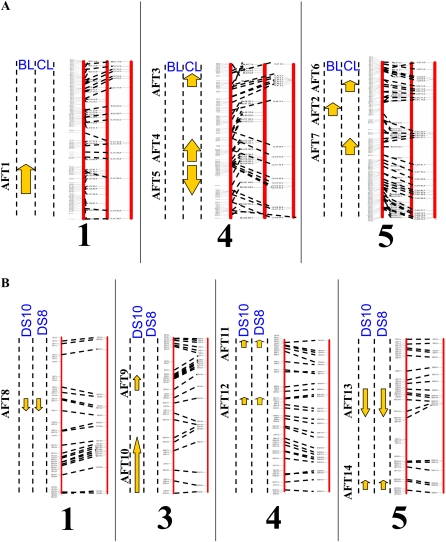
DNA was extracted from F2 plants derived from each cross grown under 14-h days to construct genetic maps and facilitate identification of QTLs that delay flowering under these conditions. Single nucleotide polymorphisms between Ler and Bs-1 or Cen-0 were identified (see “Materials and Methods”; Supplemental Table S3) and used to construct genetic maps (Supplemental Figs. S6 and S7). QTLs for flowering time under 14-h days were identified in the crosses of Bs-1 or Cen-0 to Ler (Fig. 4A; Supplemental Table S4). This analysis identified one major QTL, ALTERED FLOWERING TIME2 (AFT2), and a putative QTL, AFT1. AFT2 is located on the upper arm of chromosome 5 with a high probability of contributing to flowering time and was responsible for approximately 35% of the variation in flowering time under 14-h days. AFT1 was located on the lower arm of chromosome 1 and was responsible for only 5% of the variation in flowering time of the mapping population (Fig. 4A; Supplemental Table S4).
Figure 4.
Positions of QTLs that influence flowering time under various photoperiods. A, QTLs identified in the F2 of the cross between Bs-1 and Ler (BL) or between Cen-0 and Ler (CL) grown under 14-h days. The left panel represents QTLs located on chromosome 1, the center panel illustrates those located on chromosome 4, and the right panel represents those located on chromosome 5. Arrows between the vertical dotted lines represent QTLs identified in each cross. Arrows that point upward indicate QTLs for which the Ler allele causes early flowering. The three solid red lines in each panel represent the positions of markers located in the genomic sequence of Col (left), mapped in the BL cross (center), and mapped in the CL cross (right). Dotted black lines connect markers in the sequence to their positions in each genetic map. Long vertical black lines separate the data for each chromosome. B, QTLs identified in the F2 of the cross between Dijon and Sha grown under 10-h (DS10) or 8-h (DS8) days. The structure of each panel is as for A, except that data are shown for chromosomes 1, 3, 4, and 5. Arrows pointing upward represent those QTLs for which Sha alleles cause early flowering.
Flowering time variation under 14-h days in the F2 population derived from crossing Ler to Cen-0 was conferred by a different set of QTLs than detected in the cross of Bs-1 to Ler (Fig. 4A). In this cross, five high-probability QTLs (AFT3, AFT4, AFT5, AFT6, and AFT7) were detected. These were distributed across the upper and lower arms of chromosomes 4 and 5. The QTL located on the upper arm of chromosome 5 (AFT6) was located at a different position from AFT2, detected in the cross of Ler and Bs-1 (Supplemental Table S4). Overall, the QTLs on chromosomes 4 (AFT3, AFT4, and AFT5) and 5 (AFT6 and AFT7) in the Cen-0 × Ler cross contributed 11% and 27% of variation in flowering time, respectively (Supplemental Table S4).
These experiments identified regions in both populations that are likely to contain QTLs that delay flowering under 14-h days and showed that those present in the Bs-1 cross to Ler differ from those present in the Cen-0 cross to Ler.
Identification of QTLs Causing Enhanced Discrimination between 14-h and 16-h Daylengths
To test the heritability of the QTLs identified in the F2 mapping populations and to determine whether they had differing effects on flowering time under 14- and 16-h days, the flowering times of F3 families made by self-fertilizing selected F2 plants were scored under both daylengths. From the Bs-1 cross to Ler, five F3 families were scored in total and the range of flowering times observed in one of these, family 72, is shown in Figure 5A. Each of these families was derived from an F2 plant that was predicted to be heterozygous for the region containing the major effect QTL (AFT2) on chromosome 5. To allow comparison of the effects of QTLs under both daylengths, each F3 family was grown under 14- and 16-h days and flowering time was scored. All of the F3 families grown under 14-h days contained plants that flowered later than any of the plants grown under 16-h days, confirming that the F2 plants contained QTLs conferring late flowering under 14-h days. Plants in populations grown under both daylengths were tested with a molecular marker linked to AFT2 (Chr5_7.79). Plants homozygous or heterozygous for the Bs-1 allele at this locus were later flowering under 14-h days with higher leaf numbers than Ler plants, but they did not show a significant difference in flowering time under 16-h days (Fig. 5B). ANOVA confirmed that the difference under 14-h days but not under 16-h days was statistically significant (P < 0.05; Fig. 5B; Supplemental Table S5, A and B). However, the weaker effect QTL detected on chromosome 1 (AFT1) may also contribute, particularly because AFT2 was predicted to explain only 35% of the variation in flowering time in the F2 population (Supplemental Table S4).
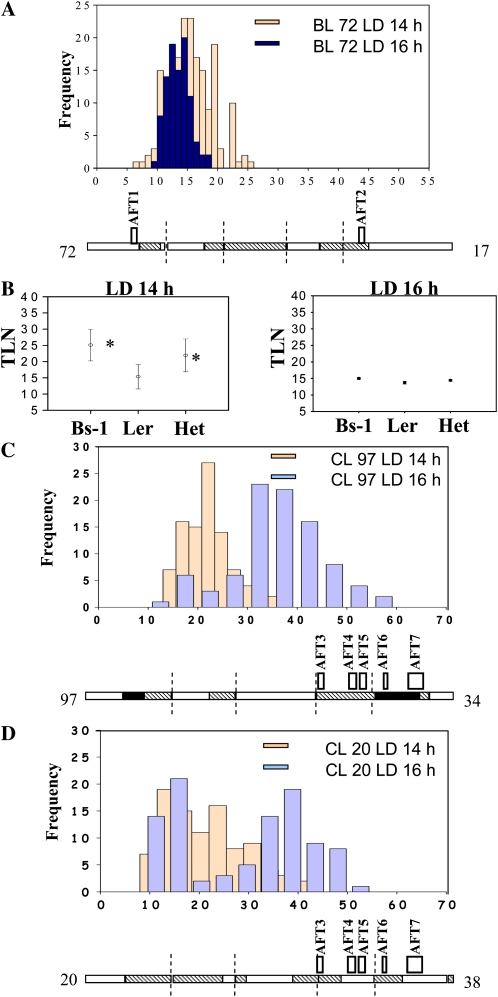
Figure 5.
Analysis of flowering time in F3 populations used to validate QTLs and test their interaction with daylength. The flowering times of plants in three F3 families grown under 14- and 16-h days are shown. In each histogram, the horizontal axis represents flowering time as leaf number. The vertical axis illustrates the number of plants. Blue indicates plants grown under 16-h days, whereas cream illustrates plants grown under 14-h days. The diagram below each histogram illustrates the genotype of the parental F2 plant. White illustrates Ler homozygous alleles, black illustrates Bs-1 or Cen-0 homozygous alleles, and hatched illustrates heterozygous alleles. The positions of the QTLs mapped in the F2 are indicated by the rectangles above each diagram, and the name of each QTL is indicated. The number to the left of the genotype indicates the number of the F2 plant whose genotype is illustrated, and the number to the right indicates the leaf number at flowering of this F2 plant. A, F3 family derived from F2 plant 72 in the Bs-1 cross to Ler (BL). B, ANOVA of the relationship between ERD2 alleles and flowering time under 14- or 16-h days in the F3 family shown in A. Left panel, analysis of data for plants grown under 14-h days; right panel, analysis of data for plants grown under 16-h days. The y axis shows flowering time represented as total leaf number (TLN). The x axis shows genotype at ERD2; the marker used was Chr5_7.79 (see “Materials and Methods”). Asterisks illustrate significant differences from Ler (P < 0.05). C, F3 family derived from F2 plant 97 in the Cen-0 cross to Ler (CL). D, F3 family derived from F2 plant 20 in the Cen-0 cross to Ler. [See online article for color version of this figure.]
Flowering time was also scored in F3 families derived from the cross of Cen-0 to Ler (Fig. 5, C and D). More QTLs were identified in the F2 of this cross (Fig. 4A), making confirmation of their effect more complex. The F2 individuals selected for F3 progeny testing were fixed or heterozygous for different combinations of QTLs. F2 plant 20 carried Cen-0 and Ler alleles at AFT3 (chromosome 4) and AFT6 (chromosome 5) and only Ler alleles at the other three loci (Fig. 5D). The F3 progeny contained severely late-flowering plants under 14 h (Fig. 5D), indicating that Cen-0 alleles at AFT3 and AFT6 interact to delay flowering under these conditions. However, the presence of Cen-0 alleles at all loci more uniformly delays flowering under 14 h (plant 97; Fig. 5C), although plants with this genotype do not exhibit a more extreme late-flowering phenotype than observed in the progeny of plant 20. Taken together, these data indicate that interactions between the Cen-0 alleles at AFT3 and AFT6 cause the most severe delays in flowering in the Cen-0 cross to Ler, but the effect of these loci under 14 h is further enhanced by Cen-0 alleles at the other loci.
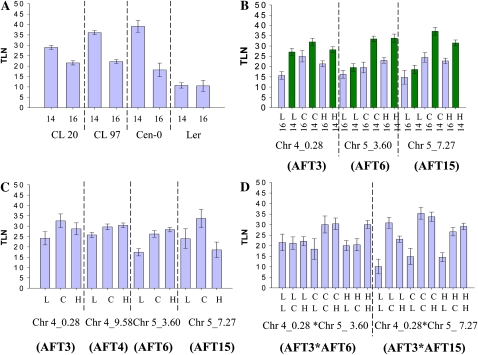
To further study the complex inheritance of flowering time variation in the Cen-0 cross to Ler, QTLs were mapped in segregating F3 populations derived from plants 97 and 20 grown under 14- and 16-h days. The mean flowering time of these families was significantly later under 14 h compared with 16 h (Fig. 6A; Supplemental Table S5, C and D). The F3 plants grown under both conditions were analyzed with 40 markers in regions of all five chromosomes that were segregating for Cen-0 and Ler alleles. Interactions between flowering time QTLs and daylength were then analyzed (Fig. 6B; Supplemental Table S5G). Homozygosity for Cen-0 alleles at AFT3 (Chr4_2.81) delayed flowering, particularly under 16 h, and reduced the effect of the daylength difference on flowering time. In contrast, homozygous Cen-0 at AFT6 (Chr5_3.60) delayed flowering under 14 h and enhanced the effect of daylength. Finally, AFT15, a QTL on chromosome 5 that was not detected in the F2 (Chr5_7.27; see below), delayed flowering under 14 h when homozygous for Cen-0 and enhanced the effect of daylength on flowering time. Taken together, these data demonstrated strong interactions between daylength and the Cen-0 allele present at AFT6 and AFT15 that would be consistent with them causing a delay in flowering under 14 h compared with 16 h, as observed in the parental accession. However, the Ler allele at AFT3 delayed flowering under 14 h but not 16 h; therefore, it did not show the effect observed in the parental Ler accession.
Figure 6.
Validation of the QTLs in the Cen-0 to Ler cross using F3 families 20 and 97, and analysis of interactions between QTLs and daylength. A, Mean flowering times of each family and parental controls under 14- and 16-h days. The y axis shows flowering time represented as total leaf number (TLN). Error bars indicate sd. The x axis shows genotypes and daylength. CL20, F3 family derived from F2 plant 20 of the Cen-0 × Ler cross; CL97, same nomenclature for F2 plant 97. B, Univariate ANOVA detection of two-way interactions between markers and changing daylength in the control of flowering time. Loci on top of chromosome 4 (4_0.28) and chromosome 5 (5_3.6 and 5_7.27) interact differently with the two daylengths. The Cen-0 allele at the chromosome 4 locus delays flowering under 16 h, reducing the difference in flowering time between 14- and 16-h days. The Cen-0 alleles at the chromosome 5 loci more strongly delay flowering under 14 h and enhance the difference in flowering time between the two daylengths. Data for families 20 and 97 were combined. The y axis shows flowering time represented by total leaf number. The x axis shows genotype and daylength. The letters correspond to allelic variation at the corresponding loci (L, Ler homozygous; C, Cen-0 homozygous; H, heterozygous alleles), while the numbers represent daylength. The positions of the markers used are indicated together with the name of the QTL mapped in the region. All markers are described in “Materials and Methods.” C, Univariate ANOVA detection of flowering time effects independently of daylength. Markers on chromosome 4 (4_0.28 and 4_9.58) and 5 (5_3.6) detect loci that delay flowering when Cen-0 alleles are homozygous or heterozygous, whereas the Cen-0 allele at Chr5_7.27 delays flowering only when homozygous. Markers are described in “Materials and Methods.” Flowering time was scored under 14 and 16 h for both families, and all data were combined for analysis. The genotype at each marker is shown: L, Ler homozygous; C, Cen-0 homozygous; H, heterozygous alleles. The position of each marker is shown together with the name of the QTL mapped in the region. D, Univariate ANOVA detection of two-way interactions between markers in determining flowering time. Allelic variation at a locus at the top of chromosome 4 (linked to marker 4_0.28) interacts with two other loci on chromosome 5 (linked to markers 5_3.6 and 5_7.27). Flowering time was scored under 14 and 16 h for both families, and all data were combined for analysis. Top letters on the x axis correspond to alleles of the chromosome 4 locus, while bottom letters correspond to alleles of the chromosome 5 loci: L, Ler homozygous; C, Cen-0 homozygous; H, heterozygous alleles. [See online article for color version of this figure.]
The flowering time data under both daylengths were then combined, and QTLs were tested for their effect on flowering time independently of daylength. Univariate ANOVA identified four regions for which the Cen-0 alleles were significantly correlated with late flowering independently of daylength (Fig. 6C; Supplemental Table S5E). The markers used for this analysis were located in regions previously shown to contain the AFT3 (marker Chr4_2.81), AFT4 (marker Chr4_9.58), and AFT6 (marker Chr5_3.60) QTLs. The region containing AFT7 was not polymorphic in either population, preventing its detection, while AFT5 was not detected. Therefore, this analysis validated three of the four QTLs identified in the F2 and segregating in the F3 material. In addition, a further QTL for flowering time (AFT15) was identified on chromosome 5 using markers at 7.27 Mb (Fig. 6C) that was not detected in the F2 population, probably due to its smaller effect.
Finally, interactions between loci on chromosomes 4 and 5 were found to have important effects on flowering time in these families (Fig. 6D; Supplemental Table S5F). The Cen-0 allele at AFT3 (Chr4_2.81) interacts with the Cen-0 allele near AFT6 (Chr5_3.6) to delay flowering in all genotypic combinations except when AFT3 is Cen-0 heterozygous and AFT6 is Cen-0 homozygous. Interaction was also observed between Cen-0 alleles near AFT3 (Chr4_0.28) and AFT15, although in this case AFT15 alone caused a severe delay in flowering (Fig. 6D).
Taken together, analysis of F3 families confirmed most of the QTLs proposed from the F2 data. These analyses also detected a strong interaction between daylength and AFT2 in delaying flowering in the Bs-1 cross to Ler and between daylength and AFT6 and AFT15 in delaying flowering in the Cen-0 cross to Ler.
Accessions Showing Enhanced Photoperiodic Flowering Response under SDs
Accessions showing differential sensitivity to SDs of less than 10 h of light were also analyzed (Fig. 2D). Flowering time of the Sha accession did not change as daylength was shortened from 10 to 6 h, while Dijon-G flowered much later under 6 h than under 10 h. To identify the loci responsible for this difference in daylength sensitivity, the two accessions were crossed and an F2 population was made. F2 plants were grown under 8- or 10-h days and flowering time was scored. In parallel, DNA was extracted from both populations and used to construct a genetic map (Supplemental Figs. S6 and S7). The flowering time data and mapping information were used to identify QTLs that influence flowering time under 8- or 10-h days (Fig. 4B). Under 8-h days, QTLs were identified on chromosomes 1 (AFT8), 4 (AFT11 and AFT12), and 5 (AFT13 and AFT14; Supplemental Table S4). Under 10-h days, the same QTLs were detected as under 8-h days, but in addition, QTLs were found on chromosome 3 (AFT9 and AFT10; Supplemental Table S4). The detection of QTLs under 10-h days but not 8-h days suggests the involvement of different genes in controlling flowering time under 10-h days compared with 8-h days. These genes may act together with or independent of the QTLs that influence flowering time under both daylengths.
Detection of Conditional Epistatic Interactions That Influence Flowering under SDs in the Cross of Dijon-G to Sha
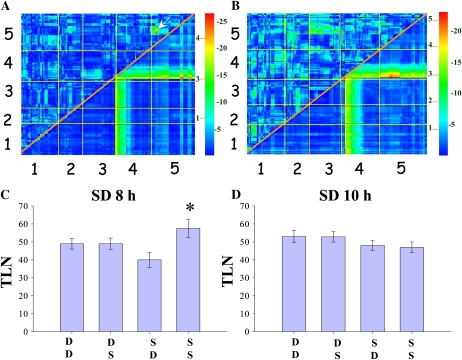
A genome-wide screen was performed to identify epistatic interactions influencing flowering time in the F2 population derived from crossing Dijon-G to Sha (Fig. 7; see “Materials and Methods”). The results are illustrated by heat maps (Fig. 7, A and B) in which the strongest interactions appear in red, as represented in the scale diagrams (see “Materials and Methods”). This analysis revealed one highly significant epistatic interaction between two loci influencing daylength perception. This interaction was between a locus at the top of chromosome 5 (1.2–2.1 Mb) and a second one in the middle of the same chromosome (13.6–17.1 Mb; Fig. 7A). The interaction was conditional on daylength because it appears only under SD of 8 h but not under SD of 10 h (Fig. 7, A and B). The phenotypic effect of this epistatic interaction is to delay or accelerate flowering by approximately 10 leaves, and it is statistically significant based on ANOVA (Fig. 7C). However, no significant difference was observed in plants grown under 10 h (Fig. 7D). These data indicate that genetic interactions with significant phenotypic effects on flowering time can be environmentally dependent and thereby contribute to daylength perception.
Figure 7.
Genome-wide detection of epistatic interactions in the mapping population created by crossing Dijon-G to Sha. A, Heat map of two-dimensional genome scan for interactions under 8-h SD. B, Heat map of two-dimensional genome scan for interactions under 10-h SD. In A and B, the numerals on the horizontal and vertical axes illustrate the five chromosomes. The top left triangle shows the epistasis log of the odds scores. The bottom right triangle illustrates the joint log of the odds scores. The color scale on the right of each panel indicates separate scales for the epistasis and joint log of the odds scores (on the left and right, respectively). The white arrow in A indicates the presence of an epistatic interaction, which is conditional, as it occurs only under 8 h, between a locus on the top of chromosome 5 (horizontal axis) and one in the middle of the same chromosome (vertical axis). C, Effect plot of the epistatic interaction under 8-h days marked in Figure 7A. Vertical axis represents flowering time as total leaf number (TLN). Horizontal axis represents genotype. Homozygous Dijon-G is indicated by D, and homozygous Sha is indicated by S. Top letters represent the locus at the top of chromosome 5, while bottom letters represent the locus in the middle of chromosome 5. The presence of Sha alleles at the top of chromosome 5 makes a difference in flowering time of around 20 leaves if the locus in the middle of chromosome 5 is also Sha. The asterisk indicates a significant difference (P < 0.05). D, The same analysis with the same loci as in C, but for plants grown under 10-h days. No significant effect of genotype on flowering time was observed.
DISCUSSION
We identified extensive quantitative variation in the photoperiodic responses of Arabidopsis accessions. The response curves of most accessions were sigmoidal, as described previously for Ws and Ler (Pouteau et al., 2008; Wilczek et al., 2009). These curves are in agreement with the classical description of Arabidopsis as a facultative long-day species (Laibach, 1951), flowering earlier under LDs than SDs. The sigmoidal curve was previously used to define two key determinants of the photoperiod response, the critical daylength and the ceiling photoperiod (Pouteau et al., 2008). The ceiling photoperiod is the longest daylength under which the accession reached the plateau in flowering time characteristic of SDs. In contrast, the critical photoperiod is the shortest photoperiod under which the accession shows the full LD response. In our data, variation in both critical photoperiod and ceiling photoperiod was observed in many accessions; however, these parameters were often difficult to score precisely because the flowering times under SDs frequently did not form a perfectly horizontal plateau, and under LDs a progressive delay in flowering time was sometimes observed as daylength was shortened. Therefore, rather than comparing these two defined positions on the photoperiod response curves, we used more general statistical approaches such as PC analysis or clustering to evaluate the general shape of the response and thereby compare the photoperiod response curves of different accessions. These approaches allowed the accessions to be placed in broad phenotypic groups that were defined relative to the mean flowering response. These groups provide a basis for genetic and molecular analysis as well as comparison with existing mutants. One group was early flowering under all photoperiods, and this included Cvi, which showed a strongly diminished response to photoperiod (Alonso-Blanco et al., 1998; El-Assal et al., 2001). This early-flowering group also included Ws, which was previously used for mutational analysis of photoperiodic response (Pouteau et al., 2008). However, the early flowering of this accession under all photoperiods reduces the absolute difference in flowering time between LD and SD and may make it more difficult to use for mutational studies. Future genetic analysis of those accessions showing enhanced responses to daylength may be particularly useful, because as well as showing an enhanced response at extreme daylengths, some of them demonstrated increased capacity to distinguish between similar daylengths. For example, Cen-0 and Bs-1 discriminated between 16- and 14-h days, whereas Ler and Col flowered at the same time under both daylengths. One interpretation of this result is that Cen-0 and Bs-1 have a longer critical daylength than Ler or Col. In addition to the three broad categories, extensive quantitative variation for photoperiodic response was detected, consistent with previous studies demonstrating tremendous variation for flowering time in Arabidopsis (Laibach, 1951; Alonso-Blanco et al., 1998; Lempe et al., 2005; Shindo et al., 2005; Werner et al., 2005a).
Altered photoperiodic responses shown by mutants or transgenic plants were compared with those of the accessions. Photoperiod-insensitive genotypes included late-flowering mutants under LDs that were impaired in the photoperiodic flowering pathway or transgenic plants overexpressing genes in this pathway that flowered early under SDs. These results were broadly consistent with previous publications and data obtained under a narrower range of daylengths (Redei, 1962; Koornneef et al., 1991; Kardailsky et al., 1999; Kobayashi et al., 1999; Onouchi et al., 2000; Mizoguchi et al., 2005). The circadian clock provides the timekeeping mechanism required to measure daylength, and genetic variation in clock function influences flowering time. Plants overexpressing the clock components LHY and CCA1 are strongly impaired in circadian clock regulation of different processes (Schaffer et al., 1998; Wang and Tobin, 1998), and in our data these plants were almost daylength insensitive, flowering much later than wild-type plants under LDs. This result is consistent with the previous observation that the mRNA of the long-day flowering pathway component CO is reduced in abundance in LHY-overexpressing plants (Suarez-Lopez et al., 2001). Similarly, lhy-11 cca1-1 double loss-of-function mutants were early flowering under all conditions tested, except in extreme SDs of 6 h of light, where flowering time was severely delayed. The early flowering of lhy-11 cca1-1 double mutants under SDs was previously proposed to be due to the earlier phase of expression of clock-regulated genes such as CO (Mizoguchi et al., 2002, 2005), so that they are expressed in the light in the double mutant under SDs but not in wild-type plants. The abrupt delay in flowering of the double mutant under 6-h SDs might be due to the expression of CO occurring in darkness under such extreme SD conditions. Alternatively, in lhy-11 cca1-1 double mutants, increased expression of repressors of flowering may be activated only under certain environmental conditions (Fujiwara et al., 2008), or metabolic defects caused by impaired clock function (Dodd et al., 2005) may indirectly delay flowering under extreme SDs.
Statistical approaches were used to compare the photoperiodic responses of the mutants and transgenic lines with those of the accessions. The extensive variation found in the mutant and transgenic lines occupied much of the phenotypic space occupied by the accessions. However, some accessions clearly showed photoperiodic responses that were not found among the mutants. The responses of some of the accessions may be caused by allelic variation at several genes, creating complex interactions that cannot be induced with single mutations in the widely used laboratory accessions Col-0, Ler, and Ws. The presence of response types in the accessions that were not observed in any of the mutants emphasizes the value of using natural genetic variation to study photoperiodic responses.
The ecological significance of the genetic variation in daylength responses that we detected is not immediately clear. Correlations between photoperiodic responses and latitude at which accessions were collected have been demonstrated for other species (Ray and Alexander, 1966). Also, the expansion of the geographical range of crop plants that occurred after domestication involved selection for photoperiod-insensitive varieties (Yano et al., 2000; Turner et al., 2005; Purugganan and Fuller, 2009). In Arabidopsis accessions, relationships have been detected between the presence of particular alleles at FRI or PHYC and the latitude at which the accessions were collected (Stinchcombe et al., 2004; Balasubramanian et al., 2006). We did not observe a strong relationship between photoperiodic response and the latitude at which the accessions were collected. In particular, the Cen-0 and Bs-1 accessions that showed a longer critical daylength than the mean response and enhanced discrimination between similar LDs were collected from France and Switzerland, at similar latitudes to many of the other accessions. Recently, the ecological significance of different flowering pathways was explored in field experiments by planting a wide range of mutant genotypes in five sites across the European range of Arabidopsis (Wilczek et al., 2009). These experiments indicated that the significance of different flowering pathways is highly dependent on germination time and that flowering of most genotypes is suppressed in winter if they germinate at an appropriate time during late summer or autumn. Therefore, the relationship between variation in photoperiodic flowering regulation and latitude may be difficult to assess in isolation but may depend on variation in the regulation of other traits, including germination. The significance of variation in Arabidopsis photoperiodic response to adaptation at different latitudes, therefore, may emerge when analyzed together with a more thorough description of the phenotypic variation in other traits in the same accessions.
In the three mapping populations, a total of 15 QTLs were detected and named AFT. None of these QTLs were mapped to high enough resolution to identify the underlying genes with certainty, but some are located in regions previously shown to contain one or more genes regulating flowering time, while others are located in regions in which no genes regulating flowering time were previously identified. Those QTLs that interacted with daylength and contribute to the increased discrimination in daylength response between 14 and 16 h or between 10 and 8 h were studied. In the Bs-1 cross to Ler, AFT2 and putatively AFT1 were detected as causing late flowering under 14 h in the F2. Furthermore, phenotypic and genotypic testing of F3 progeny strongly indicated that AFT2 interacts with daylength, conferring a delay in flowering between 14- and 16-h days. The location of AFT2 on chromosome 5 is close to the flowering time genes FRL1, HUA2, PRR5, and FPF1, which were previously described (Kania et al., 1997; Chen and Meyerowitz, 1999; Michaels et al., 2004; Doyle et al., 2005; Nakamichi et al., 2007; Wang et al., 2007). In addition, AFT1 had a small effect on flowering time in this cross and is located on chromosome 1 in a broad region containing two genes that promote flowering within the photoperiod pathway (FKF1 and FT; Kardailsky et al., 1999; Kobayashi et al., 1999; Nelson et al., 2000) and a general repressor of flowering (FLM; Ratcliffe et al., 2001; Scortecci et al., 2001). Strong loss-of-function alleles in the photoperiod pathway similar to PpdH-1 of barley (Hordeum vulgare; Turner et al., 2005) or Hd-1 of rice (Oryza sativa; Yano et al., 2000) were not expected to be detected, because accessions showing a severe late-flowering phenotype under 16-h LDs were excluded from the analysis. Nevertheless, AFT1 may represent in Bs-1 a weak loss-of-function allele of FKF1 or FT or a strong allele of FLM.
The inheritance of flowering time variation was more complex in the Cen-0 cross to Ler, and a total of six QTLs were detected in the F2 and F3 generations. Of these, AFT6 and AFT15 showed strong interactions with daylength in F3 families, delaying flowering more strongly under 14 h than under 16 h. AFT6 is located on chromosome 5 in a region containing FLC. Furthermore, AFT6 delayed flowering more strongly when combined with AFT3, a QTL located on chromosome 4 close to the position of FRI. Therefore, AFT6 and AFT3 may represent FLC and FRI, two loci previously shown to contribute much of the variation for flowering time among Arabidopsis accessions (Lempe et al., 2005, Shindo et al., 2005). Nevertheless, FLC is not expected to show such a strong interaction with daylength, because it is a central component of the vernalization pathway, and both parental accessions, Cen-0 and Ler, harbor weak alleles of FLC. AFT15, the other locus that showed a strong interaction with daylength in this cross, is located on chromosome 5 close to AFT2 and, therefore, to FRL1, HUA2, PRR5, and FPF1 (Kania et al., 1997; Chen and Meyerowitz, 1999; Michaels et al., 2004; Doyle et al., 2005; Nakamichi et al., 2007).
The cross between Dijon-G and Sha identified seven QTLs influencing flowering time under SDs of 10 or 8 h. Most of these did not show an interaction with daylength and were detected in mapping populations grown under 10- and 8-h days. However, AFT9 and AFT10 delayed flowering only under 10-h days and were not detected under 8-h days. These two QTLs are located in a region of chromosome 3 not previously shown to contain genes that affect flowering time.
The experiments reported here demonstrated that Arabidopsis accessions are a rich source of quantitative phenotypic and genetic variation in the photoperiodic response. Furthermore, the QTL analysis showed that most of this variation is genetically tractable and in some cases allowed candidate genes to be proposed. Construction of near isogenic lines as well as more detailed genetic mapping and molecular analysis should allow identification of some of the genes underlying these QTLs and examination of the mechanisms by which they contribute to the photoperiodic response.
MATERIALS AND METHODS
Flowering Time Analysis
For the flowering time analysis, 72 accessions of Arabidopsis (Arabidopsis thaliana) from the Altmann and the Nordborg collection and 31 mutants and transgenic lines were selected (Supplemental Tables S1 and S2). The accessions were taken through one generation of single seed selection to further reduce variation. The flowering time was scored in six different daylengths, three of which are SDs of 6, 8, and 10 h of light, and the remaining ones are LDs of 12, 14, and 16 h of light. The analysis was performed in controlled-environment growth chambers at 22°C after stratification for 3 d at 4°C. A population of 18 individuals represented each accession. Flowering time parameters such as (1) number of total leaves, (2) days until bolting at the stage of 1 cm, and (3) days until anthesis were monitored. The experimental design was a randomized complete block. For the vernalization treatment, 10-d-old seedlings, grown under SD of 8 h at 22°C, were exposed to 4°C for 1 month under the same photoperiod and then moved to 22°C under the desired daylength for flowering time scoring. For the genetic analysis, 100 to 150 individuals were used for each F2 population, and populations of 50 individuals were used per F3 family and condition.
Statistical Analysis
For the statistical analysis, SigmaStat version 3 was used. A two-way ANOVA was performed with accessions and daylengths as the two factors and for parameters such as total leaf number and bolting time for flowering. Demonstration of the results was performed with SigmaPlot version 10.
The hierarchical clustering shown in Figure 2A and the PC analysis were performed using Cluster version 3. The raw data were mean centered across the different daylengths and accessions in order to provide an internal control for comparisons. Self-organizing maps were calculated for both the accessions (100,000 iterations) and the daylength (20,000 iterations). Both factors were then clustered according to the complete linkage clustering method using Euclidean distance as similarity metric. TreeView version 1.6 was used to demonstrate the results. PC analysis was performed using Cluster.
QTL mapping was performed using the MapQTL program. Linkage maps were created using JoinMap. A permutation test defined the log of the odds threshold for each population. First, an interval mapping was performed, and subsequently, multiple QTL mapping with automatically selected cofactors was used. SPSS version 13 was used for the univariate ANOVA in the F3 families of Bs-1 × Ler and Cen-0 × Ler crosses.
The genetic relationship between the accessions (Supplemental Fig. S1) was calculated with the MEGA software using 149 polymorphic single nucleotide polymorphisms (www.naturalvariation.org). Genome-wide genetic interactions were performed with the J/qtl software.
Analysis of genome-wide genetic interactions (Fig. 7A) was performed in R (http://www.R-project.org/). Interactions were calculated with the function “scantwo” using 4,000 permutations. The interactions were visualized with J/qtl software.
DNA Isolation and Genotyping
DNA isolation was performed with 100 mg of fresh tissue using either the cetyl-trimethyl-ammonium bromide method (for a small no. of samples) or the semiautomated method for DNA extraction using the Biosprint robot (Qiagen) for the mapping populations. Genotyping in the F2 populations was performed in collaboration with Sequenom. Polymorphic markers were selected for each cross after parental screening of a pool of 360 markers (Supplemental Table S3; for further information, see www.naturalvariation.org). For validation and further genotyping, a standard PCR protocol was used, after which the PCR fragments were analyzed on 3% agarose gels after electrophoresis in 100 V for approximately 40 min. The Chr5_7.79 marker used for the validation in the F3 of AFT2 in the Bs-1 × Ler cross and shown in Figure 5 was a dCAPS marker cleaved with HinfI. The marker utilized the primers 5′-TCCACCGCCTTCACAATCATTAACAACTCGAC-3′ and 5′-GACAATTTGATCACCCTGCAC-3′.
Markers used for validation in the F3 of the QTL in the Cen-0 × Ler cross and shown in Figure 6 were as follows. Chr4_2.81 is a dCAPS marker cleaved with HindIII, and the primers used were 5′-GGCTGCTTTCTTAGCATCAGATGATTCTTCTTACATCACTGGAGAAGC-3′ and 5′-AAGTATCCAATGGCCTCGTG-3′. The other three markers used in Figure 6 are described at www.naturalvariation.org. Chr4_9.58 is AtMSQT_NW_173, Chr5_3.60 is AtMSQT_NW_208, and Chr5_7.27 is AtMSQT_NW_216.
Expression Analysis
Total RNA isolation was performed in 100 mg of fresh tissue using the RNAeasy kit (Qiagen). A total of 3 mg of RNA was tested on formaldehyde gel in order to evaluate the quality of the RNA. A total of 5 mg of total RNA was DNase treated with the turbo DNase kit (Ambion). The following primer pairs were used in order to perform real-time PCR after evaluation of the correct PCR condition in a gradient reaction: for actin, forward primer 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and reverse primer 5′-AACGACCTTAATCTTCATGCTGC-3′; for FLC, forward primer 5′-ACGCATCCGTCGCTCTTCT-3′ and reverse primer 5′-GCATGCTGTTTCCCATATCGA-3′.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A diagram illustrating the genetic relatedness of the accessions used in this project.
Supplemental Figure S2. Graphs depicting the flowering responses of mutants, transgenic lines, and accessions under a range of daylengths.
Supplemental Figure S3. Comparison of daylength responses of accessions, mutants, and transgenic lines by PC analysis.
Supplemental Figure S4. Daylength loadings of the PC analysis.
Supplemental Figure S5. Histograms illustrating the segregation of delayed flowering in F2 populations grown under intermediate LDs and the effect of vernalization.
Supplemental Figure S6. Detailed descriptions of the three linkage maps made in the course of this work.
Supplemental Figure S7. Alignment of the three linkage maps to the physical map of the Arabidopsis genome.
Supplemental Table S1. Description of the accessions used in this work and their flowering times under different daylengths.
Supplemental Table S2. Description of the mutants and transgenic lines used in this work together with their flowering times under different daylengths.
Supplemental Table S3. Description of the polymorphic markers used for genotyping of the three populations.
Supplemental Table S4. Summary of the QTLs detected in the three F2 mapping populations.
Supplemental Table S5. Statistical data used to validate QTLs as summarized in Figures 5 and 6.
Supplementary Material
Acknowledgments
We are grateful to Thomas Altmann (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for providing the seeds of the accessions and to Detlef Weigel (Max Planck Institute for Developmental Biology, Tübingen, Germany) for suggesting that we use Sequenom for the mapping as well as for providing Sequenom with the marker primers. We are also indebted to Maarten Koornneef (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for his advice in designing these experiments and for discussions. Amaury de Montaigu and Maria von Korff (both at Max Planck Institute for Plant Breeding Research, Cologne, Germany) made valuable comments on the text.
This work was supported by a core grant from the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: George Coupland (coupland@mpiz-koeln.mpg.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso-Blanco C, El-Assal SED, Coupland G, Koornneef M (1998) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167 [DOI] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043 [DOI] [PubMed] [Google Scholar]
- Borthwick HA, Parker MW (1939) Photoperiodic responses of several varieties of soybean. Bot Gaz 101 341–365 [Google Scholar]
- Burn JE, Smyth DR, Peacock WJ, Dennis ES (1993) Genes conferring late flowering in Arabidopsis thaliana. Genetica 90 147–155 [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD (2004) Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, Meyerowitz EM (1999) HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell 3 349–360 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Dean C (1994) Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet 242 81–89 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Bizzell CM, Keller MR, Michaels SD, Song JD, Noh YS, Amasino RM (2005) HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J 41 376–385 [DOI] [PubMed] [Google Scholar]
- El-Assal SED, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29 435–440 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Oda A, Yoshida R, Niinuma K, Miyata K, Tomozoe Y, Tajima T, Nakagawa M, Hayashi K, Coupland G, et al (2008) Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1F-BOX protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297 [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347 [DOI] [PubMed] [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S (1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D (2007) Move on up, it's time for change: mobile signals controlling photoperiod-dependent flowering. Genes Dev 21 2371–2384 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J 6 911–919 [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229 57–66 [DOI] [PubMed] [Google Scholar]
- Laibach F (1951) Uber sommer und winterannuelle Rasse von Arabidopsis thaliana (L.) Heynh: Ein Beitrag zur Atiologie der Blutenbildung. Beitr Biol Pflanz 28 173–210 [Google Scholar]
- Le Corre V, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19 1261–1271 [DOI] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM (1994) The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6 903–909 [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Roycewicz P, Smith E, Borevitz JO (2006) Genetics of local adaptation in the laboratory: flowering time quantitative trait loci under geographic and seasonal conditions in Arabidopsis. PLoS One 1 e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He YH, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T (2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48 822–832 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340 [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the GIGANTEA gene. Science 285 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pouteau S, Carre I, Gaudin V, Ferret V, Lefebvre D, Wilson M (2008) Diversification of photoperiodic response patterns in a collection of early-flowering mutants of Arabidopsis. Plant Physiol 148 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ (2009) The nature of selection during plant domestication. Nature 457 843–848 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol 126 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PM, Alexander WE (1966) Photoperiodic adaptation to latitude in Xanthium strumarium. Am J Bot 53 806–816 [Google Scholar]
- Redei GP (1962) Supervital mutants of Arabidopsis. Genetics 47 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM (2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J 26 229–236 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung SB, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159–164 [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue B (1997) Photoperiodism in Plants, Ed 2. Academic Press, San Diego
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59 573–594 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein and the mechanism of photoperiodic flowering. Science 303 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V (2007) HUA2 caused natural variation in shoot morphology of A. thaliana. Curr Biol 17 1513–1519 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005. a) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D (2005. b) Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA 102 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, et al (2009) Effects of genetic perturbation on seasonal life history plasticity. Science 323 930–934 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.