Abstract
Pseudomonas syringae pv tomato DC3000 (Pst DC3000), which causes disease in tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana), produces coronatine (COR), a non-host-specific phytotoxin. COR, which functions as a jasmonate mimic, is required for full virulence of Pst DC3000 and for the induction of chlorosis in host plants. Previous genetic screens based on insensitivity to COR and/or methyl jasmonate identified several potential targets for COR and methyl jasmonate. In this study, we utilized Nicotiana benthamiana and virus-induced gene silencing to individually reduce the expression of over 4,000 genes. The silenced lines of N. benthamiana were then screened for altered responses to purified COR. Using this forward genetics approach, several genes were identified with altered responses to COR. These were designated as ALC (for altered COR response) genes. When silenced, one of the identified genes, ALC1, produced a hypersensitive/necrosis-like phenotype upon COR application in a Coronatine-Insensitive1 (COI1)-dependent manner. To understand the involvement of ALC1 during the Pst DC3000-host interaction, we used the nucleotide sequence of ALC1 and identified its ortholog in Arabidopsis (Thylakoid Formation1 [THF1]) and tomato (SlALC1). In pathogenicity assays performed on Arabidopsis thf1 mutant and SlALC1-silenced tomato plants, Pst DC3000 induced accelerated coalescing necrotic lesions. Furthermore, we showed that COR affects ALC1 localization in chloroplasts in a COI1-dependent manner. In conclusion, our results show that the virus-induced gene silencing-based forward genetic screen has the potential to identify new players in COR signaling and disease-associated necrotic cell death.
In nature, plants come in contact with numerous microbes that are potential pathogens. Active plant defense mechanisms, in general, involve a complex network of three genetically distinct signaling pathways, known as the salicylic acid (SA), jasmonic acid (JA), and ethylene pathways (Kunkel and Brooks, 2002; Glazebrook, 2005). Pathogens, in turn, have coevolved by developing mechanisms that suppress plant defense pathways by secreting virulence factors. Several pathovars of Pseudomonas syringae produce phytotoxins. In plants, these phytotoxins generally induce chlorosis (e.g. coronatine [COR], phaseolotoxin, and tabtoxin; Mitchell, 1976; Gnanamanickam et al., 1982; Levi and Durbin, 1986) or necrosis (e.g. syringomycin and syringopeptin; Paynter and Alconero, 1979; Iacobellis et al., 1992). Bacterial toxins act as virulence factors and contribute to increased disease severity by facilitating bacterial movement in planta (Patil et al., 1974), lesion size (Bender et al., 1987; Xu and Gross, 1988), pathogen multiplication (Bender et al., 1987; Feys et al., 1994; Mittal and Davis, 1995), and suppression of plant defense (Uppalapati et al., 2007, 2008).
COR, a phytotoxin produced by P. syringae pv tomato DC3000 (Pst DC3000), is induced in the presence of the plant host metabolites such as malic, citric, shikimic, and quinic acids, which are present in leaf extracts and apoplastic fluids of tomato (Solanum lycopersicum; Li et al., 1998). COR contributes to the virulence of Pst DC3000 in Arabidopsis (Arabidopsis thaliana), tomato, collard (Brassica oleracea var viridis), and turnip (Brassica rapa var utilis; Brooks et al., 2004; Elizabeth and Bender, 2007; Uppalapati et al., 2007). It has been shown that COR has structural and functional resemblance to 12-oxo-phytodienoic acid, methyl jasmonate (MeJA), and related derivatives known as the jasmonates (JAs; Feys et al., 1994; Weiler et al., 1994; Uppalapati et al., 2005). MeJA is a plant growth hormone that plays a key role in plant defense response to biotic and abiotic stress (Howe et al., 1996; McConn et al., 1997; Vijayan et al., 1998; Truman et al., 2007).
During a compatible interaction with a host, Pst DC3000 infection results in the activation of the JA signaling pathway (Zhao et al., 2003; Laurie-Berry et al., 2006; Uppalapati et al., 2007). This causes the suppression of the SA pathway owing to its antagonistic relation with the JA pathway (Kloek et al., 2001; Kunkel and Brooks, 2002; Zhao et al., 2003; Uppalapati et al., 2007). The suppression of the SA pathway during the Pst DC3000-host interaction is thought to be caused by COR, which functions as a molecular mimic of JAs (Feys et al., 1994; Bender et al., 1999; Staswick and Tiryaki, 2004).
Pst DC3000 causes disease on several plant species including tomato and Arabidopsis. A typical symptom on tomato leaves is bacterial speck, which includes necrosis surrounded by a chlorotic halo (Mittal and Davis, 1995; Zhao et al., 2003). In Arabidopsis, the infected area exhibits water-soaked lesions accompanied by diffused chlorosis (Mittal and Davis, 1995; Brooks et al., 2004). Pst DC3000 infection also causes chlorosis in other plants belonging to the Brassicaceae family, such as collard and turnip (Elizabeth and Bender, 2007). In addition to chlorosis, Pst DC3000-infected collard plants exhibit water-soaked lesions and anthocyanin, suggesting that Pst DC3000 elicits unique responses in different plants. Studies have shown that tomato plants inoculated with a COR-defective mutant of Pst DC3000 did not develop typical chlorotic symptoms; furthermore, COR contributed to pathogen fitness and disease development in a SA-independent manner (Uppalapati et al., 2007). Tomato leaf tissues treated with purified COR show chlorosis (Gnanamanickam et al., 1982; Uppalapati et al., 2005, 2007). Unlike tomato, purified COR does not elicit chlorosis on Arabidopsis leaves (Bent et al., 1992; Mach et al., 2001). However, in Arabidopsis, COR is required for full disease symptom development and pathogen fitness in a SA-dependent manner (Kloek et al., 2001; Brooks et al., 2004). These results suggest that COR functions as an important virulence factor in tomato and Arabidopsis, although it functions differently in these hosts.
More recently, we have demonstrated a role for COR-induced effects on photosynthetic machinery and reactive oxygen species (ROS) in modulating necrotic cell death during bacterial speck disease of tomato (Ishiga et al., 2009a). Despite our present understanding of COR function, it is not clear how chlorosis impacts or benefits pathogen virulence. Furthermore, the identity of host molecular targets for COR and the downstream signaling cascades that ensue are not well understood. Based on similarities between COR and JAs in terms of structure and function (Feys et al., 1994; Uppalapati et al., 2005), it seems likely that COR and JA interact with at least one common host receptor (Katsir et al., 2008). Thus, in addition to furthering our understanding of disease development, studies aimed at understanding the molecular mechanism of COR may provide information on JA-mediated plant defense.
In an effort to identify plant proteins that are the molecular targets of COR, we used a tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) as a fast-forward genetics tool (Liu et al., 2001a, 2001b; Anand et al., 2007) to screen a Nicotiana benthamiana cDNA library for genes that are involved in the response to COR. We identified a N. benthamiana gene, ALC1 (for altered COR response), that when silenced displayed an unexpected hypersensitive/necrosis-like phenotype rather than a typical chlorotic phenotype in response to COR application. ALC1 has homology to an Arabidopsis gene, Thylakoid Formation1 (THF1; Wang et al., 2004). The pathogenicity assays performed in this study indicate that loss of ALC1/THF1 leads to accelerated cell death in response to Pst DC3000 infection in both tomato and Arabidopsis.
RESULTS
Application of Purified COR on N. benthamiana Leaves Results in Chlorosis
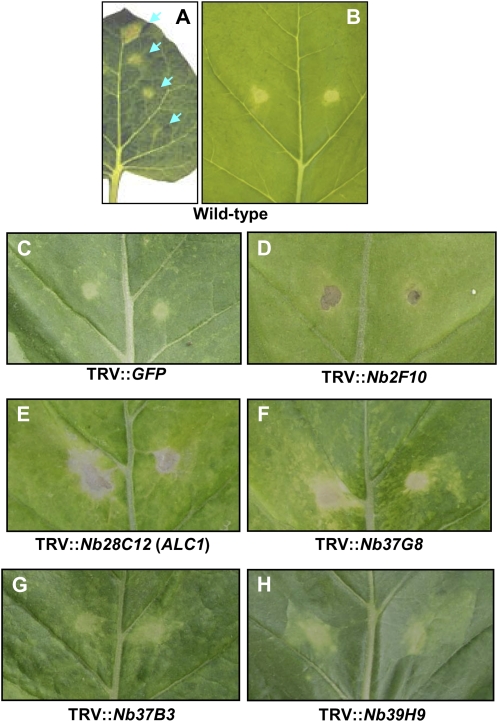
Unlike tomato, the efficiency of VIGS is quite uniform in N. benthamiana; therefore, this host is suitable for large-scale fast-forward screening studies (Lu et al., 2003; del Pozo et al., 2004; Anand et al., 2007). Purified COR, when spotted onto N. benthamiana leaves at different concentrations (0.002–2 nm in 2-μL aliquots), produced a visible, confined chlorosis in a dose-dependent manner (Fig. 1A). The 0.2 nm concentration, which produces a confined chlorosis phenotype, was used for screening (Fig. 1B). Based on these results, we concluded that a VIGS-based approach in N. benthamiana was suitable for screening silenced plants that show an altered chlorosis phenotype upon COR application; therefore, this approach was used to identify genes involved in COR-mediated signaling.
Figure 1.
COR induces visible chlorosis on N. benthamiana leaves. A, Purified COR when applied to N. benthamiana leaves in 2-μL aliquots (arrows) at different concentrations (0.002, 0.02, 0.2, and 2 nm; from bottom to top part of the leaf); a visible chlorotic zone was observed at 4 dpi. B, Chlorosis induced by 0.2 nm COR (per inoculation site) on N. benthamiana. C to H, Responses of silenced lines of N. benthamiana leaves to 2 nm COR. COR was applied 3 weeks post agroinoculation to silenced lines of N. benthamiana. In response to COR, leaves of silenced lines showed necrosis (D) or a necrosis-like phenotype (E and F). Some lines exhibited an enhanced chlorosis (G and H). Photographs were taken 7 d after COR application.
VIGS-Based Screening Identifies Several N. benthamiana Genes with Altered COR-Induced Response
To identify plant genes that are involved in COR signaling, a normalized N. benthamiana cDNA library cloned in pTRV2 and transformed into Agrobacterium tumefaciens GV2260 was used (Anand et al., 2007). N. benthamiana plants were individually inoculated with Agrobacterium containing TRV2 cDNA clones, along with an Agrobacterium strain containing TRV1, in duplicate, to silence their corresponding genes in N. benthamiana (Anand et al., 2007). Two weeks after TRV inoculation, COR (0.2 nm) was spotted on the leaves of silenced plants, and the phenotypes were recorded 5 to 7 d after COR application.
After screening approximately 4,000 cDNA clones, we identified five nonredundant cDNA clones that when silenced resulted in the ALC phenotype upon exogenous application of COR (Fig. 1, D–H). The application of COR to wild-type (Fig. 1B) or TRV″GFP-inoculated (mock control [Fig. 1C]; the GFP sequence does not have any homology to plant DNA and therefore will not cause gene silencing) N. benthamiana plants resulted in a defined chlorotic halo. The silenced lines with the ALC phenotype exhibited either hypersensitive response (HR)-like necrosis (Fig. 1, D–F) or increased chlorosis (Fig. 1, G and H) in response to COR. For example, Nb28C12- and Nb2F10-silenced plants exhibited a severe HR-like necrosis that is confined or extended beyond the area of COR application (Fig. 1, D and E), whereas Nb37G8-silenced lines (Fig. 1F) displayed a less severe necrotic phenotype. Nb37B3- and Nb39H9-silenced plants displayed slightly diffused chlorosis (Fig. 1, G and H). In brief, the VIGS-mediated fast-forward genetic approach proved to be an effective tool to identify plant genes involved in COR-induced responses in N. benthamiana.
ALC1 Is an Ortholog of a Gene Encoding THF1 Protein
Since the silencing of Nb28C12 produced the most dramatic phenotype in response to COR application, the Nb28C12-silenced line was selected for further study. The phenotype of Nb28C12-silenced plants was similar to that of control plants (TRV″GFP) up to 4 weeks postsilencing. However, after the 5th week, leaves of the Nb28C12-silenced plants turned slightly pale green in color (Supplemental Fig. S1A). At 5 weeks postinoculation, portions of some leaves showed a variegated gray coloration (Supplemental Fig. S1A). To confirm the suppression of the Nb28C12 mRNA in the silenced plants, quantitative real-time reverse transcription (qRT)-PCR was performed. The relative expression ratio of the Nb28C12 gene in the silenced line versus the control plant was 0.023, indicating that the transcript level was approximately 40-fold lower than in the control plant (Supplemental Fig. S2A).
We termed Nb28C12 as ALC1. The sequence information was then analyzed to predict gene function. A BLASTn search against The Institute for Genomic Research database using the NbALC1 sequence revealed 77% identity to an Arabidopsis gene named THF1 (GenBank accession no. AY899908); 92% identity to a potato (Solanum tuberosum) gene that encodes a light-regulated chloroplast-localized protein (StTHF1; GenBank accession no. AY342161); 81% identity to a rice (Oryza sativa) gene encoding inositol phosphatase-like protein (GenBank accession no. AY224446); and 79% identity to a wheat (Triticum aestivum) gene encoding Ptr ToxA-binding protein (GenBank accession no. AY377991). To facilitate a more comprehensive comparative analysis of ALC1, we designed a primer pair to clone the full-length ALC1 gene based on the sequence of a tobacco (Nicotiana tabacum) ortholog (GenBank accession no. TC10126). The cloned gene was then sequenced and the translated amino acid sequence was aligned with orthologous plant protein sequences using ClustalW (http://www.ebi.ac.uk/clustalw/). As shown in Supplemental Figure S3A, N. benthamiana ALC1 shows strong sequence identity with orthologs from other species. N. benthamiana ALC1 also displays a higher degree of evolutionary relatedness with the tobacco ortholog when compared with other plant orthologs that were analyzed (Supplemental Fig. S3B).
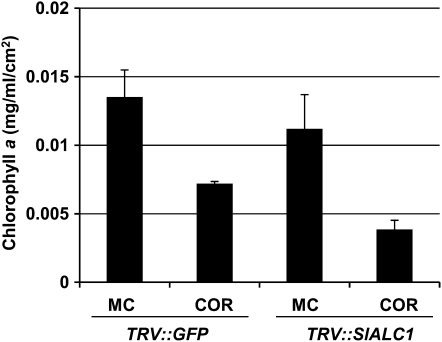
The silencing of Nb28C12 resulted in a necrotic phenotype upon COR treatment without a visible chlorosis (Fig. 1D). To test if the COR-induced altered necrotic phenotype is associated with chlorophyll degradation, we quantified chlorophyll a (Chl a) levels in vector control and NbALC1-silenced plants. Application of COR resulted in greater reduction of Chl a levels in NbALC1-silenced plants (65% reduction over mock treatment) when compared with the vector control (47% reduction over mock treatment; Fig. 2). These results suggested that the necrotic phenotype in NbALC1-silenced plants is associated with COR-induced chlorophyll degradation.
Figure 2.
COR induces chlorophyll degradation in vector control (TRV″GFP) and NbALC1-silenced (TRV″NbALC1) N. benthamiana plants. Two leaf discs (0.78 cm2) were collected 6 dpi with 2 nm COR and were analyzed for Chl a content to monitor chlorosis. MC, Mock control.
ALC1-Silenced Tomato Plants Show Accelerated Necrosis in Response to COR in a COI1-Dependent Manner
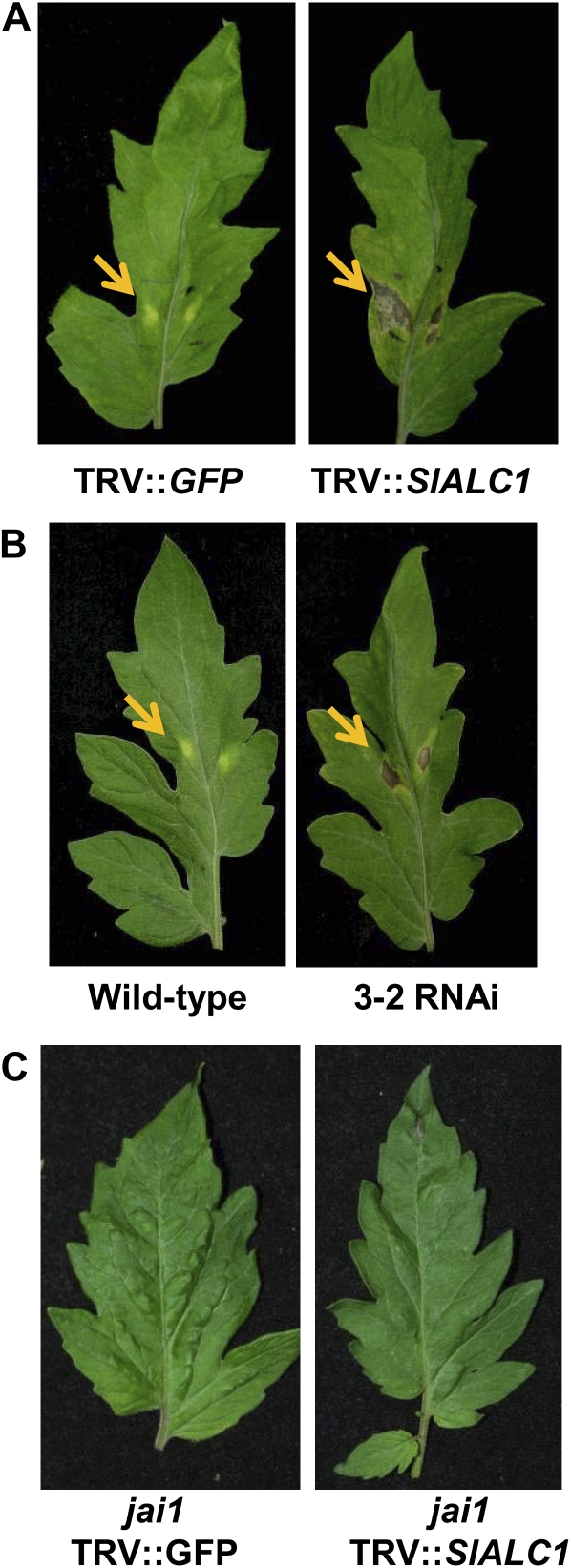
To understand the potential role of ALC1 in Pst DC3000-mediated disease development, we used the host plant tomato. Using primers specific to the tomato ortholog (The Institute for Genomic Research accession no. TC162724), we cloned the tomato ALC1 (see “Materials and Methods”), which we refer to as SlALC1. A fragment of SlALC1 was subcloned into pTRV2 and used for VIGS in tomato. qRT-PCR analysis of silenced tomato plants revealed that the expression of SlALC1 in the silenced plant was about 5-fold lower than control plants (Supplemental Fig. S2B). However, the majority of the leaves in ALC1-silenced tomatoes did not exhibit any obvious phenotype (Supplemental Fig. S1B, middle), and some of the older leaves (6 weeks after TRV inoculation) showed variegated coloration on the leaf surface (Supplemental Fig. S1B, right). When purified COR (2 nm) was exogenously applied, the silenced line showed a necrosis-like phenotype, whereas the control plants showed a typical confined chlorosis as expected (Fig. 3A).
Figure 3.
Silencing of ALC1 in tomato displays a necrosis-like phenotype in a COI1/JAI1-dependent manner in response to (2 nm) COR at 7 d posttreatment. Transient (TRV″SlALC1; A) and stably silenced (3-2; B) tomato (cv Glamour) showed necrosis in response to COR, whereas vector control (TRV″GFP) and transiently silenced (TRV″SlALC1) jai1 mutants (cv Castlemart; C) showed no chlorosis or necrosis. Photographs were taken 7 d after COR application. Arrows indicate COR response.
Although we were fairly successful in transiently silencing the tomato ALC1 gene, a uniform and pronounced silencing, such as that observed in N. benthamiana, is often difficult to achieve in tomato (Ekengren et al., 2003; Ryu et al., 2004). Therefore, to achieve stable and uniform silencing and to confirm the necrosis phenotype induced by COR and Pst DC3000 on SlALC1-silenced tomato lines, we generated SlALC1 RNA interference (RNAi) lines. We assayed three independent transgenic RNAi lines, and all responded similarly to COR application and Pst DC3000 infection. Here, we discuss the data for one of the transgenic lines, 3-2. Results obtained from qRT-PCR indicated that the transcript levels of SlALC1 were 20-fold less in RNAi line 3-2 when compared with wild-type tomato plants (Supplemental Fig. S2B). When COR (2 nm) was spotted on the leaves of the RNAi line 3-2, necrosis appeared 5 d post inoculation (dpi; Fig. 3B, right).
COI1/Jasmonic Acid Insensitive1 (JAI1), an F-box protein, is shown to be required for COR signaling in tomato and Arabidopsis (Feys et al., 1994; Zhao et al., 2003; Katsir et al., 2008). Using VIGS, we transiently silenced SlALC1 in jai1 mutant tomato plants (Supplemental Fig. S4) to know whether the necrosis we observed in SlALC1-silenced tomato plants upon COR treatment (Fig. 3, A and B) is COI1/JAI1 dependent. When purified COR (2 nm) was exogenously applied, the SlALC1-silenced jai1 plants showed no visible necrosis (Fig. 3C). Furthermore, silencing of COI1 and NbALC1 in N. benthamiana abolished ALC1-mediated COR-induced necrosis (Supplemental Fig. S9). These results confirmed that COI1/JAI1 is required for the altered chlorosis phenotype in ALC1-silenced tomato and N. benthamiana plants.
ALC1-Silenced Tomato Plants Show Accelerated Necrosis in Response to Pst DC3000
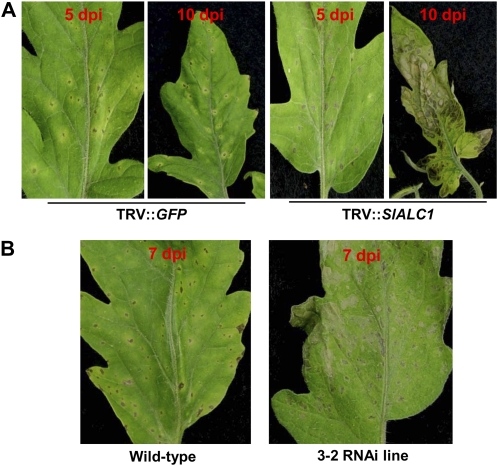
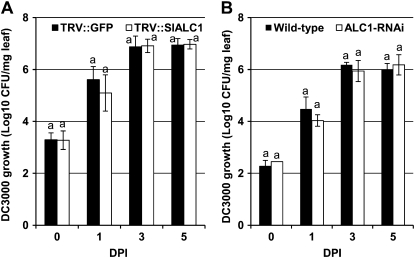
To study the influence of SlALC1 on the virulence of Pst DC3000 in tomato, SlALC1-silenced and control (TRV″GFP) tomato plants were spray inoculated with Pst DC3000 (5 × 106 colony-forming units [cfu] mL−1). Control (TRV″GFP) plants showed typical bacterial speck symptoms at 5 dpi, which consisted of necrotic lesions surrounded by chlorotic halo (Fig. 4A, left). At 5 dpi, the leaves of SlALC1-silenced plants showed necrosis with little or no chlorosis (Fig. 4A, right). At 10 dpi, the necrosis observed on the silenced plants was severe (Fig. 4A, right). Similarly, when line 3-2 was spray inoculated with Pst DC3000 (5 × 106 cfu mL−1), leaves developed severe coalescing necrotic lesions without the visible chlorotic halos surrounding the necrotic lesions as seen on the wild type (Fig. 4B, right), further confirming that ALC1 plays a role in bacterial speck symptom development.
Figure 4.
Response of SlALC1-silenced tomato lines to Pst DC3000. A, Response of control and transiently silenced tomato lines to Pst DC3000. Pst DC3000 (5 × 106 cfu mL−1) was spray inoculated on control (TRV″GFP; left panels) and SlALC1-silenced (by VIGS; right panels) tomato plants. Photographs were taken 5 and 10 dpi. B, SlALC1-silenced transgenic RNAi line 3-2 and wild-type tomato plants were also spray inoculated with Pst DC3000. Photographs were taken 7 dpi.
To determine if the severe necrosis could be explained by a higher amount of bacterial growth in the silenced lines, the population of Pst DC3000 was monitored at 1, 3, and 5 dpi. Interestingly, the bacterial population on the transiently or stably silenced SlALC1 plants was not significantly different from that on the inoculated control plants (Fig. 5). These results suggest that silencing of ALC1 does not have a significant effect on the growth of the bacteria in tomato plants during the early stage of infection.
Figure 5.
Silencing of ALC1 in tomato has no effect on the growth of Pst DC3000. Leaf samples from Pst DC3000-inoculated SlALC1-silenced VIGS plants (A) and SlALC1-silenced RNAi line 3-2 (B) and their corresponding control plants (TRV″GFP and the wild type; described in Fig. 3) were collected at various days after inoculation and homogenized in water, and dilutions were plated onto King's B medium to determine cfu. Error bars represent sd. All experiments were repeated at least twice with several biological replicates, and the data shown are representative of the experiments. Growth measurements with the same letters showed no significant differences based on Fisher lsd values (P < 0.005).
The Arabidopsis thf1 Mutant Displays Severe Necrosis upon Pst DC3000 Inoculation
As mentioned above, ALC1 is closely related to an Arabidopsis gene called THF1 (Supplemental Fig. S3A). An Arabidopsis thf1 mutant was previously identified and shown to have variegated leaves (Supplemental Fig. S1C) that lacked normal chloroplast development in the variegated regions (Wang et al., 2004). We obtained the Arabidopsis thf1 mutant and reconfirmed the mutation by ascertaining the insertion of T-DNA in THF1 (data not shown). Unlike N. benthamiana and tomato, exogenous application of COR on Arabidopsis leaves does not induce chlorosis. Instead, Arabidopsis seedlings are shown to respond to COR by displaying a strong purple hue indicative of anthocyanin accumulation (Bent et al., 1992; Laurie-Berry et al., 2006). To further characterize the thf1 mutant line, we germinated seeds of Arabidopsis ecotype Columbia (Col-0), the thf1 mutant line, the complemented line, and the THF1-overexpressing line on half-strength Murashige and Skoog (MS) medium containing 2 nm COR (Laurie-Berry et al., 2006). As expected, Col-0 seedlings showed anthocyanin accumulation within 10 d after germination (Fig. 6A). Strikingly, the thf1 mutant showed hypersensitivity to COR by displaying a severe growth defect and more anthocyanin accumulation than Col-0 (Fig. 4A). The growth phenotypes of the thf1-complemented and the overexpressing lines were similar to that of the wild type (Fig. 6A).
Figure 6.
The Arabidopsis thf1 mutant exhibits hypersensitivity in the presence of COR and produces necrosis with no chlorosis upon Pst DC3000 inoculation. A, Arabidopsis wild-type Col-0, the thf1 mutant line, the complemented line of thf1 (Comp), and the THF1 overexpression line (35S-Thf1) were germinated on half-strength MS medium containing 2 nm COR (top row). These lines were also germinated on half-strength MS medium alone without COR (controls; bottom row). Photographs were taken 10 d after germination. B, Foliar parts of 4-week-old Arabidopsis lines mentioned above either dipped in a Pst DC3000 culture suspension (108 cfu mL−1; top row) or infiltrated with 106 cfu mL−1 Pst DC3000 using a needleless syringe (bottom row). Photographs were taken 6 dpi.
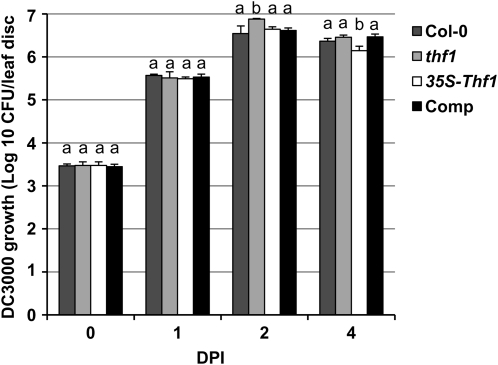
To determine whether THF1 has an effect on Pst DC3000-induced disease symptoms on Arabidopsis, we dip inoculated (108 cfu mL−1) or syringe infiltrated (106 cfu mL−1) the wild-type Col-0 and thf1 mutant with Pst DC3000. As expected, Col-0 showed water-soaked necrotic lesions accompanied by chlorosis (Fig. 6B). However, the thf1 mutant plants exhibited accelerated necrotic lesions without visible chlorosis (Fig. 6B). Complemented lines of the thf1 mutant and THF1-overexpressing plants displayed disease symptoms similar to the wild-type Col-0 after inoculation with Pst DC3000 (Fig. 6B). Interestingly, when the growth of Pst DC3000 was monitored at 0, 1, 2, and 4 dpi, no significant fold differences in the bacterial growth were observed between the wild-type Col-0, the thf1 mutant, the complemented line of thf1, and the THF1 overexpression line (Fig. 7). However, unlike in tomato, thf1 mutants supported slightly increased (1.5-fold) bacterial growth than the wild type (Fig. 7). These results suggest that THF1 does not significantly contribute to the pathogen growth in Arabidopsis, at least for the duration of time the bacterial growth was monitored.
Figure 7.
The mutation in Arabidopsis THF1 has no effect on the growth of Pst DC3000. Arabidopsis leaves of the lines described in Figure 5 were syringe infiltrated with Pst DC3000 (106 cfu mL−1), collected at intervals after inoculation, homogenized in water, and plated on KB medium to determine cfu. Error bars represent sd. The experiments were conducted at least three times, and the data shown are representative of each experiment. Growth measurements with the same letters showed no significant differences based on Fisher lsd values (P < 0.005). Comp, Complemented line of thf1.
Necrosis occurred much earlier on Pst DC3000-infected thf1 leaves than on leaves of the wild-type Col-0 (data not shown). Therefore, we investigated whether the thf1 mutant had a weaker defense response and was more susceptible to biotic and abiotic stress because of defects in thylakoid formation (Wang et al., 2004). To investigate this, leaves of Col-0 and the thf1 mutant were infiltrated with two nonhost pathogens that do not infect Arabidopsis, P. syringae pv tabaci and P. syringae pv glycinea, and growth and symptoms were compared with a COR-producing P. syringae pv maculicola, which is pathogenic to Arabidopsis (Dong et al., 1991; Cuppels and Ainsworth, 1995; Mishina and Zeier, 2006). As expected, the population of P. syringae pv maculicola increased approximately 100-fold on both Col-0 and thf1 leaves by 3 dpi; however, neither P. syringae pv glycinea nor P. syringae pv tabaci multiplied to a significant level on Col-0 or thf1 plants (Supplemental Fig. S5A).
Arabidopsis Col-0 and the thf1 mutant were also monitored for symptom development in response to inoculation with P. syringae pvs maculicola, glycinea, and tabaci and the soft rot pathogen Erwinia carotovora subspecies carotovora. P. syringae pv maculicola induced chlorosis on Col-0 but not on the thf1 mutant line (Supplemental Fig. S5B). Neither Col-0 nor thf1 plants developed visible symptoms in response to P. syringae pvs tabaci or glycinea (Supplemental Fig. S5B). E. carotovora subspecies carotovora induced soft rot on both Col-0 and thf1, with no apparent difference in phenotypic response between the wild-type and the mutant line (Supplemental Fig. S5B). Infiltration of leaves with cell death-inducing agents such as NaCl (500 mm) or hydrogen peroxide (3%; Peart et al., 2002; Kang et al., 2004) caused similar cell death responses on both Col-0 and the thf1 mutant line (data not shown). In the above experiments, only P. syringae pv maculicola induced a unique response on thf1 when compared with Col-0. This response was similar to the one induced by Pst DC3000. These results indicate that the early necrotic cell death of infected leaves in the thf1 mutant is specific to the COR-producing pathogens of Arabidopsis, Pst DC3000, and P. syringae pv maculicola.
The JA Pathway Appears Intact in Arabidopsis thf1 Mutant Plants after Pst DC3000 Inoculation
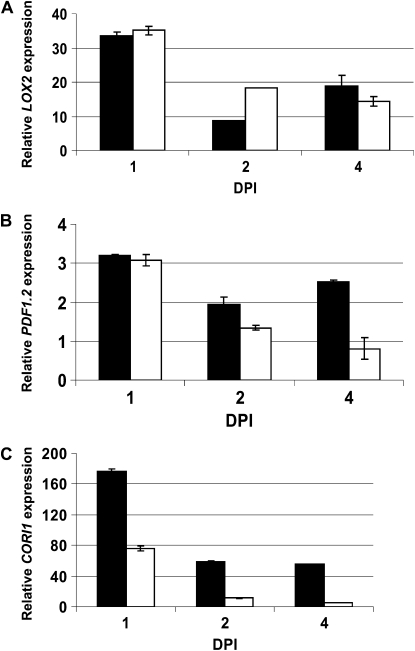
COR functions as a mimic of JAs and mediates signaling via JA perception machinery in tomato and Arabidopsis (Feys et al., 1994; Zhao et al., 2003). Thus, it remained possible that the absence of chlorosis was due to disruption of the JA-dependent signaling pathway. Therefore, we used qRT-PCR to analyze transcript levels of Lipoxygenase2 (LOX2) and Plant Defensin1.2 (PDF1.2). Transcripts of LOX2 and PDF1.2 were induced in both Col-0 and thf1 in response to Pst DC3000. Although expression of both genes was lower in the thf1 mutant line, especially at 4 dpi, the JA pathway appears to be functional (Fig. 8, A and B) at the time points analyzed.
Figure 8.
JA-dependent signaling in thf1 mutants in response to Pst DC3000. Transcripts of LOX2, PDF1.2, and CORI1 were quantified by real-time qRT-PCR in both wild-type Col-0 (black bars) and thf1 mutant (white bars) after Pst DC3000 inoculation. Four-week-old Col-0 and the thf1 mutant line were syringe infiltrated with either Pst DC3000 (106 cfu mL−1) or buffer (mock control). Total RNA was extracted from the leaves of the infected plants collected 1, 2, and 4 dpi, and cDNA was synthesized for qRT-PCR analysis. The transcript levels were normalized against EF1α, which was used as an endogenous control as described by Pfaffl (2001). The transcript levels were quantified relative to the transcript levels on the mock control, which was assigned a value of 1. JA pathway genes (represented by LOX2 and PDF1.2; A and B) and the chlorophyllase-encoding gene CORI1 (C) were activated in Pst DC3000-infected Col-0 and the thf1 mutant. All experiments were repeated at least three times. The data shown represent averages of three biological replicates and three technical replicates, with the sd values shown as error bars.
Chlorosis occurs due to the degradation of proteins in the chloroplast (Quirino et al., 2000), and the Arabidopsis CORI1 gene (encoding chlorophyllase) is induced upon COR or MeJA application (Benedetti et al., 1998), resulting in chlorophyll degradation (Benedetti and Arruda, 2002). The lack of chlorosis in thf1 could be due to repression of CORI1 as a result of loss of THF1 function. Thus, we analyzed CORI1 transcript levels in Pst DC3000-inoculated Col-0 and thf1 plants. CORI1 expression in Col-0 and thf1 was up-regulated approximately 175-fold and approximately 75-fold, respectively, at 1 dpi (Fig. 8C). These results further suggest that the chlorophyllase activity and JA-dependent pathway are not severely affected in the thf1 mutant. Although we did not notice any visible chlorosis in COR- or Pst DC3000-inoculated tissues (Fig. 6B), the COR-induced chlorophyllase activity suggests some degree of chlorophyll degradation in COR- or Pst DC3000-inoculated thf1 plants. Similarly, significant levels of COR-induced chlorophyll degradation were observed in NbALC1-silenced plants (Fig. 2). Taken together, these results suggest that THF1 may be operating downstream of COR-induced JA signaling and chlorosis.
COR Affects the Localization of ALC1 in a COI1-Dependent Manner in N. benthamiana Plants
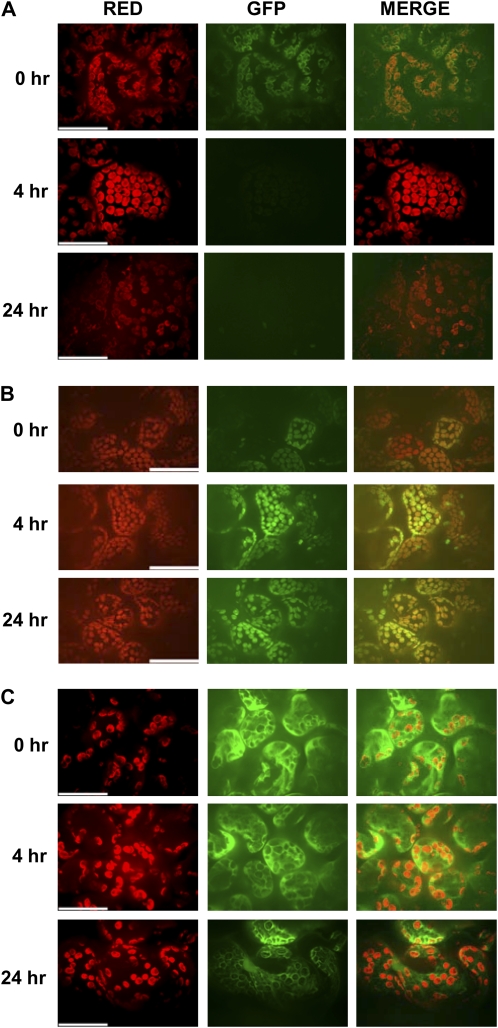
To determine if COR directly affected ALC1, we monitored the effect of COR treatment on the localization/stability of ALC1. An Agrobacterium strain containing a binary plasmid that included 35S″ALC1 fused to GFP within its T-DNA was infiltrated into N. benthamiana plants as described in “Materials and Methods.” COR was spotted on four to five marked regions on the Agrobacterium-infiltrated leaf, and leaf samples were monitored for localization of GFP-ALC1 at various time intervals after COR inoculation (Fig. 9). As reported previously for THF1, GFP-ALC1 localized to the chloroplast (Fig. 9A). Interestingly, within 4 h of COR application, ALC1 was destabilized/degraded, as shown by the loss of GFP fluorescence (Fig. 9A). ALC1 florescence was not detected even after 24 (Fig. 9A), 48, and 72 h (the time at which chlorosis is visible on the leaf; data not shown). It is also noteworthy that the destabilization/degradation of GFP-ALC1 is seen only at the site of application of COR and the nearby region, but the leaf areas away from the region of COR application remain unaffected even after 24 and 48 h (Supplemental Fig. S6). To rule out the possibility that COR application is leading to alterations in chloroplast structure, therefore resulting in nonspecific effects on ALC1, we tested COR effects on GFP-RecA (Kohler et al., 1997), another chloroplast-localizing protein (Supplemental Fig. S7). Interestingly, COR application did not result in destabilization/degradation of GFP-RecA or GFP alone (Fig. 9C; Supplemental Fig. S7).
Figure 9.
COI1-dependent effect of COR on localization or stability of ALC1in N. benthamiana. A and B, Localization of ALC1-GFP in the wild-type (A) and COI1-silenced (TRV″COI1; B) N. benthamiana leaves treated with COR was observed by fluorescence microscopy at 0, 4, and 24 h posttreatment. C, Localization of GFP in N. benthamiana leaves treated with COR. Red, Autofluorescence of the chloroplast by excitation at 647 nm and emission in the cyan/far-red channel; GFP, GFP fluorescence of the tagged protein by excitation at 488 nm and emission in the green channel. All images were magnified using a 63× water-immersion objective. Bars = 5 μm.
The COI1-dependent nature of COR-induced alterations in ALC1 localization was further confirmed in COI1-silenced N. benthamiana plants (Fig. 9B). Interestingly, in COI1-silenced plants, an increased signal intensity of 35S″ALC1 GFP was observed following COR application (Fig. 9B). It is not clear if this is due to lack of COR activity upon COI1 silencing or a COI1-independent activity of COR on ALC1. Silencing of COI1 did not affect the expression levels of 35S″ALC1 GFP florescence (Supplemental Fig. S8). However, silencing of COI1 abolished COR-induced destabilization/degradation of 35S″ALC1 GFP florescence (Fig. 9B). Consistent with these results, COI1 and NbALC1 double-silenced plants showed no chlorosis/necrosis phenotypes upon COR application (Supplemental Fig. S9). Taken together, these results suggested that direct effects of COR or COR-induced effects on chloroplast/ALC1 directly alter ALC1 localization or stability in a COI1-dependent manner and therefore may affect its function.
DISCUSSION
Our observation that COR could induce chlorosis on N. benthamiana and our ability to do VIGS in N. benthamiana provided an excellent strategy to identify plant genes that play a role in COR signaling. Here, we have shown that loss of the N. benthamiana gene NbALC1 and its orthologs, SlALC1 in tomato and AtTHF1 in Arabidopsis, results in a necrotic phenotype in response to COR and/or Pst DC3000. Spray inoculation of Pst DC3000 on ALC1-silenced tomato plants induced accelerated necrotic lesions without visible chlorosis on the majority of the leaves instead of typical bacterial speck symptoms with a chlorotic halo (Mittal and Davis, 1995; Zhao et al., 2003). Furthermore, necrosis spread beyond the region where COR was applied as early as 10 dpi, which is similar to the runaway cell death phenotype reported earlier in the Arabidopsis lsd1 mutant (Fig. 2B; Jabs et al., 1996).
To determine the role of ALC1 in the development of symptoms in response to COR or Pst DC3000, we chose Arabidopsis, since it is genetically tractable and is a host of Pst DC3000. The ortholog of ALC1 in Arabidopsis, known as THF1, is a single-copy gene with no closely related sequences in the Arabidopsis genome (Wang et al., 2004). Expression of the Arabidopsis THF1 gene is positively regulated by light, and the gene product is a chloroplast-localized protein. A T-DNA knockout mutant of THF1 has been identified previously (Wang et al., 2004). The mutant line, thf1, is stunted and has variegated leaves. The chloroplasts in the thf1 mutant contain shorter stacks of thylakoids in the green sector of the leaves and show accumulation of membrane vesicles but no thylakoids within the intact chloroplast membrane in the white sector of the leaves (Wang et al., 2004).
The exogenous application of COR (2 nm) on Arabidopsis leaves did not produce any chlorosis. However, consistent with earlier observations (Bent et al., 1992; Feys et al., 1994), our study showed that Arabidopsis seedlings grown on MS medium supplemented with COR accumulated anthocyanin. Interestingly, anthocyanin accumulation was significantly elevated in the thf1 mutant as compared with the wild-type plants. Therefore, the THF1 mutation has a positive effect on anthocyanin accumulation in Arabidopsis. These results are consistent with an earlier observation that COR induces different phenotypes in Arabidopsis and tomato (Mach et al., 2001; Uppalapati et al., 2005).
Similar to ALC1-silenced tomato plants, inoculation of thf1 with Pst DC3000 did not result in a typical chlorotic phenotype around the water-soaked lesion. Analysis of the Pst DC3000 population dynamics in both ALC1-silenced tomato plants and the thf1 mutant line indicated that there was no difference in the bacterial population dynamics when compared with wild-type plants. Interestingly, Pst DC3000-inoculated leaves of ALC1-silenced tomato and the Arabidopsis thf1 mutant line died earlier than corresponding wild-type lines. These results suggested that Pst DC3000 may tightly regulate the chloroplast homeostasis and the levels of THF1 for controlled necrosis during infection to help pathogen dissemination and spread. We have recently demonstrated that COR-induced effects on the photosynthetic machinery result in light-dependent ROS generation in tomato seedlings (Ishiga et al., 2009a, 2009b). We speculate that in COR-treated or Pst DC3000-inoculated ALC1-silenced tomato and in the Pst DC3000-inoculated Arabidopsis thf1 mutant, the necrosis/HR-like cell death phenotype may appear because the effect of ROS supersedes the detoxifying capacity of antioxidants. Current evidence suggests that THF1 may have multiple functions in the biogenesis of PSII and sugar signaling (Keren et al., 2005; Huang et al., 2006). THF1 was also identified as an interactor of the G-protein GPA1 in Arabidopsis and was shown to play a role in far-red irradiation-preconditioned cell death (Huang et al., 2006; Wei et al., 2008). Our results using GFP-tagged ALC1 suggest that COR has direct effects on ALC1 and might target ALC1 to degradation in a COI1-dependent manner (Fig. 8). Based on these results, it is tempting to speculate if ALC1/THF1, localized in the chloroplast membrane, may directly interact with COR. Interestingly, a chloroplast protein in wheat, ToxABP1 (an ortholog of THF1; Supplemental Fig. S2), directly interacts with the Pyrenophora tritici-repentis protein ToxA (Manning et al., 2007). ToxA is a determinant of virulence in P. tritici-repentis, a pathogen that causes the tan spot of wheat. Therefore, it is possible that COR may interact directly with ALC1/THF1 during the Pst-host interactions. Thus, we speculate that ALC1 and THF1 (Wang et al., 2004), which is localized on the chloroplast, is somehow involved in the maintenance of ROS homeostasis and, therefore, that Arabidopsis thf1 mutants and ALC1-silenced tomato leaves are more sensitive to COR/pathogen-induced ROS, leading to accelerated cell death (necrosis) in tomato and Arabidopsis.
In conclusion, we have developed a VIGS-based forward genetic screen for identification of new targets involved in COR signaling. Although we set out to identify genes involved in COR-induced chlorosis, we identified a gene, THF1, that when silenced causes necrosis upon COR application. We are now screening a COR-responsive N. benthamiana cDNA library to identify components involved in COR-induced chlorosis. Although the precise role of THF1 in the COR signaling pathway could be argued and needs further confirmation, our results present a new role for chloroplast-localized THF1 in bacterial speck disease development.
MATERIALS AND METHODS
Plant Materials, Bacterial Cultures, and Plant Infections
Nicotiana benthamiana plants were maintained in the greenhouse with conditions as described previously (Senthil-Kumar et al., 2007). Seeds of tomato (Solanum lycopersicum ‘Glamour’) were obtained from Stokes Seeds. Seeds of the Arabidopsis (Arabidopsis thaliana) thf1 T-DNA mutant, its complemented lines, and overexpression lines were kindly provided by Dr. Ken Korth (University of Arkansas). Tomato jai1 mutants (cv Castlemart) were obtained from Dr. Gregg Howe (Michigan State University). Agar and broth cultures of Pseudomonas syringae pvs tomato (DC3000), glycinea, maculicola, and tabaci were grown on King's B medium with appropriate antibiotics (King et al., 1954). Agrobacterium tumefaciens and Escherichia coli cultures were grown on Luria-Bertani medium (1% yeast extract, 0.5% bacto-tryptone, 1% NaCl, and 1.5% agar for plates) with appropriate antibiotics. For pathogen infection assays on silenced tomato lines, plants were inoculated with a bacterial suspension as described (Uppalapati et al., 2007). Bacterial suspensions (optical density at 600 nm [OD600] = 0.1) were prepared in sterile distilled water containing 0.0025% Silwet L-77 (OSI Specialties) and sprayed on plants using a Paasche VL airbrush (Paasche Airbrush Co.). To infect Arabidopsis, 4-week-old plants were either infiltrated (OD600 = 0.2) with bacterial suspensions into the leaves using a needleless syringe or were dipped into the bacterial suspension (OD600 = 0.002). The plants were then placed in trays, covered with transparent lids, and incubated in growth chambers for the rest of the experimental period. Inoculated leaves were harvested, ground in 10 mm MgCl2, and cfu per leaf disc (1-cm diameter) was determined by serial dilution of leaf extracts. The bacterial growth data were subjected to ANOVA using Biostat 2008 (Analystsoft). Significant differences in the means of the treatments were obtained based on Fisher's lsd set at P < 0.005.
VIGS-Mediated Forward Genetic Screening
A N. benthamiana cDNA library cloned in a TRV-VIGS vector (Anand et al., 2007) was used to screen and identify plant genes involved in COR responses. Agroinoculation for VIGS was performed using the toothpick method as described previously (Anand et al., 2007). About 3 weeks postinoculation, 2 μL of COR (0.2 nm) was placed on either side of the midrib of two fully expanded leaves per plant. Using N. benthamiana inoculated with TRV″GFP as a control, altered phenotypes in response to COR were recorded 5 to 7 d after COR application.
Cloning a Full-Length ALC1 Gene
To clone the full-length N. benthamiana ALC1 gene, a tobacco (Nicotiana tabacum) full-length sequence of ALC1 (226 bp) homolog was obtained from the J.C. Venter Institute's plant genome database (accession no. TC10126; www.tigr.org). Based on the tobacco sequence, PCR primers were designed (forward, 5′-CAACTCCATTCTCTAAAGCAAC-3′; reverse, 5′-GTCAATGAGGTCCAAGCAGG-3′) at approximately 70 bp upstream and 40 bp downstream of the putative coding region. PCR was performed using the above primer sets on a cDNA mixture obtained from N. benthamiana leaves to amplify a full-length N. benthamiana ALC1. The PCR product obtained was then cloned into the pGEMT Easy vector (Promega) and transformed into JM109-competent cells. The insert was confirmed by sequencing.
Construction of pTRV″SlALC1 and VIGS in Tomato
The vectors pTRV1 and Gateway-ready pTRV2 (Liu et al., 2002b) were kindly provided by Dr. S.P. Dinesh-Kumar (Yale University). An antisense sequence of SlALC1 consisting of a 324-bp fragment (The Institute for Genomic Research accession no. TC212458) was PCR amplified from tomato (Glamour) by RT-PCR using primers SlALC1attB1 (5′-ggggacaagtttgtacaaaaaagcaggctTTCCACCTCTCGCTTTGTCG-3′) and SlALC1attB2 (5′-ggggaccactttgtacaagaaagctgggtGCATCAGCTCTGTATTGCTC-3′; lowercase letters indicate the Gateway adapters) and was cloned into Gateway-ready pTRV2 (Liu et al., 2002b). The construct, pTRV2-SlALC1, was then introduced into A. tumefaciens strain GV2260 by electroporation.
For gene silencing in wild-type or jai1 mutant tomato plants, A. tumefaciens strains containing pTRV1 and pTRV2″SlALC1 were mixed in a 1:1 ratio (OD600 = 1.0) in a buffer containing 10 mm MES, 10 mm MgCl2, and 100 μm acetosyringone and incubated at room temperature for 3 to 4 h. Two-week-old tomato seedlings with fully expanded cotyledons were removed from the pots and were completely submerged in an Agrobacterium mixture and vacuum infiltrated for 2 min as described earlier by Uppalapati et al. (2007). The seedlings were then transplanted into Professional Blend potting mixture (Sun Gro). To improve the silencing efficiency, the remaining Agrobacterium culture was dispensed around the seedlings using the Agrodrench method (Ryu et al., 2004). Inoculated potted seedlings were then maintained in growth chambers for 2 d with a 12-h photoperiod (22°C day, 18°C night). Then the plants were moved to a greenhouse and maintained at 14 h of daylight at 25°C and 22°C at night for the next 10 to 14 d.
Generation of Tomato ALC1 Transgenic RNAi Lines
For the generation of a tomato ALC1 RNAi line, the SlALC1 fragment (described above) was introduced into the Gateway-ready binary RNAi vector pK7GWIWG2(I) (Karimi et al., 2002) to generate the SlALC1 RNAi construct that was later transformed into A. tumefaciens strain GV2260 by electroporation. For the transformation of tomato plants, a tomato tissue culture method developed by Frary and Van Eck (2005) was followed with a slight modification in that no tobacco feeder cells were used. Cotyledons of 7- to 8-d-old tomato seedlings (Glamour) were dissected and maintained on KCMS (KC Biological MS medium; Frary and Van Eck, 2005) for 24 h. Tomato cotyledons were then cocultivated with A. tumefaciens cultures carrying the RNAi construct and maintained in darkness for 48 h. For all subsequent steps, the transformation protocol described by Frary and Van Eck (2005) was followed.
Measurement of Chlorophyll Content
The chlorophyll content of leaf discs was measured as described by Arnon (1949) and Ishiga et al. (2009a). Two leaf discs (0.78 cm2 each) were isolated 6 dpi from leaves treated with water (mock control) or 2 μL of COR (2 nm) and then macerated in liquid nitrogen, placed in 6 mL of acetone, and incubated at 4°C in the dark for 12 h. Aliquots of total chlorophyll dissolved in acetone were mixed with hexane and 10 mm KOH at a ratio of 4:6:1 (v/v). Chl a was quantified spectrophotometrically using the formula described by Arnon (1949).
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from leaves of N. benthamiana, tomato, and Arabidopsis plants using TRIZOL reagent (Invitrogen). The first-strand cDNA was synthesized using oligo(dT)15 primer and the Omniscript RT kit (Qiagen). For quantitative analysis of transcripts, primer pairs were designed using the Primer Express software (Applied Biosystems) to amplify the target sequences. qRT-PCR was performed with an ABI HT7900 machine using the SYBR Green method (Applied Biosystems). PCR efficiency was determined using the linear regression software LinRegPCR (Ramakers et al., 2003). In order to normalize the data, parallel reactions were run using the Elongation Factor-α (EF1α) primers as the endogenous control for Arabidopsis and actin or tubulin primers as the endogenous control for N. benthamiana and tomato (Supplemental Table S1). The relative transcript levels were quantified as described previously (Pfaffl, 2001).
Subcellular Localization of ALC1
To transiently express ALC1 in N. benthamiana, the Gateway-ready pMDCC83 was used as a vector to generate a GFP fusion (Curtis and Grossniklaus, 2003). Full-length ALC1 sequence was amplified from N. benthamiana cDNA using the following gene-specific primers: ALC1attB1 (5′-ggggacaagtttgtacaaaaaagcaggcttcATGGCGGCAGTTACTTCG-3′) and ALC1attB2 (5′-ggggaccactttgtacaagaaagctgggtcCCTCCCAGCATATTGGTAATCT-3′; lowercase letters indicate the Gateway adapters). The amplified sequence was cloned into the donor vector pDONR 207 (Invitrogen), and the resulting clone was then transformed into E. coli DH5α-competent cells (Invitrogen). The full-length gene was further subcloned into pMDC83, and pmDC83-ALC1 was then introduced into A. tumefaciens GV2260 by electroporation. To generate pMDC83 empty vector that can replicate in A. tumefaciens GV2260 without killing the host (Dao-Thi et al., 2005), the vector was restriction digested with KpnI to remove the ccdB9 (for controller of cell division or death) region. The open ends were then ligated with T4 DNA ligase. pMDC83:ΔccdB was then introduced into A. tumefaciens GV2260 by electroporation. pCAMBIA1390-RecA in Agrobacterium C58C1 (Kohler et al., 1997) was obtained from Dr. Elison Blancaflor (Noble Foundation).
To test the COI-dependent ALC1 localization, N. benthamiana leaves were syringe inoculated with a 1:1 mixture of TRV1 and pTRV2″NbCOI1 (Ekengren et al., 2003) as described by Ryu et al. (2004). Wild-type and COI1-silenced leaves at 2 weeks postinoculation were used for ALC1-GFP localization studies. Purified COR at a concentration of 2.0 nm was spotted on the leaves of wild-type or COI1-silenced N. benthamiana plants that were infiltrated, 3 d prior, with Agrobacterium containing ALC1-GFP, RecA-GFP, or GFP alone. Imaging of COR-treated or untreated cells was conducted using a Perkin-Elmer UltraView ERS spinning-disc confocal system coupled to a Zeiss Observer D1 inverted microscope equipped with a 63× water-immersion objective (numerical aperture 1.2). GFP was excited with the 488-nm line of the argon-kyrpton laser, and emission was detected at 510 nm. To image chloroplast autofluorescence, leaf samples were excited with the 647-nm line of the argon-kyrpton laser, and emission was detected at 680 nm.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU106046.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of the Arabidopsis thf1 line, ALC1-silenced N. benthamiana, and ALC1-silenced tomato share similarities.
Supplemental Figure S2. Determination of silencing efficiency of ALC1.
Supplemental Figure S3. ALC1 has homologs in several plants.
Supplemental Figure S4. Determination of silencing efficiency of SlALC1 in COR-treated mock and jai1 tomato mutants.
Supplemental Figure S5. Response of the Arabidopsis thf1 mutant line to pathogens (P. syringae pv maculicola and Erwinia carotovora subspecies carotovora) and nonhost pathogens (P. syringae pvs tabaci and glycinea).
Supplemental Figure S6. Effect of COR on localization or stability of ALC1 near and away from the site of inoculation zone in N. benthamiana leaf samples.
Supplemental Figure S7. Effect of COR on localization of a chloroplast-localized RecA protein in N. benthamiana leaf samples.
Supplemental Figure S8. Effect of COI1 silencing on expression and localization of ALC1 in N. benthamiana leaf samples.
Supplemental Figure S9. COR-induced chlorosis and NbALC1-mediated necrotic phenotype are COI1 dependent in N. benthamiana.
Supplemental Table S1. Primers used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Ken Korth for providing the Arabidopsis thf1 mutant and overexpresser lines, Drs. Greg Martin and Olga Del-Pozo for providing the TRV-NbcDNA library, Dr. S.P. Dinesh-Kumar for providing the Gateway-ready TRV-VIGS vectors, Stacy Allen for assistance with the qRT-PCR experiments, Janie Gallaway for maintaining the plants, and Dr. Steve Marek for critical reading of the manuscript.
This work was supported by the Samuel Roberts Noble Foundation, in part by the Oklahoma Center for Advancement of Science and Technology (grant no. PSB09–021 to S.R.U.), and by the National Science Foundation (grant no. IOB–0620469 to C.L.B. and grant no. DBI–0722635 for purchase of the spinning-disk confocal microscope).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kirankumar S. Mysore (ksmysore@noble.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Anand A, Vaghchhipawala Z, Ryu CM, Kang L, Wang K, del-Pozo O, Martin GB, Mysore KS (2007) Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol Plant Microbe Interact 20 41–52 [DOI] [PubMed] [Google Scholar]
- Arnon DT (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CL, Alarcon-Chaidez F, Gross DC (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CL, Stone HE, Sims J, Cooksey DA (1987) Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol Mol Plant Pathol 30 272–283 [Google Scholar]
- Benedetti CE, Arruda P (2002) Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol 128 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti CE, Costa CL, Turcinelli SR, Arruda P (1998) Differential expression of a novel gene in response to coronatine, methyl jasmonate, and wounding in the Coi1 mutant of Arabidopsis. Plant Physiol 116 1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5 372–378 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chaidez F, Sreedharan A, Rangaswamy V, Penaloza-Vazquez A, Bender CL, Kunkel BN (2004) Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17 162–174 [DOI] [PubMed] [Google Scholar]
- Cuppels DA, Ainsworth T (1995) Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl Environ Microbiol 61 3530–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao-Thi MH, Van Melderen L, De Genst E, Afif H, Buts L, Wyns L, Loris R (2005) Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol 348 1091–1102 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB (2004) MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36 905–917 [DOI] [PubMed] [Google Scholar]
- Elizabeth SV, Bender CL (2007) The phytotoxin coronatine from Pseudomonas syringae pv. tomato DC3000 functions as a virulence factor and influences defence pathways in edible brassicas. Mol Plant Pathol 8 83–92 [DOI] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Van Eck J (2005) Organogenesis from transformed tomato explants. Methods Mol Biol 286 141–150 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Gnanamanickam SS, Starratt AN, Ward EWB (1982) Coronatine production in vitro and in vivo and its relation to symptom development in bacterial blight of soybean. Can J Bot 60 645–650 [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA (1996) An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobellis N, Lavermicocca SP, Grgurina I, Simmaco M, Ballio A (1992) Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol 40 107–116 [Google Scholar]
- Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL (2009. a) The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol 181 147–160 [DOI] [PubMed] [Google Scholar]
- Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL (2009. b) Involvement of coronatine-inducible reactive oxygen species in bacterial speck disease of tomato. Plant Signal Behav 4 237–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273 1853–1856 [DOI] [PubMed] [Google Scholar]
- Kang L, Tang X, Mysore KS (2004) Pseudomonas type III effector AvrPto suppresses the programmed cell death induced by two nonhost pathogens in Nicotiana benthamiana and tomato. Mol Plant Microbe Interact 17 1328–1336 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB (2005) Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17 2768–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44 301–307 [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26 509–522 [DOI] [PubMed] [Google Scholar]
- Kohler RH, Cao J, Zipfel WR, Webb WW, Hanson MR (1997) Exchange of protein molecules through connections between higher plant plastids. Science 276 2039–2042 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19 789–800 [DOI] [PubMed] [Google Scholar]
- Levi C, Durbin RD (1986) The isolation and properties of a tabtoxin-hydrolyzing aminopeptidase from the periplasm of Pseudomonas syringae pv. tabaci. Physiol Mol Plant Pathol 28 345–352 [Google Scholar]
- Li XZ, Starratt AN, Cuppels DA (1998) Identification of tomato leaf factors that activate toxin gene expression in Pseudomonas syringae pv. tomato DC3000. Phytopathology 88 1094–1100 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002. a) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002. b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30 415–429 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning VA, Hardison LK, Ciuffetti LM (2007) Ptr ToxA interacts with a chloroplast-localized protein. Mol Plant Microbe Interact 20 168–177 [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RE (1976) Isolation and structure of a chlorosis-inducing toxin of Pseudomonas phaseolicola. Phytochemistry 15 1941–1947 [Google Scholar]
- Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 8 165–171 [DOI] [PubMed] [Google Scholar]
- Patil SS, Hayward AC, Emmons R (1974) An ultraviolet-induced non-toxigenic mutant of Pseudomonas phaseolicola of altered pathogenicity. Phytopathology 64 590–595 [Google Scholar]
- Paynter VA, Alconero R (1979) A specific fluorescent antibody for detection of syringomycin in infected peach tree tissues. Phytopathology 69 493–496 [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DA, Xiao S, Coleman MJ, et al (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5 278–282 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Ryu CM, Anand A, Kang L, Mysore KS (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse solanaceous species. Plant J 40 322–331 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Govind G, Kang L, Mysore KS, Udayakumar M (2007) Functional characterization of Nicotiana benthamiana homologs of peanut water deficit-induced genes by virus-induced gene silencing. Planta 225 523–539 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL (2005) The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J 42 201–217 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 20 955–965 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Urbanczyk-Wochniak E, Ishiga T, Mysore KS, Bender CL (2008) Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: phenotypic and gene expression analyses of the virulence function of coronatine. Mol Plant Microbe Interact 21 383–395 [DOI] [PubMed] [Google Scholar]
- Vijayan P, Shockey J, Levesque CA, Cook RJ, Browse J (1998) A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA 95 7209–7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL (2004) Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol 136 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Zhou W, Hu G, Wei J, Yang H, Huang J (2008) Heterotrimeric G-protein is involved in phytochrome A-mediated cell death of Arabidopsis hypocotyls. Cell Res 18 949–960 [DOI] [PubMed] [Google Scholar]
- Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F (1994) The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett 345 9–13 [DOI] [PubMed] [Google Scholar]
- Xu GW, Gross DC (1988) Evaluation of the role of syringomycin in plant pathogenesis by using Tn5 mutants of Pseudomonas syringae pv. syringae defective in syringomycin production. Appl Environ Microbiol 54 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J 36 485–499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.