Abstract
In an approach to study lateral root development in monocots, genome-wide searches for homologs of the Gibberellic Acid Stimulated Transcript-like (GAST-like) gene family in rice (Oryza sativa) and maize (Zea mays) were carried out. Six novel GAST-like genes in rice and 10 members of the gene family in maize, which were designated ZmGSL (for Z. mays Gibberellic Acid Stimulated-Like), were identified. The ZmGSL family encodes small proteins of 75 to 128 amino acids, which are characterized by a conserved 59 to 64 amino acid C-terminal domain. Within this domain, 17 amino acids, including 12 cysteines, are perfectly conserved. The transcript of the ZmGSL1 gene is differentially spliced into the alternative variants ZmGSL1a and ZmGSL1b, the latter of which is translated into a premature protein that lacks the C-terminal domain. The presence of an additional N-terminal cleavable signal sequence in eight of the 10 ZmGSL proteins suggests that they are secreted into the extracellular matrix. In-depth root-specific gene expression analyses carried out in the wild type and the lateral root mutants lrt1 and rum1 suggest a role for ZmGSL genes in early lateral root development, which is likely regulated by gibberellic acid. Expression patterns of ZmGSL1a and ZmGSL1b propose antagonistic functions of these splice variants during early lateral root formation.
The morphological and histological organization of root systems in monocotyledonous plants differs extensively from that of dicotyledonous species, such as Arabidopsis (Arabidopsis thaliana; Hochholdinger and Zimmermann, 2008). Cereals, for example, maize (Zea mays) or rice (Oryza sativa), are characterized by a complex root architecture that is elaborated in two successive stages of development (Hochholdinger, 2009). During embryogenesis, only a single primary root and a variable number of seminal roots are laid down that emerge shortly after germination (Hochholdinger, 2009). During vegetative development, this embryonic root system becomes functionally replaced by a complex shoot-borne root stock that is initiated from nodal tissue of the postembryonic shoot (Hochholdinger, 2009). As a common feature, all main root types of maize form lateral roots (Hochholdinger et al., 2004). While in Arabidopsis lateral roots are formed from pericycle cells adjacent to xylem poles, maize lateral roots are initiated from pericycle cells in close vicinity to phloem poles (Hochholdinger and Zimmermann, 2008). In maize, lateral roots constitute the major part of the water and nutrient absorbing surfaces and therefore are crucial for vigor and anchorage in one of the agronomically most outstanding crop species. In contrast to Arabidopsis, the molecular mechanisms underlying lateral root formation in maize are largely unknown, and genes involved have not been described so far. The analysis of lateral root mutants (Hochholdinger and Feix, 1998; Hochholdinger et al., 2001; Woll et al., 2005), however, has shed some light on the genetic control of lateral root development in maize. All mutants identified so far affect lateral root formation only in the embryonic root system, while postembryonic root development remains unaffected: In the monogenic recessive lateralrootless1 mutant (lrt1; Hochholdinger and Feix, 1998), lateral roots are not initiated at the primary root, whereas the recessive mutant rootless with undetectable meristems1 (rum1; Woll et al., 2005) fails to initiate lateral roots at the primary root and at seminal roots.
Phylogenetic analyses of related gene families can serve as a starting point to a deeper understanding of how a particular developmental process is accomplished in crop species and how it has diversified from dicots during evolution. Arabidopsis COBRA Like9 and ROOTHAIRLESS3, two members of the COBRA gene family in Arabidopsis (Jones et al., 2006; Brady et al., 2007) and maize (Hochholdinger et al., 2008), for example, are involved in different aspects of root hair formation. Phylogenetic reconstructions, however, have revealed that they are orthologs and therefore share a common ancestor.
In dicot species, numerous genes have been described to be involved in the molecular network of lateral root formation. Among these, Root System Induced1 (RSI1) was one of the first genes identified to mark early lateral root formation in tomato (Solanum lycopersicum; Taylor and Scheuring, 1994). RSI1 is closely related to the tomato Gibberellic Acid Stimulated Transcript1 (GAST1) gene (Shi et al., 1992) and to 15 GAST-like genes from Arabidopsis designated AtGASA1 to AtGASA14 (for Gibberellic Acid Stimulated Arabidopsis; Roxrud et al., 2007) and AtGAST1-like (GenBank accession no. AK229086). Among these 15 genes, AtGASA13, however, is most likely a pseudogene (Roxrud et al., 2007). The GAST-like gene superfamily encodes small polypeptides that are characterized by a C-terminal domain that contains 12 perfectly conserved Cys residues. The N-terminal domain of GAST-like proteins, by contrast, is highly individual and contains a signal sequence that is characteristic of proteins secreted in the extracellular matrix (Roxrud et al., 2007). Due to the absence of structural and biochemical data, the dedicated function of both domains in GAST-like proteins is still unclear to date. A variety of results from different plant species suggest that GAST-like genes are mainly involved in cell elongation and cell division. Expression data and mutant analyses in petunia (Petunia hybrida), for example, have suggested that the Gibberellic Acid Induced Petunia1 (GIP1) gene is involved in the elongation of the stem and the corolla (Ben-Nissan and Weiss, 1996; Ben-Nissan et al., 2004). Similarly, expression of tomato GAST1 has been associated with stem elongation (Shi et al., 1992), although plants either over- or underexpressing GAST1 did not display an altered phenotype (Shi and Olszewski, 1998). Plants constitutively expressing the gerbera GAST-like gene Gerbera hybrida homolog of GAST1 display reduced cell expansion in the corolla and carpels, hence suggesting a regulatory role for this gene in shaping these organs (Kotilainen et al., 1999). Petunia GIP4 and GIP5 (Ben-Nissan et al., 2004) and Arabidopsis GASA4 (Aubert et al., 1998), on the other hand, were suggested to act in cell division based on their expression in meristematic tissue. Furthermore, for AtGASA4, a role in directly controlling seed size and indirectly regulating seed yield by affecting inflorescence branching and seed set has been demonstrated (Roxrud et al., 2007). Last but not least, in monocots, it has been suggested that OsGASR1 and OsGASR2 are involved in panicle differentiation (Furukawa et al., 2006). These results demonstrate that analyses of GAST-like gene functions to date have mainly focused on aboveground plant development. Thus far, expression of tomato RSI1 in lateral root primordia (Taylor and Scheuring, 1994) and of AtGASA4 in primary roots and lateral root primordia (Aubert et al., 1998) are the only clues on a function of GAST-like genes in root development. The goal of this study, therefore, was to unravel the role of the GAST-like gene family in monocot root development with emphasis on lateral root formation.
RESULTS
The Maize GSL Gene Family Consists of at Least 10 Members and Is Characterized by a Conserved Cys-Rich Domain
In order to study their potential role in maize development, 10 maize genes (ZmGSL1–ZmGSL10) were identified based on homology to the tomato RSI1 gene and to members of the GASA family from the dicot model system Arabidopsis as a reference (Taylor and Scheuring, 1994; Herzog et al., 1995) in a comprehensive screen of different maize genomic databases (see “Materials and Methods”). GenBank accession numbers of the 10 newly identified maize genes are summarized in Table I. To obtain a more comprehensive overview on monocot GSL genes, the rice genome was also searched for RSI1 homologs as a reference. This led to the identification of six novel gene family members in addition to the previously published genes OsGSR1 (Wang et al., 2009), OsGASR1, and OsGASR2 (Furukawa et al., 2006; Table I). The novel rice genes were numbered according to their closest maize relatives (OsGSL1, OsGSL5, OsGSL7, OsGSL8, and OsGSL9) or in one instance, when no maize ortholog was available, according to the closest Arabidopsis relative (OsGSL12).
Table I.
Overview of the maize and rice GSL gene family indicating database accession numbers, chromosomal location, protein length, predicted N-terminal signal peptide size, and C-terminal Cys-rich domain size
aa, Amino acids.
| Gene Name | TIGR/PlantGDB Accession No. | Maizesequence.org BAC Accession No. (Position Start/Stop Codon) | Map Position (Chromosome) | Protein Length (aa) | N-Terminal Signal Peptide (Size in aa)a | C-Terminal Cys-Rich Domain (aa) |
|---|---|---|---|---|---|---|
| Maize | ||||||
| ZmGSL1a/b | ZmGSStuc11-12-04.38482 | AC206247 (178712389–178713120) | 6 | 128/75 | No (–) | 59 |
| ZmGSL2 | ZmGSStuc11-12-04.2491.3 | AC190635 (2633783–2634605) | 10 | 129 | Yes (24) | 59 |
| ZmGSL3 | AZM5_102125 | AC208906 (142281–141826) | 9 | 114 | Yes (24) | 59 |
| ZmGSL4 | AZM5_20008 | AC206348 (72910–73479) | 2 | 115 | Yes (24) | 60 |
| ZmGSL5 | AZM5_100491 | AC204384 (118093783–118094355) | 8 | 94 | Yes (31) | 59 |
| ZmGSL6 | AZM5_15753 | AC191416 (1896419–1896948) | 2 | 127 | Yes (24) | 59 |
| ZmGSL7 | AZM5_6379 | AC206643 (332058393–332058770) | 1 | 100 | Yes (32) | 64 |
| ZmGSL8 | AZM5_96128 | AC212758 (124160–124474) | 5 | 80 | No (–) | 61 |
| ZmGSL9 | AZM5_5532 | AC208899 (262273623–262274009) | 2 | 96 | Yes (26) | 60 |
| ZmGSL10 | AZM5_2270 | AC215857 (64607–64998) | 5 | 96 | Yes (30) | 62 |
| Rice | ||||||
| OsGSR1 | AY604180 | – | 6 | 110 | Yes (28) | 59 |
| OsGASR1 | AB192574 | – | 3 | 93 | Yes (27) | 62 |
| OsGASR2 | AB192575 | – | 4 | 105 | Yes (23) | 60 |
| OsGSL1 | Os05g0376800 | – | 5 | 152 | Yes (34) | 59 |
| OsGSL5 | Os05g0432200 | – | 5 | 92 | Yes (29) | 59 |
| OsGSL7 | Os03g0607200 | – | 3 | 94 | Yes (28) | 62 |
| OsGSL8 | Os06g0729400 | – | 6 | 133 | Yes (43) | 61 |
| OsGSL9 | Os07g0592000 | – | 7 | 102 | Yes (28) | 60 |
| OsGSL12 |
Os09g0414900 |
– |
9 |
112 |
Yes (27) |
59 |
Determined by SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP).
ZmGSL1 to ZmGSL10 map to seven of the 10 maize chromosomes with chromosome 2 harboring three and chromosome 5 harboring two loci (Table I). Similarly, the nine rice GSL homologs are distributed over six of the 12 rice chromosomes with chromosomes 3, 5, and 6 accommodating two members. The maize and rice GSL loci encode small proteins of 80 to 152 amino acids (Table I), which are characterized by a conserved C-terminal domain of 59 to 64 amino acids (Table I; Fig. 1A). Within this C-terminal domain, 17 amino acids, including 12 Cys residues, are perfectly conserved (Fig. 1A). In this respect, the ZmGSL1 locus displays a particular scenario as it gives rise to a full-length protein of 128 amino acids (ZmGSL1a) and also to a truncated protein of 75 amino acids (ZmGSL1b), which lacks the conserved C-terminal domain. ZmGSL1a und ZmGSL1b will be discussed in more detail later.
Figure 1.
A, Sequence alignment of the conserved C-terminal Cys-rich region of the 10 members of the maize ZmGSL gene family. Identical residues that are not only conserved in maize but also in the rice and Arabidopsis GSL families are highlighted in black. Maize and rice C-terminal GSL domains are separated by a black line. The amino acids included in the alignment are indicated with reference to their position in the full-length proteins in parenthesis. B, Unrooted phylogenetic tree generated with the neighbor-joining algorithm including the two founding members of this gene family from tomato (SlRSI1 and SlGAST1), all 14 GASA proteins encoded in the genome of the dicot model plant Arabidopsis, all nine rice, and 10 maize GAST-like proteins encoded in these monocotyledonous model organisms.
In addition to the C-terminal domain, all rice and maize GSL proteins except ZmGSL1a/b and ZmGSL8 contain an individual stretch of 24 to 43 amino acids at the N terminus, which, according to the SignalP.v3.0 software, is a cleavable signal sequence characteristic of proteins secreted into the extracellular matrix (Table I). In contrast to the high sequence similarity in the respective C-terminal domains, all monocot GSL proteins share only little similarity in their N-terminal domains.
Phylogenetic Analyses Reveal Rice Orthologs for Eight of the 10 ZmGSL Proteins, While It Is Challenging to Determine Distinct Dicot Homologs Based on Sequence Similarity
Phylogenetic analyses were based on the full-length protein sequences of all known members of the Arabidopsis, tomato, and rice GAST-like families and the newly identified GSL proteins from maize and rice. The AtGASA13 protein was not included in this analysis because it most likely represents a pseudogene (Roxrud et al., 2007). The resulting phylogenetic tree (Fig. 1B) is organized into three clades in which ZmGSL1 to ZmGSL3 belong to clade 1, ZmGSL4 to ZmGSL6 constitute clade 2, and ZmGSL7 to ZmGSL10 represent clade 3. Remarkably, from the nine GASR/GSR/GSL proteins from rice and the 10 maize GSL proteins from maize, eight form pairwise orthologs (Fig. 1B). In this respect, the previously published OsGSR1 is most closely related to ZmGSL3, and OsGASR1 and OsGASR2 are orthologous to ZmGSL4 and ZmGSL10, respectively. While the close relatedness of the pairwise orthologs is reflected by only 4% to 18% sequence identity on the level of the full-length proteins (Supplemental Table S2a, nos. shaded in gray), the conserved C-terminal domains of these proteins display 68% to 92% sequence identity (Supplemental Table S2b, nos. shaded in gray). The Cys-rich domains of ZmGSL7 and OsGSL7, by contrast, share less identity (23%), and ZmGSL2 and ZmGSL6 cannot be assigned to a corresponding rice ortholog.
With respect to the relatedness of eudicot members in the tree, it is often not possible to assign the newly identified GSL proteins to distinct homologs from Arabidopsis or tomato. One exception is OsGSL12, which is closely related to GASA12 from Arabidopsis. Beyond this, only AtGASA7 in the third clade is positioned adjacent to OsGSL9 and ZmGSL9. In the same clade, the pairwise orthologs ZmGSL7/OsGSL7 and ZmGSL8/OsGSL8 form a distinct branch and thus are likely to represent monocot-specific functions.
Members of the ZmGSL Gene Family Are Widely Expressed throughout Aboveground Maize Development
Massively Parallel Signature Sequencing (MPSS) technology (Brenner et al., 2000) allows for the spatiotemporal quantification of the relative abundance (parts per million) of unique transcripts during development (Christensen et al., 2003). In order to gain a first insight into the expression characteristics of the 10 ZmGSL genes, 61 different MPSS libraries representing different stages of kernel (Supplemental Table S3a), vegetative shoot (Supplemental Table S3b), and generative development (Supplemental Table S3c) of the maize inbred line B73 were surveyed in DuPont's proprietary MPSS database. Apart from ZmGSL6, which is not expressed in any of the tissues analyzed and ZmGSL7, which is only weakly expressed in embryos (Supplemental Table S3a), all ZmGSL genes are active in at least two of the three categories of aboveground development. A summary of overall aboveground expression levels and expression levels in kernels is provided in Supplemental Figure S1.
Expression of Some ZmGSL Genes Is Affected in the Lateral Root Mutants lrt1 and rum1
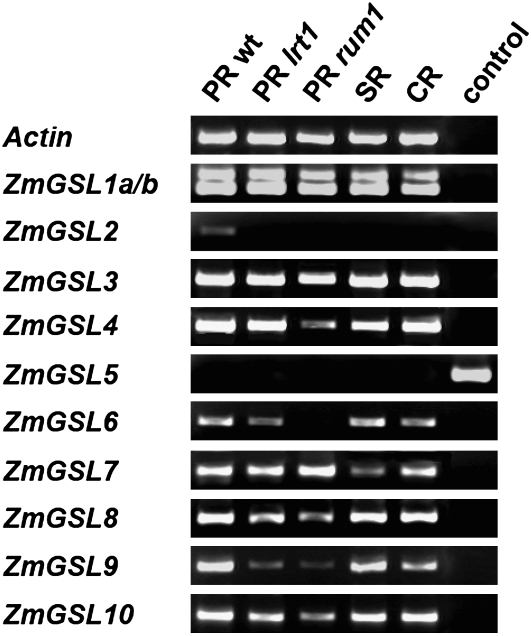
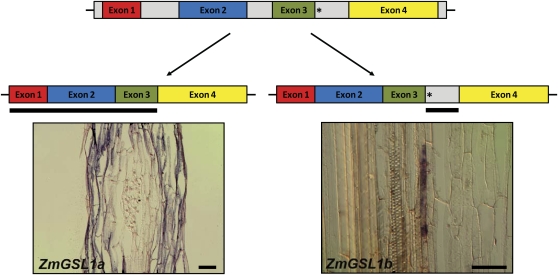
While MPSS provides extensive expression data for aboveground development in maize, it yields only limited information on developmental aspects of root formation. Therefore, as a first step in elaborating on the potential function of the ZmGSL gene family in lateral root development, ZmGSL gene expression (Fig. 2) was surveyed in primary (PR), seminal, and crown roots of the wild type by semiquantitative (sq) PCR and compared to expression in primary roots of the lateral root mutants lrt1 (PR lrt1; Hochholdinger and Feix, 1998) and rum1 (PR rum1; Woll et al., 2005). For any PCR product, amplification of the correct cDNA fragment was verified by cloning and sequencing of the corresponding amplicon. As shown in Figure 2, ZmGSL5 is the only gene not expressed in roots. All other genes are transcribed in every wild-type root tissue tested with the exception of ZmGSL2, which is only active in primary roots. The absence of ZmGSL5 expression in roots was confirmed using genomic DNA in a control reaction that resulted in amplification of the expected PCR product. In all other instances, amplified PCR products were verified not to result from DNA contamination by performing a control reaction without template (see control reactions in Fig. 2). sq reverse transcription (RT)-PCR carried out for ZmGSL1 yielded two bands of different size and expression level (Fig. 2). Sequencing of the corresponding PCR fragments revealed that they represent different splice variants of the ZmGSL1 gene (ZmGSL1a and ZmGSL1b). Splice variant ZmGSL1b is characterized by the retention of the third intron leading to an increased transcript size compared to ZmGSL1a (Fig. 4A). The upper band for ZmGSL1a/b in Figure 2 therefore represents splice variant ZmGSL1b, while the lower band corresponds to splice variant ZmGSL1a. Due to the presence of the third intron, a premature stop codon is introduced into the ZmGSL1b transcript, which results in the complete loss of the conserved Cys-rich domain in ZmGSL1b (Fig. 4A).
Figure 2.
sqRT-PCR expression analysis of ZmGSL gene family members in primary (PR), seminal (SR), and crown roots (CR). Expression in primary roots was surveyed in wild-type seedlings, and the mutants lrt1 and rum1, which do not initiate lateral roots. Control reactions were for all ZmGSL members, except ZmGSL5, negative controls without template. For ZmGSL5, which is not expressed in roots, a positive control with genomic DNA was performed. Actin was used as a constitutively expressed control gene.
Figure 4.
Differential splicing of the ZmGSL1 gene. The ZmGSL1 gene contains four exons (colored) and three introns (gray) and encodes for two alternatively spliced variants, ZmGSL1a and ZmGSL1b. ZmGSL1b retains the third intron, which introduces a premature stop codon at the 5′ end of the intron (asterisks). ZmGSL1a and ZmGSL1b display complementary expression in longitudinal sections of the maize primary root. While ZmGSL1a is preferentially expressed in cells surrounding lateral root primordia, ZmGSL1b is specifically expressed in pericycle cells that initiate lateral roots. Hybridization probes are indicated by a black bar under the schematic transcript structures. Bars = 200 μm. [See online article for color version of this figure.]
sqRT-PCR studies demonstrated that expression of neither ZmGSL1a nor ZmGSL1b is significantly affected in the lrt1 or rum1 mutant background. For several other genes, however, expression was reduced or completely lacking in primary roots of lrt1 and/or rum1. ZmGSL9, for example, is strongly expressed in wild-type primary roots but only weakly transcribed in lrt1 and rum1. ZmGSL6 is expressed in wild-type and primary roots of lrt1 but not in primary roots of the rum1 mutant. Similarly, ZmGSL4 is strongly expressed in wild-type and lrt1 primary roots but only transcribed at low levels in primary roots of rum1. Finally, weak ZmGSL2 expression detected in primary roots of the wild type is abolished in the lrt1 as well as the rum1 mutant background.
In Situ Expression Experiments Confirm a Role for ZmGSL Genes in Lateral Root Development
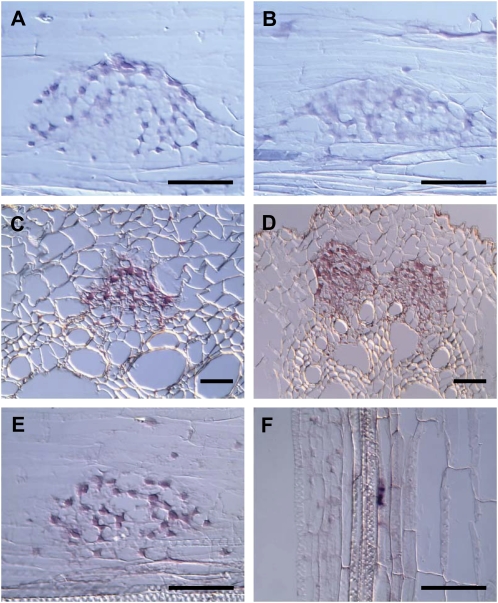
The finding that ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 gene expression is reduced or abolished in lrt1 and/or rum1 mutant background suggests that these genes play a role in lateral root formation. To gain a more detailed insight into the potential role of these genes in root development, cell-type-specific transcription patterns were studied by in situ hybridization experiments. As a result, expression of ZmGSL2 (Fig. 3A), ZmGSL4 (Fig. 3C), ZmGSL6 (Fig. 3E), and ZmGSL9 (Fig. 3D) indeed was detected at different intensities in emerging lateral root primordia of wild-type primary roots. While ZmGSL4, ZmGSL6, and ZmGSL9 are ubiquitously expressed at moderate levels in transverse tissue sections through a lateral root primordium, ZmGSL2 expression is only weak in corresponding longitudinal sections (Fig. 3A). However, this weak ZmGSL2 expression intensity is still distinguishable from the result for a longitudinal section through a lateral root primordium emanating from a crown root (compare to Fig. 3B). These findings are in accordance with previous results from sqRT-PCR analyses (compare to Fig. 2), which also show weak ZmGSL2 expression in the primary root but lack of expression in seminal or crown roots. In addition to ubiquitous expression in lateral root primordia as detected for ZmGSL2, ZmGSL4, and ZmGSL9, ZmGSL6 transcription could also be verified to occur in a small subdomain of pericycle precursor cells as shown for a longitudinal section through a primary root in Figure 3F.
Figure 3.
In situ gene expression analysis of members of the ZmGSL gene family in emerging lateral roots. A, Expression of ZmGSL2 in an emerging lateral root primordium of a primary root (longitudinal section). B, Lack of ZmGSL2 expression in a lateral root primordium emerging from a crown root (longitudinal section) is consistent with the lack of ZmGSL2 expression in this root type (see Fig. 2). C, Expression of ZmGSL4 in an emerging lateral root primordium of a primary root (transverse section). D, Expression of ZmGSL9 in two emerging lateral root primordia of a primary root (transverse section). E, Expression of ZmGSL6 in a transverse section of a lateral root primordium emerging from a primary root. F, Expression of ZmGSL6 in pericycle precursor cell that will divide and generate a lateral root in a transverse section of a lateral root primordium emerging from a primary root. Expression in the pericycle cell appears to be limited to the vicinity of the nucleus. Bars = 100 μm. [See online article for color version of this figure.]
In conclusion, in situ hybridization experiments were found to coincide with the results from previous sqRT-PCR studies and confirmed that ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 contribute to lateral root development. In this respect, ZmGSL6 expression is already detectable in single cells of the pericycle prior to the histological emergence of a lateral root primordium. ZmGSL6 therefore might already be involved in the initial steps of lateral root formation.
Expression Patterns for the Two ZmGSL1 Splice Variants Suggest Complementary Expression Domains in Primary Roots
sqRT-PCR analyses had revealed that the ZmGSL1 gene is expressed in two alternative splice forms during root development (Fig. 2). To analyze the potential function of ZmGSL1a and ZmGSL1b in more detail, in situ hybridization experiments were performed on tissue sections through wild-type primary roots. As described before, splice variant ZmGSL1b differs from ZmGSL1a by the retention of the third exon of the ZmGSL1 locus (Fig. 4A). Designing an in situ probe directed against intron three of the ZmGSL1 locus (see black line under the right schematic drawing in Fig. 4B) therefore allowed us to specifically detect transcripts of the ZmGSL1b splice variant. In situ hybridizations on longitudinal sections through a wild-type primary root detected ZmGSL1b expression in cells of the pericycle (see bottom right picture in Fig. 4B). In addition, expression was weakly detectable in emerging lateral root primordia (data not shown). For ZmGSL1a, it was not possible to design a specific in situ hybridization probe (see schematic drawings in Fig. 4B). Nevertheless, taking into account that it would cross-hybridize with transcripts of the ZmGSL1b splice variant, a second in situ probe was designed and directed against the first three exons and part of the fourth exon of the ZmGSL1a transcript variant (see black line in Fig. 4B). As a result, strong expression was detected in cortex cells surrounding a lateral root primordium as exemplified for a longitudinal section through a lateral root primordium in the left picture of Figure 4B. In addition, weak ZmGSL1a expression was detected in lateral root primordia adjacent to cortical cells of a primary root. This faint expression was comparable to the result obtained for the ZmGSL1b hybridization probe. Comparing the in situ transcription patterns detected by the ZmGSL1a and ZmGSL1b probes suggests that strong expression detected in cortex cells surrounding lateral root primordia most likely is related to ZmGSL1a, while strong expression in specific cells of the pericycle and weak expression in emerging lateral root primordia presumably reflects transcript abundance of the ZmGSL1b splice variant. We therefore concluded that ZmGSL1a and ZmGSL1b are both associated with lateral root development and that strong ZmGSL1a expression in cortical cells is organized in a domain that is oriented complementary to the weak ZmGSL1b expression domain detectable in developing lateral root primordia.
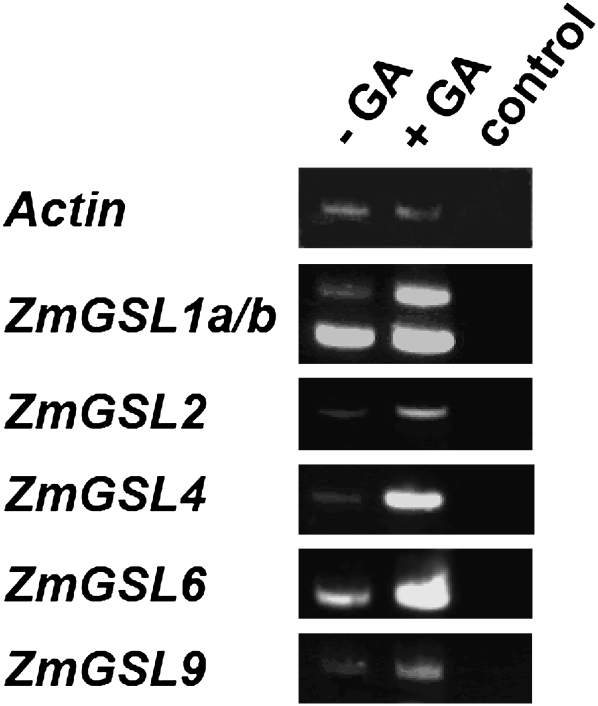
GA3 Induces Expression of ZmGSL Genes
It has been demonstrated previously that GA3 is capable of stimulating the expression of numerous GASA genes in Arabidopsis (Aubert et al., 1998), tomato (Shi and Olszewski, 1998), rice (Furukawa et al., 2006), and petunia (Ben-Nissan et al., 2004). Therefore, GA3 inducibility of ZmGSL1, ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 was tested in 7-d-old primary roots (Fig. 5). As a result, expression of all five genes was induced at different levels in primary roots that had been incubated in GA3 for 4 d. While expression of ZmGSL4 and ZmGSL6 was strongly induced after GA3 treatment, expression of ZmGSL2 and ZmGSL9 had elevated to only moderate levels. Interestingly, with regard to the two ZmGSL1 splice variants, expression of ZmGSL1b was consistently found to be induced stronger than ZmGSL1a (Fig. 5).
Figure 5.
GA3 induction of ZmGSL genes that are expressed in lateral root primordia. All genes for which expression in lateral root primordia or pericycle precursor cells has been demonstrated in Figure 3 and 4 are inducible by GA3. Note that splice variant ZmGSL1b, which is expressed in pericycle precursor cells, displays a stronger induction by GA3 than ZmGSL1a, which is expressed in cells surrounding the lateral root primordium.
DISCUSSION
The GASA/GAST Family of Angiosperms Is Organized in Three Distinct Phylogenetic Clades
In this study, comprehensive genome scans of the monocot GASA/GAST-like gene family were performed in the recently sequenced maize and rice genomes. Besides the newly identified maize GAST-like genes ZmGSL1 to ZmGSL10, six novel rice genes were identified, bringing the total number of GASA/GAST-like members from rice to nine. Phylogenetic reconstructions involving the previously identified rice (Furukawa et al., 2006; Wang et al., 2009), tomato (Shi et al., 1992; Taylor and Scheuring, 1994), and Arabidopsis (Roxrud et al., 2007) full-length GAST-like protein sequences grouped the newly identified monocot members into three distinct phylogenetic clades. The comparison of monocot and dicot GASA/GAST-like proteins in our study therefore reveals that during evolution eudicot members of the GASA/GAST-like family have developed along three different directions.
Among the newly identified members of the monocot GASA/GAST-like family, eight form pairwise orthologs. No putative ortholog from rice, Arabidopsis, or tomato could be identified for ZmGSL2 and ZmGSL6. ZmGSL2 and ZmGSL6 therefore might encode functions that are specific to maize development. Among the pairwise orthologs, three GAST-like proteins from rice, namely, OsGSR1 as the ortholog of ZmGSL3, OsGASR2 as the closest relative of ZmGSL4, and OsGASR1 as being most closely related to ZmGSL10, have been characterized in more detail. While OsGSR1 is involved in the cross talk between GAs and brassinosteroids by directly activating brassinosteroid biosynthesis and positively regulating GA signaling (Wang et al., 2009), OsGASR1 and OsGASR2 were shown to confer cell division in the context of panicle differentiation (Furukawa et al., 2006). MPSS and root-specific expression analyses for ZmGSL3, ZmGSL4, and ZmGSL10 are generally in line with the expression data published for OsGSR1, OsGASR2, and OSGASR1, respectively. This may suggest that ZmGSL3, ZmGSL4, and ZmGSL10 in maize may exert functions that are analogous to those of OsGSR1, OsGASR1, and OsGASR2 in rice.
ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 Are Dependent on the Activity of LRT1 and/or RUM1
Results from sqRT-PCR studies in combination with expression patterns obtained from in situ hybridization analyses suggest that besides ZmGSL1, which will be discussed in a separate section, ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 contribute to lateral root primordium formation. Misexpression of these genes in the lrt1 and/or rum1 mutant backgrounds supports this notion and furthermore helps to define their putative role in the molecular framework of lateral root development in maize. ZmGSL2 is the only member of the ZmGSL gene family that is expressed in a root-type-specific way. In the wild type, weak ZmGSL2 expression is detectable in primary roots but not in seminal or crown roots. In primary roots of the lateral root mutants lrt1 (Hochholdinger and Feix, 1998) and rum1 (Woll et al., 2005), however, ZmGSL2 gene expression is abolished. Together with the fact that no corresponding ortholog can be assigned to ZmGSL2 in phylogenetic analyses, these results propose that ZmGSL2 represents a maize-specific checkpoint of lateral root formation in primary roots that is likely to depend on the activity of LRT1 and RUM1. Similar to ZmGSL2, ZmGSL9 expression also is affected in the two maize lateral root mutants lrt1 and rum1. However, since ZmGSL9 gene expression is not completely lost in the lrt1 and rum1 mutant backgrounds, its function in primary roots presumably is only in part controlled by LRT1 and RUM1. According to MPSS analyses, ZmGSL6 is the only ZmGSL gene that is not expressed in aboveground tissues of maize, while RT-PCR and in situ hybridization experiments carried out for roots suggest distinct roles for ZmGSL6 during lateral root formation: Besides expression in emanating lateral root primordia, ZmGSL6 transcripts can already be detected in prospective lateral root precursor cells of the pericycle. In comparison with ZmGSL2, ZmGSL4, and ZmGSL9, ZmGSL6 is likely to contribute to the initial events of lateral root formation. Expression of ZmGSL6 in primary roots of the mutant lrt1 but not in rum1 furthermore suggests this early function is independent of LRT1 but dependent on RUM1. As for ZmGSL2, this function probably is specific to maize, as no discrete rice ortholog can be assigned in phylogenetic reconstructions for ZmGSL6. In summary, RT-PCR expression analyses in wild-type and the lrt1 and rum1 mutants in combination with in situ expression patterns detected in primary roots of the wild type suggest different roles for ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 during lateral root formation, which are either controlled by LRT1 or RUM1 or both.
ZmGSL1 Reveals Novel Aspects in the Regulation of GASA/GAST-Like Genes
ZmGSL1 is the only GAST-like gene described to date that exhibits alternative splicing. This implies that among the members of the eudicot GASA/GAST-like family known to date, alternative splicing represents an evolutionary event that has occurred only recently in maize. Alternative splicing is an important mechanism of gene regulation that increases the variety of transcripts from the same gene (Reddy, 2007). Comprehensive surveys suggested that approximately 21% to 22% of all unique cDNAs in rice and Arabidopsis represent alternative splicing events (Wang and Brendel, 2006) and that in maize, alternative splicing is even more frequent than in Arabidopsis (Alexandrov et al., 2009). Remarkably, the two splice variants of ZmGSL1 display complementary in situ expression patterns in primary roots. While ZmGSL1b is expressed in pericycle precursor cells that give rise to emerging lateral roots, ZmGSL1a is transcribed in parenchyma cells surrounding root primordia that are not related to meristematic activity. Thus far, only a few examples for tissue-specific expression patterns of different splice variants from the same gene have been published in plants (Macknight et al., 2002). Since both ZmGSL1a and ZmGSL1b originate from the same promoter, their distinct in situ expression patterns presumably reflect activity of different splice factors for the ZmGSL1 transcription in different tissues. ZmGSL1b differs from ZmGSL1a by the retention of the third intron (Fig. 4A). In conclusion, factors that confer splicing of the third intron in the ZmGSL1 transcript in cells surrounding a lateral root primordium might either be not present or inhibited in cells of the lateral root primordium itself. The retention of the third intron in ZmGSL1b leads to a premature stop codon and hence to the complete loss of the highly conserved Cys-rich domain in cells of an emerging lateral root. This implies that the C-terminal domain in ZmGSL1a either functions in inhibiting lateral root development or promoting nonlateral root development in cells of the root cortex.
GA3 Inducibility
According to the results from sqRT-PCR studies, ZmGSL1b, ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 are all induced by GA3. GA3 is involved in manifold developmental processes, including germination, stem elongation, and the transition from vegetative to reproductive development (Olszewski et al., 2002). In Arabidopsis, it has been demonstrated that the growth-promoting effect of auxin on primary roots is largely mediated by enhancing the GA-triggered inactivation of the proteins GIBBERELLIC ACID INSENSITIVE and REPRESSOR OF GA1 (Fu and Harberd, 2003). This pathway could also be active in lateral roots (Hardtke, 2003). GA1, which catalyzes the first step in GA3 biosynthesis, is highly expressed in lateral root meristems (Silverstone et al., 1997), which could suggest a role of ZmGSL1b, ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9 in GA3-dependant lateral root formation. Remarkably, splice variant ZmGSL1b, which is associated with expression in the pericycle and thus lateral root formation, is very strongly induced by GA3, while ZmGSL1a transcript abundance is only slightly increased by GA3 in parenchyma cells. This may suggest that GA3 has a negative effect on the activity of splice factors that normally are responsible for splicing of the third intron in the ZmGSL1 precursor.
In summary, the comprehensive phylogenetic and expression analysis of the newly identified ZmGSL genes in maize suggest distinct roles for several members of the gene family (ZmGSL1b, ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9) in GA3-regulated lateral root formation, which is either dependent or independent of LRT1 and/or RUM1.
MATERIALS AND METHODS
Phylogenetic Reconstructions
The databases at The Institute for Genomic Research (TIGR) and PlantGDB were screened for sequences homologous to the characteristic C-terminal domain of the tomato (Solanum lycopersicum) protein SlRSI1 (Taylor and Scheuring, 1994) and of members of the Arabidopsis (Arabidopsis thaliana) GASA family (Herzog et al., 1995). GenBank accession numbers for Arabidopsis AtGASA1 to AtGASA12 and AtGASA14 have been summarized by Roxrud et al. (2007). In addition, Arabidopsis AtGAST1-like (AK229086), tomato SlRSI1 (AAA20130), and tomato SlGAST (CAA44807) protein sequences were retrieved from GenBank. Phylogenetic and molecular evolutionary analyses were conducted using the neighbor-joining algorithm in MEGA version 4 (Tamura et al., 2007) considering 1,000 replications with bootstrap analyses. Coding regions and corresponding protein sequences for ZmGSL1 to ZmGSL10 were retrieved from TIGR (http://maize.jcvi.org) and MaizeGDB (www.maizegdb.org) databases. The genomic context and the chromosomal location were determined based on sequences from the maize (Zea mays) genome sequencing project (www.maizegenome.org). Accession numbers of all maize and rice GSL genes are summarized in Table I. The C-terminal Cys-rich domains of the nine GSL sequences are summarized in Figure 1A. The novel members of the rice gene family identified in this study are OsGSL1, OsGSL5, OsGSL7, OsGSL8, OsGSL9, and OsGSL12. Pairwise calculations of protein similarities among all maize and rice GSL full-length proteins and conserved Cys-rich C-terminal domains (Supplemental Table S3) were performed with MegAlign software (DNAstar).
Survey of MPSS
The Solexa MPSS technology (Brenner et al., 2000) permits quantification of 17-bp sequences that start with the signature sequence GATC in populations of 2 × 105 to 2 × 106 cDNAs of a defined developmental stage of a maize organ. These 17-bp signature sequences almost always correspond to unique cDNAs, thus allowing for the quantification of the abundance of a particular cDNA in a sample representing a particular organ and developmental stage. MPSS data were normalized and filtered according to the Solexa protocol. All analyzed libraries were generated from tissue of the inbred line B73.
RT-PCR Analyses
All RT-PCR experiments were based on pools of primary, seminal, and crown root samples from three independent biological replicates. For root-type-specific expression studies in wild-type and lrt1 and rum1 mutant backgrounds, separate pools of primary roots with a length of at least 4 cm were selected. At this stage, lateral root formation is clearly manifested in all main root types of maize according to the Feulgen-staining procedure (data not shown).
For GA3 induction experiments, primary roots were germinated in paper rolls in distilled water for 3 d under standard conditions (Woll et al., 2005). Pairs of germinated seedlings that had the same root length were selected to constitute two equivalent populations that were subsequently grown either in distilled water or in 0.1 mg L−1 GA3 (Sigma-Aldrich). Roots from both pools were harvested 4 d after incubation, frozen in liquid nitrogen, and processed immediately for total RNA isolation.
Total RNA was extracted using Trizol (Invitrogen), incubated with RNAse free DNAseI (Invitrogen) overnight, and reverse transcribed employing SuperScriptII (Invitrogen) according to the manufacturer's protocols. PCR was performed using LA-Taq Polymerase (TaKaRa) following the manufacturer's guidelines. The PCR temperature scheme was adjusted according to the oligonucleotide primers employed for the experiments. All PCR experiments were repeated several times to confirm reproducibility of results.
In Situ Hybridization Experiments
Root samples were fixed in 4% formaldehyde in phosphate-buffered saline overnight, dehydrated in an increasing ethanol series, and embedded in paraffin wax (Paraplast plus; Sigma-Aldrich). Tissue sections (7 μm) were taken using a Jung rotary microtome and coated on Superfrost Plus slides (Microm). Templates for all in situ hybridization probes were cloned into the pGem-T Easy vector (Promega) and amplified using a combination of a gene-specific and a M13 forward or M13 reverse primer, respectively. The template for ZmGSL1b was chosen to correspond to the third intron of the ZmGSL1 locus (compare Fig. 4) which is not present in ZmGSL1a. All probes were transcribed using an in vitro transcription kit containing digoxigenin-labeled UTP (Roche). In situ hybridizations were performed according to Jackson (1991). Gene-specific primers used for RT-PCR and the generation of templates for in situ probes are summarized in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Summary of MPSS expression analyses of the 10 ZmGSL family members in aboveground tissues.
Supplemental Table S1. Oligonucleotide primer sequences used for RT-PCR and in situ hybridization experiments.
Supplemental Table S2. Identity between full-length protein sequences deduced from maize and rice GSL family members.
Supplemental Table S3 MPSS expression profiles of the ZmGSL gene family in kernel, shoot, and generative tissues.
Supplementary Material
This project was supported by a DuPont Crop Science research grant and the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 446 “Mechanisms of cell behavior in eukaryotes.”
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Frank Hochholdinger (frank.hochholdinger@zmbp.uni-tuebingen.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alexandrov NN, Brover VV, Freidin S, Troukhan ME, Tatarinova TV, Zhang H, Swaller TJ, Lu YP, Bouck J, Flavell RB, et al (2009) Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol 69 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M (1998) Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol 36 871–883 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Lee JY, Borohov A, Weiss D (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J 37 229–238 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Weiss D (1996) The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol 32 1067–1074 [DOI] [PubMed] [Google Scholar]
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN (2007) Combining expression and comparative evolutionary analysis. The COBRA gene family. Plant Physiol 143 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Christensen TM, Veijlupkova Z, Sharma YK, Arthur KM, Spatafora JW, Albright CA, Meeley RB, Duvick JP, Quantrano RS, Fowler JE (2003) Conserved subgroups and developmental regulation in the monocot rop gene family. Plant Physiol 133 1791–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes root growth by modulating gibberellin response. Nature 421 740–743 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Sakaguchi N, Shimada H (2006) Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst 81 171–180 [DOI] [PubMed] [Google Scholar]
- Hardtke CS (2003) Gibberellin signaling: GRASs growing roots. Curr Biol 13 R366–R367 [DOI] [PubMed] [Google Scholar]
- Herzog M, Dorne AM, Grellet F (1995) GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol 27 743–752 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Feix G (1998) Early post-embryonic root formation is specifically affected in the maize mutant lrt1. Plant J 16 247–255 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Feix G (2001) Cooperative action of SLR1 and SLR2 is required for lateral root specific cell-elongation in maize. Plant Physiol 125 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K (2004) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Wen TJ, Zimmermann R, Chimot-Marolle P, da Costa e Silva O, Bruce W, Lamkey KR, Wienand U, Schnable PS (2008) The maize (Zea mays L.) roothairless 3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J 54 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Zimmermann R (2008) Conserved and diverse mechanisms in root development. Curr Opin Plant Biol 11 70–74 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F (2009) The maize root system: morphology, anatomy and genetics. In J Bennetzen, S Hake, eds, The Handbook of Maize. Springer, New York, pp 145–160
- Jackson DP (1991) In situ hybridisation in plants. In DJ Bowles, SJ Gurr, M McPhereson, eds, Molecular Plant Pathology: A Practical Approach, Oxford University Press, Oxford, pp 163–166
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45 83–100 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, Teeri TH (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Duroux M, Laurie R, Dijkwel P, Simpson G, Dean C (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signalling: biosynthesis, catabolism and response pathways. Plant Cell (Suppl) 14 S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58 267–294 [DOI] [PubMed] [Google Scholar]
- Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG (2007) GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol 48 471–83 [DOI] [PubMed] [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE (1992) Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J 2 153–159 [PubMed] [Google Scholar]
- Shi L, Olszewski NE (1998) Gibberellin and abscisic acid regulate GAST1 expression at the level of transcription. Plant Mol Biol 38 1053–1060 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Chang CW, Krol E, Sun TP (1997) Developmental regulation of gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12 9–19 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Taylor BH, Scheuring CF (1994) A molecular marker for lateral root initiation: the RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia. Mol Gen Genet 243 148–57 [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57 498–510 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.