Abstract
Plants growing in different environments develop with different photosynthetic capacities—developmental acclimation of photosynthesis. It is also possible for fully developed leaves to change their photosynthetic capacity—dynamic acclimation. The importance of acclimation has not previously been demonstrated. Here, we show that developmental and dynamic acclimation are distinct processes. Furthermore, we demonstrate that dynamic acclimation plays an important role in increasing the fitness of plants in natural environments. Plants of Arabidopsis (Arabidopsis thaliana) were grown at low light and then transferred to high light for up to 9 d. This resulted in an increase in photosynthetic capacity of approximately 40%. A microarray analysis showed that transfer to high light resulted in a substantial but transient increase in expression of a gene, At1g61800, encoding a glucose-6-phosphate/phosphate translocator GPT2. Plants where this gene was disrupted were unable to undergo dynamic acclimation. They were, however, still able to acclimate developmentally. When grown under controlled conditions, fitness, measured as seed output and germination, was identical, regardless of GPT2 expression. Under naturally variable conditions, however, fitness was substantially reduced in plants lacking the ability to acclimate. Seed production was halved in gpt2− plants, relative to wild type, and germination of the seed produced substantially less. Dynamic acclimation of photosynthesis is thus shown to play a crucial and previously unrecognized role in determining the fitness of plants growing in changing environments.
It has long been recognized that when plants are grown under a particular set of conditions they adjust their photosynthetic capacity to match those conditions (for review, see Walters, 2005). For example, early work from Bjorkman and Holmgren (1963) showed that plants of Solidago virgaurea had different photosynthetic capacities when grown either in sun or shade. In spite of its long history, however, neither the mechanism nor the significance of this response is understood (Walters, 2005). Work from Murchie and Horton (1997) showed that there is substantial variation between species in their ability to acclimate, with plants from semishaded habitats having the greatest variation in photosynthetic capacity, suggesting that there is both a benefit and cost of acclimation. Neither benefit nor cost has been demonstrated.
Photosynthetic acclimation can be observed at levels ranging from whole-plant morphology to the detailed stoichiometry of the photosynthetic apparatus (Boardman, 1977; Walters, 2005). Plants grown at low light tend to invest more in leaves than in roots and to have thinner leaves. They have more chlorophyll-containing light-harvesting proteins relative to light-using enzymes involved in electron transport and metabolism, meaning that photosynthesis saturates with light at a lower irradiance. Plants can also adjust the relative proportions of the different photosystems to suit the light quality they experience (Chow et al., 1990; Walters and Horton, 1995a, 1995b).
Most studies that have examined the acclimation of plants have done so by making measurements on material that has experienced only one set of conditions—e.g. either high or low light. Differences between plants therefore reflect the conditions experienced as the leaves develop, with leaf morphology and composition being optimized for the conditions seen. Plants do not, however, exist in static environments. Even for a plant growing in an unshaded location, the light incident on a leaf can vary by an order of magnitude from second to second, day to day, and week to week depending on the weather conditions. This variation will typically be accompanied by variation in the temperature, which will also impact on metabolic capacity.
When plants are exposed to light at irradiances that are above saturating for photosynthesis, which may result from increases in light or from environmental conditions (e.g. cold, drought) restricting metabolism, they are liable to suffer from stress (Demmig-Adams and Adams, 1992). Specifically, excess light can give rise to reactive oxygen species (Asada, 2006). The damaging effects of this can be limited by investing in antioxidant systems; however, these are metabolically expensive with, for example, substantial amounts of individual antioxidants such as ascorbic acid, being found in the chloroplast (Asada, 2006). It seems likely therefore that the ability of a plant to minimize stress, by adjusting photosynthetic capacity to suit as well as possible the prevailing conditions, will benefit the plant and increase overall fitness.
In this study, we have investigated photosynthetic acclimation of the model plant Arabidopsis (Arabidopsis thaliana). Starting with a microarray analysis, we have identified a gene that is essential for acclimation to increases in irradiance. We further show that the ability to acclimate to changes in light has a major role in determining fitness under naturally variable light conditions.
RESULTS
Transfer from Low to High Light Results in Photosynthetic Acclimation in Some, But Not All, Accessions of Arabidopsis
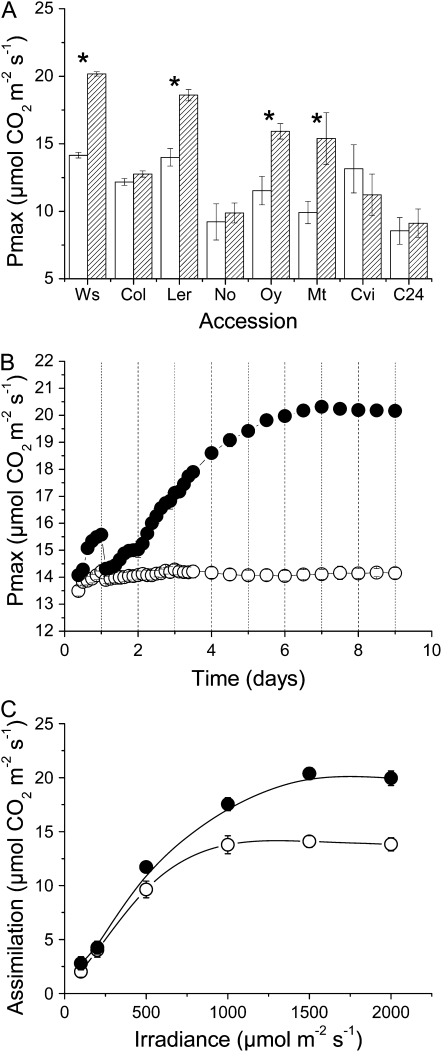
Plants of eight accessions of Arabidopsis were grown from seed under controlled conditions, at an irradiance of 100 μmol m−2 s−1 white light on an 8-h d for 8 weeks. Under these growth conditions, Arabidopsis, a short-day plant, forms a mature rosette of leaves before flowering. Mature plants were transferred from 100 to 400 μmol m−2 s−1 and, after 9 d, their photosynthetic capacity measured under conditions of high light and CO2 (Fig. 1A). There was significant variation seen in the ability of different accessions to acclimate to the increase in light. For example, while the Wassilewskija-2 (Ws-2) accession increased its photosynthetic capacity by approximately 40%, no significant increase was seen in Columbia-0 (Col-0). Variation was seen not only among the widely used laboratory strains (Ws, Col, Landsberg erecta) but also in accessions that might be regarded as true wild-type plants.
Figure 1.
Dynamic photosynthetic acclimation in Arabidopsis. A, Maximum photosynthetic capacity, measured at 1,500 μmol m−2 s−1 white light and 2,000 μL L−1 CO2 in eight accessions of Arabidopsis grown for 8 weeks at an irradiance of 100 μmol m−2 s−1 and then maintained in the same conditions (white bars) or transferred to 400 μmol m−2 s−1 light (hatched bars) for 9 d. Accessions measured were Ws-2, Col-0, Landsberg erecta (Ler), Nossen (No), Oystese (Oy), Martuba (Mt), Cape Verdi Island (Cvi), and C24. Asterisk (*) indicates that treated plants differ significantly from their respective control (P < 0.05). B, Changes in maximum rate of photosynthesis in Ws following a step increase in light from 100 to 400 μmol m−2 s−1 (black circles) compared to plants maintained at 100 μmol m−2 s−1 (white circles). C, Net CO2 assimilation of Ws as a function of irradiance, in plants grown at 100 μmol m−2 s−1 (white circles) or transferred to 400 μmol m−2 s−1 (black circles) for 9 d. All data represent the mean (±se) of at least three replicates.
The dynamics of acclimation were examined in more detail in the accession Ws. Previous work on Ws has shown that growth from seed under a range of different irradiances results in clear developmental acclimation of photosynthesis, with different responses being seen in different irradiance ranges (Bailey et al., 2001). Yin and Johnson (2000) observed dynamic acclimation in Ws. When plants of Ws were transferred from 100 to 400 μmol m−2 s−1, there was a small (approximately 10%) increase in photosynthetic capacity within the first photoperiod; however, this difference was no longer apparent by the following day (Fig. 1B). A second photoperiod at the higher irradiance resulted in a similar difference in photosynthetic capacity; however, this was not reversed overnight. From the 3rd to the 6th d at light there was a steady increase in the photosynthetic capacity of the plants, reaching a value approximately 40% higher than the control after a week at high light.
Examination of the irradiance sensitivity of photosynthesis showed that while the maximum photosynthetic capacity of plants had increased after 9 d at high light, the rate of photosynthesis under light-limiting conditions was not significantly changed (Fig. 1C). Measurements of leaf chlorophyll content showed that while the total chlorophyll per unit area did not change during acclimation, the ratio of chlorophyll a/b was slightly, but significantly, altered (Table I). The changes in chlorophyll seen were similar to those observed by Bailey et al. (2004) under conditions of developmental acclimation.
Table I.
Chlorophyll content and composition following acclimation
Leaves of Arabidopsis were grown for 8 weeks at 100 μmol m−2 s−1 light and then either maintained at 100 μmol m−2 s−1 for a further 9 d (LL) or transferred to an irradiance of 400 μmol m−2 s−1 (HL). Data represent the mean ± se of eight replicates. Significance of results was tested using a Student's t test.
| Parameter | LL | HL | P |
|---|---|---|---|
| Total chlorophyll (mg mL−1) | 262.9 ± 1.2 | 261.1 ± 0. 8 | 0.067 |
| Chlorophyll a/b ratio |
2.87 ± 0.03 |
3.03 ± 0.05 |
0.034 |
Dynamic Acclimation to Increased Irradiance Requires the Expression of a Glc-6-P/Phosphate Translocator GPT2
Data in Figure 1B suggest that events occurring on the first and subsequent days of acclimation to high light are distinct. We performed a microarray analysis on leaf material taken from plants exposed to 400 μmol m−2 s−1 for 4 h on the 1st and 3rd d of acclimation. On the 1st d of acclimation, a total of 666 genes were found to have expression that was altered significantly relative to the control (≥2-fold up- or down-regulated; P < 0.01; mean expression level greater than 100 in at least one condition). Of these, 315 genes were up-regulated, 351 repressed. By day 3, 285 genes had significantly altered expression levels, only 59 up-regulated. Of the genes with altered expression on day 1, 210 were also altered on day 3. The most up- and down-regulated genes observed are listed in Table II, a full list is available in the supporting information (Supplemental Table S1). The most up-regulated gene on day 1 of acclimation was At1g61800, encoding a Glc-6-P/phosphate translocator, GPT2. Previous work on the GPT2 gene product has shown it to be able to translocate a range of sugar phosphates in exchange for phosphate with similar efficiency (Knappe et al., 2003); however, a study examining GPT2 knockout plants in a Col background was unable to identify a specific function for this gene (Niewiadomski et al., 2005).
Table II.
Top 10 most up- and down-regulated genes observed in plants exposed to increased light
Plants of Ws were grown for 8 weeks at 100 μmol m−2 s−1 and then transferred from 100 to 400 μmol m−2 s−1. Plants were sampled at the midpoint of the 1st and 3rd d of treatment. The gene annotation used was derived from The Arabidopsis Information Resource (http://www.arabidopsis.org/). For a full set of data, see Supplemental Table S1. AGI, Arabidopsis Genome Initiative.
| AGI Code | Gene Annotation | Fold Change—Day 1 | Fold Change—Day 3 |
|---|---|---|---|
| Up-regulated | |||
| At1g61800 | Glc-6-P/phosphate translocator 2 | 34.0 | 12 |
| At4g15210 | β-Amylase 1 | 13.7 | 12.2 |
| At1g64780 | Ammonium transporter protein | 10.0 | 6.7 |
| At5g49480 | Encodes a novel Ca2+-binding protein | 9.7 | 3.7 |
| At2g27420 | Putative Cys proteinase | 9.2 | 5.5 |
| At1g32900 | Starch synthase | 8.0 | 3.4 |
| At4g16590 | Cellulose synthase | 7.6 | 6.3 |
| At1g56650 | Myeloblastosis protein 75 | 6.2 | 6.0 |
| At4g01080 | Unknown protein | 5.4 | 3.2 |
| At1g57590 | Putative pectinacetylesterase | 4.6 | 3.8 |
| Down-regulated | |||
| At5g59080 | Unknown protein | −9.2 | −4.0 |
| At2g18700 | Trehalose-6-P synthase 11 | −9.5 | −4.0 |
| At2g22980 | Ser carboxypeptidase-like 13 precursor | −9.6 | −4.0 |
| At2g25900 | Putative Cys-3 His zinc-finger protein | −9.8 | −6.8 |
| At1g23390 | Kelch repeat-containing F-box family protein | −10.2 | −4.5 |
| At1g70290 | Trehalose-6-P synthase 8 | −10.4 | −5.7 |
| At3g15450 | Unknown protein | −12.1 | −12.4 |
| At5g48490 | Protease inhibitor/seed storage/lipid transfer protein | −15.4 | −9.2 |
| At2g40610 | Expansin A8 | −16.2 | −6.0 |
| At1g74670 |
Gibberellin-responsive protein |
−22.4 |
−7.2 |
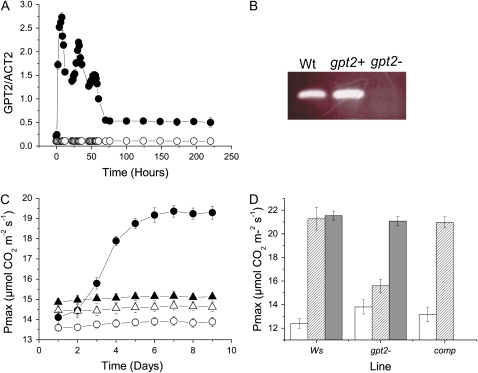
Expression of GPT2 was examined in more detail using quantitative reverse transcription (RT)-PCR (Fig. 2A). Expression was seen to rise rapidly following exposure of plants to high light—transcript levels were greater than control within 2 h of transfer to high light and continued to rise throughout the photoperiod. Transcript levels fell during the dark period but rose again in the second photoperiod. There was an overall trend for GPT2 transcript levels to fall as acclimation progressed, these reaching a steady value after the 3rd d at high light, which was, however, significantly greater than the control.
Figure 2.
GPT2 expression in photosynthetic acclimation. A, GPT2 expression in Ws following transition from 100 to 400 μmol m−2 s−1 light. Expression was quantified using real-time RT-PCR and normalized to ACT2 (At3g18780). B, Expression of GPT2 transcript estimated using RT-PCR, in plants grown for 8 weeks at 100 μmol m−2 s−1 light and then transferred to 400 μmol m−2 s−1 for 4 h. Wt, Ws wild type; gpt2−, plants from line FLAG_326E03 with a homozygous insertion in the gene At1g61800; gpt2+, plants from the same seed stock, characterized as lacking a T-DNA insertion in the GPT2 gene. C, Acclimation in plants from line FLAG_326E03 characterized as homozygote wild type (circles) or carrying a homozygote insertion in the GPT2 gene (triangles) and either maintained at 100 μmol m−2 s−1 (white symbols) or transferred to 400 μmol m−2 s−1 for 9 d (black symbols). D, Photosynthetic capacity of plants grown for 8 weeks at 100 μmol m−2 s−1 (white bars), grown at 100 μmol m−2 s−1 and then transferred to 400 μmol m−2 s−1 (hatched bars) or grown from seed at 400 μmol m−2 s−1 (gray bars) in plants of Ws wild type, gpt2− homozygote mutant, and in the mutant complemented with the wild-type GPT2 gene (comp). Quantitative data show the mean (±se) of at least three replicates. [See online article for color version of this figure.]
Examination of the Versailles INRA collection of T-DNA insertion mutants (Bechtold et al., 1993; Bouchez et al., 1993), which are in a Ws background, lead to the identification of a line, FLAG_326E03, with a T-DNA insertion in the GPT2 gene. Homozygote plants carrying this insertion were verified, using RT-PCR, to lack any detectable GPT2 transcript (Fig. 2B). Plants of the gpt2− knockout mutant were then grown at 100 μmol m−2 s−1 before being exposed to 400 μmol m−2 s−1, as before. At low light, the gpt2− mutant had a marginally higher photosynthetic capacity than did plants from the same seed stock identified as being homozygote for the wild-type gene. When gpt2− plants were transferred to high light, they did not undergo acclimation of photosynthesis (Fig. 2C). To confirm that this phenotype was due to the GPT2 gene, mutant plants were complemented with a copy of the gene under the control of its own promoter; this restored acclimation (Fig. 2D). It is concluded therefore that GPT2 is essential for dynamic acclimation.
When plants were grown from seed at 400 μmol m−2 s−1 light, gpt2− had a significantly higher photosynthetic capacity than when grown at low light and did not differ significantly from wild type (Fig. 2D). GPT2 is therefore not essential for developmental acclimation, implying that the signaling pathways for developmental and dynamic acclimation are distinct.
Dynamic Acclimation Increases Fitness under Naturally Variable Growth Conditions
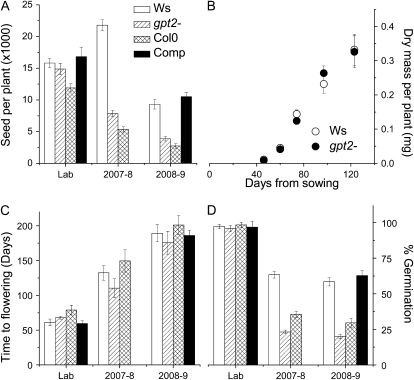
Previous work on GPT2 has failed to identify any specific role for this protein (Niewiadomski et al., 2005). It functions in vitro to transport both Glc-6-P and triose-P in exchange for phosphate (Knappe et al., 2003). Reported expression levels have typically been low, though it has been identified as being up-regulated in response to sugar feeding (Gonzali et al., 2006; Li et al., 2006) and during leaf senescence (Pourtau et al., 2006). In the Col-0 ecotype, which is unable to acclimate dynamically but which expresses the GPT2 gene, it was reported that gpt2− mutants grow normally (Niewiadomski et al., 2005). To determine whether this is also the case in a Ws background, we examined the growth and seed production of Ws gpt2− plants grown in controlled conditions (Fig. 3A). No significant difference could be seen from wild-type Ws. At the same time, Col plants were observed to have a significantly lower seed production than the Ws lines.
Figure 3.
Plant fitness in controlled and variable growth environments. A, Seed output per plant, estimated as the product of silique number and mean number of seeds per silique of plants of Ws (white bars), Ws gpt2− (hatched bars), Col (Col0; cross-hatched bars), and of the Ws gpt2− complemented mutant (Comp; black bars) grown under laboratory conditions (Lab) or in the greenhouse during the winters 2007 to 2008 and 2008 to 2009. B, Changes in total aboveground biomass of Ws (white symbols) and gpt2− (black symbols) during growth in the greenhouse in the winter of 2007 to 2008. C, Time to flowering in Ws, gpt2−, Col, and the complemented mutant grown under laboratory conditions and in the greenhouse in 2007 to 2008 and 2008 to 2009 (bar shading as in A). D, Germination of seeds of Ws and gpt2− originating from plants grown under laboratory conditions or in the greenhouse during the winter 2007 to 2008 or 2008 to 2009 (bar shading as in A). All data show the mean (±se) of at least 15 plants.
To examine whether dynamic acclimation has a role under naturally variable conditions, plants of Col, Ws, wild type, and Ws gpt2− were grown in an unheated greenhouse without supplemental lighting during the winters of 2007 to 2008 and 2008 to 2009. Environmental monitoring in the greenhouse showed substantial variation in both irradiance and temperature during the growth period with irradiances during the winter period peaking at around 800 μmol m−2 s−1. The 2008 to 2009 winter was colder than that of 2007 to 2008 and the mean time to flowering was increased from 75 to 156 d. In the 2007 to 2008 season, the growth rate of plants was seen not to be different between the Ws wild type and gpt2− (Fig. 3B); however, in both seasons, the gpt2− plants flowered slightly sooner than the Ws wild type (Fig. 3C). In controlled-environment conditions gpt2− flowered after Ws wild type. Col flowered later than either Ws or gpt2− in all conditions. Under greenhouse conditions, seed production was found to be significantly lower in all lines in the 2007 to 2008 winter than in 2008 to 2009 (Fig. 3A). In both years, the gpt2− plants had significantly lower seed production than Ws wild type, with Col having lower seed output still (Fig. 3A). These data suggest that seed production in both Ws lacking gpt2− and in Col, which has an expressed GPT2 gene but lacks acclimation, is compromised by the failure to acclimate.
In addition to examining the seed output, we also examined the germination of the seed produced (Fig. 3D). Seeds originating from plants grown in the laboratory germinated readily after 2 d preincubation at 4°C, with no significant difference being seen between Ws wild type and the gpt2− knockout. In plants grown in the greenhouse, seed germination was substantially and significantly lower in Col and gpt2−.
DISCUSSION
Although the concept of photosynthetic acclimation has been known for over 40 years, our understanding of the processes involved remains sparse. Although often referred to as a single simple concept, photosynthetic acclimation involves multiple processes that are, to a greater or lesser extent, distinct. Both light quality and quantity control leaf and chloroplast development and it is clear that these effects are independent (Walters and Horton, 1995a, 1995b). As has been previously noted (Piippo et al., 2006) responses to increases in light need to be distinguished from the effects of extreme excess light liable to cause oxidative stress. In our study, conditions were selected that did not induce stress or, indeed oversaturate photosynthesis (Yin and Johnson, 2000), and few commonly described stress-responsive genes were altered in expression (see Supplemental Table S1). Rather, we have focused specifically on dynamic changes in photosynthetic capacity to nonstressing irradiances. We have demonstrated that this response is distinct from developmental acclimation seen during leaf growth. Furthermore, we have shown that the ability to undergo dynamic acclimation is a major determinant of plant fitness under naturally fluctuating conditions, with seed production being more than 2-fold greater in plants able to acclimate. Given that the ability to acclimate has already been shown to be deficient in important crop species, including rice (Oryza sativa; Murchie et al., 2005), this process may provide a route through which crop yield can be enhanced, especially in the context of changing weather patterns.
The observation that the GPT2 protein is required for dynamic photosynthetic acclimation is, to our knowledge, the first evidence for a specific function of this protein. Previous studies have shown that this gene is induced under conditions of sugar feeding and during sugar-induced senescence (Gonzali et al., 2006; Li et al., 2006; Pourtau et al., 2006). In vitro evidence shows that this gene product is a functional sugar phosphate/phosphate translocator, with activity for triose- and hexose-Ps (Knappe et al., 2003). It might be thought therefore that expression of this protein is necessary to allow high rates of carbon export during photosynthesis at high light; however, the observation that plants that develop at high light are able to achieve the same photosynthetic rate as wild-type plants tends to exclude such a general role. Also, the decline in transcript levels toward the end of the acclimation suggests that its major role is during the acclimation process itself, rather than for supporting steady-state photosynthesis. GPT2 is probably therefore functioning in a signaling role, perhaps affecting the partitioning of sugar phosphates between the chloroplast stroma and the cytosol or shifting the phosphate balance of the cell.
The ability to acclimate varies not only between species (Murchie and Horton, 1997) but also, as shown here, among different accessions of a single species, Arabidopsis. This variation suggests that in addition to the advantages observed here, the ability to acclimate must carry with it some cost. The nature of that cost is not yet clear. It may be that for some species, e.g. fast-growing ruderal species, more efficient acclimation is achieved by developing new leaves. For plants from particularly extreme environments, development of stress-tolerance traits may be more important than optimizing photosynthesis. The observation that the widely used Col accession is unable to acclimate may reflect the history of this particular accession, rather than the source ecotype. Col does express GPT2 at least to some extent (Niewiadomski et al., 2005); however, this does not result in a dynamic acclimation response (Fig. 1A), suggesting that Col is impaired in acclimation downstream of GPT2. This needs to be borne in mind when reviewing work on Col examining acclimation.
Since Col and Ws gpt2− both show a lack of dynamic acclimation and both show reduced fitness under naturally fluctuating conditions, we can conclude that the acclimation process is responsible for optimizing fitness in nature. That this effect is seen both in the gpt2− plants and in Col, where the GPT2 gene is transcribed, implies that it is not expression of the GPT2 protein per se that affects fitness. Across the two winters measured, there was a significant difference in seed production in all the plant types considered and this difference probably reflects the overall weather conditions seen. The 2008 to 2009 winter was markedly colder than 2007 to 2008, with a higher incidence of frost and snow, which may have impacted on plant growth. Nevertheless, the penalty associated with lack of acclimation was clear in both seasons. The lack of acclimation did not significantly affect the vegetative growth of plants and there was no consistent effect on time to flowering, but the resources partitioned into seeds were clearly reduced in the absence of acclimation.
If previously fully developed leaves are to acclimate, they must do so in the context of constraints placed upon them by the anatomy of the leaf (Oguchi et al., 2003, 2005). Where changes in leaf morphology occur, as will be the case during developmental acclimation, the cost of producing thicker high-light leaves needs to be balanced against the benefits of the additional photosynthetic carbon gain (Oguchi et al., 2008). In our experiments, however, dynamic acclimation responses were only occurring at the subcellular level (increased chloroplast number or changing chloroplast content), with there being no evidence of changes in leaf morphology of the mature leaves. In such cases, the metabolic cost of acclimation still needs to be weighed against the benefit gained and this may be related to the longevity of the leaf. Variation in, for example, extent of herbivory or other forms of leaf damage may be crucial in determining whether dynamic acclimation is beneficial. Comparing species or ecotypes, the plant phenology may also be key; for example, plants growing as spring ephemerals, with much shorter growth periods, may benefit less from dynamic acclimation.
Previously, the concept of dynamic acclimation has been poorly recognized and has not been shown to be distinct from other forms of acclimation. The data presented here show not only that it is a separate response from developmental acclimation but that it is a key determinant of plant fitness. The capacity to acclimate has not been systematically examined in crop species; however, the work that is available suggests that it may be limited. In our hands, barley (Hordeum vulgare) does not show dynamic acclimation (G.N. Johnson, unpublished data) and published work on rice has suggested that this response is only partial (Murchie et al., 2005). These observations suggest that an appreciation of acclimation potential and possibly also selection for acclimation traits may be important in developing crops with improved yield that will be required to support growing world populations into the future.
MATERIALS AND METHODS
Plant Material
Seeds were sown on peat-based compost and chilled for 2 to 3 d to promote germination. Plants were then transferred to a growth cabinet (E.J. Stiell) at an irradiance of 100 μmol m−2 s−1 (low light) provided by high-frequency fluorescent lamps on an 8-h d to suppress flowering, daytime temperature 20°C, night 16°C. After 2 weeks, seedlings were pricked out into individual 3-inch pots and grown on for a further 6 weeks. As appropriate, plants were then transferred to a different shelf in the same growth cabinet providing 400 μmol m−2 s−1 light (high light).
For fecundity measurements, plants were either grown in controlled-environment chambers at 100 μmol m−2 s−1 as above or in unheated greenhouses in Manchester, UK, without supplementary lighting during the periods of October 2007 to February 2008 and October 2008 to March 2009. For tests of germination, seeds were placed on plates containing 1% agar maintained at 4°C for 48 h and then transferred to the same growth conditions as above.
Measurements of Photosynthetic Parameters
Plant capacity for CO2 fixation was measured on fully expanded leaves using a CIRAS 1 infrared gas analyzer (PP Systems) with light provided from a Schott KL-1500 lamp. All measurements were performed in an atmosphere of 2,000 μL L−1 CO2. To measure chlorophyll, the same leaves were then ground in 80% acetone and chlorophyll estimated using the method of Porra et al. (1989).
Microarray Analyses
At each sampling point, a single leaf was detached from each of five separate plants in each treatment and flash frozen to produce each biological replicate. Samples were prepared from three separate batches of plants, analyzed on separate microarrays. RNA was extracted using an RNeasy plant mini kit (Qiagen). The biotinylated cDNA produced from the total RNA of the nine samples was synthesized and hybridized to an Arabidopsis (Arabidopsis thaliana) ATH1-121501 (Affymetrix) oligonucleotide array according to the manufacturer's instructions. The arrays were read by means of an Agilent GeneArray scanner 3000 7G using Affymetrix GCOS (GeneChipOperating Software) V1.4. The technical quality control of the arrays was performed to check for outliers using dChip software (Li and Wong, 2001). The normalization and expression analysis was carried out using robust multichip average (Bolstad et al., 2003). Principal components analysis using a covariance dispersion matrix was performed by means of maxdView (http://umber.sbs.man.ac.uk/microarray/maxd/maxdView/index.html). Differential expression was assessed with a modified t test on logarithmically scaled data using Cyber-T (Baldi and Long, 2001). Since there were three conditions, three Cyber-T tests were performed to cover all three two-way comparisons. Gene lists of differentially expressed genes were generated using criteria of Cyber-T P value less than 0.01, mean fold-change greater than 2, and mean expression level greater than 100 in one condition. The gene annotation used was derived from The Arabidopsis Information Resource (http://www.arabidopsis.org/). The Affymetrix chip analysis was performed at the Microarray Facility of the Faculty of Life Sciences in the University of Manchester (Manchester, UK).
Real-Time RT-PCR
Primers used in real-time RT-PCR were designed using Primer Express version 1.0 (Applied Biosystems). For a list of primers, see Supplemental Table S2. The resulting design primers, optimized for SYBR green assays, were BLASTed against genomic databases available at the National Centre for Biotechnology Information to verify the primer's specificity. All primers were purchased from Eurogentec. Real-time RT-PCR was performed with a Eurogentec one-step RT-quantitative PCR MasterMix for SYBR green I kit using an Applied Biosystems ABI Prism 7000 instrument. Data analysis was performed using the software ABI Prism 7000 sequence detection system version 1.7 (Applied Biosystems). Each sample was analyzed in triplicate with at least three biological replicates being analyzed for each data point. Expression levels were measured relative to three housekeeping genes: ACT2 (At3g18780), UBC (At5g25760), and CBP20 (At5g44200).
Isolation of gpt2− Knockout
A T-DNA insertion line (FLAG_326E03) was obtained from the Versailles T-DNA collection (INRA, Versailles, France; Samson et al., 2002). Plants carrying a homozygote insertion in the GPT2 gene were identified using PCR with primers listed in Supplemental Table S2. To verify that GPT2 expression was disrupted in the insertion line, plants were grown for 8 weeks at 100 μmol m−2 s−1 light and then transferred for 4 h to 400 μmol m−2 s−1. Leaves were flash frozen under growth conditions and total RNA extracted as above.
Complementation of gpt2− Knockout
To restore photosynthetic acclimation in response to high light the gpt2− knockout was complemented using the entire GPT2 gene with 1.8 kb of upstream sequence to include native promoter elements and 0.4 kb of downstream terminator sequence. A 4.8-kb fragment was amplified from genomic Ws DNA using primers genomic-GPT2-F1 and genomic-GPT2-R1 (see Supplemental Table S2) with the Phusion PCR system (Finnzyme) optimized using the provided GC-rich buffer, 3% dimethyl sulfoxide, and 1.75 mm MgCl2. From this, a 4.5-kb fragment was amplified using primers attB1-GPT2-natP and attB2-GPT2-natT with Gateway adaptor sequences and first, BP cloned into pDONR201 (Invitrogen), and second, LR cloned into the binary destination vector pHGW,0 (Karimi et al., 2002) according to Invitrogen protocols. The construct was introduced to the gpt2− knockout by the floral-dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101∷pMP90. Transformants were selected for resistance to hygromycin.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Genes with significantly altered expression during acclimation.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We would like to thank Dr. Leo Zeef (University of Manchester) and the University of Manchester Microarray Facility for help in the design and analysis of the microarray experiment, and Dr. Patrick Gallois (University of Manchester) for his assistance with the quantitative PCR.
This work was supported by a studentship from the United Kingdom Natural Environment Research Council (to B.C.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Giles N. Johnson (giles.johnson@manchester.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Horton P, Walters RG (2004) Acclimation of Arabidopsis thaliana to the light environment: the relationship between photosynthetic function and chloroplast composition. Planta 218 793–802 [DOI] [PubMed] [Google Scholar]
- Bailey S, Walters RG, Jansson S, Horton P (2001) Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta 213 794–801 [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17 509–519 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III Sci Vie 316 1194–1199 [Google Scholar]
- Bjorkman O, Holmgren P (1963) Adaptability of photosynthetic apparatus to light intensity in ecotypes from exposed and shaded habitats. Annu Rev Plant Physiol Plant Mol Biol 28 355–377 [Google Scholar]
- Boardman NK (1977) Comparative photosynthesis of sun and shade plants. Annu Rev Plant Physiol 28 355–377 [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 17 185–193 [DOI] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M (1993) A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C R Acad Sci III Sci Vie 316 1188–1193 [Google Scholar]
- Chow WS, Melis A, Anderson JM (1990) Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA 87 7502–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WWI (1992) Photoprotection and other responses of plants to light stress. Annu Rev Plant Physiol Plant Mol Biol 43 599–626 [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P (2006) Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J Plant Res 119 115–123 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Knappe S, Flugge UI, Fischer K (2003) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Horton P (1997) Acclimation of photosynthesis to irradiance and spectral quality in British plant species: chlorophyll content, photosynthetic capacity and habitat preference. Plant Cell Environ 20 438–448 [Google Scholar]
- Murchie EH, Hubbart S, Peng S, Horton P (2005) Acclimation of photosynthesis to high irradiance in rice: gene expression and interactions with leaf development. J Exp Bot 56 449–460 [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flugge UI, Schneider A (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi R, Hikosaka K, Hirose T (2003) Does the photosynthetic light-acclimation need change in leaf anatomy? Plant Cell Environ 26 505–512 [Google Scholar]
- Oguchi R, Hikosaka K, Hirose T (2005) Leaf anatomy as a constraint for photosynthetic acclimation: differential responses in leaf anatomy to increasing growth irradiance among three deciduous trees. Plant Cell Environ 28 916–927 [Google Scholar]
- Oguchi R, Hikosaka K, Hiura T, Hirose T (2008) Costs and benefits of photosynthetic light acclimation by tree seedlings in response to gap formation. Oecologia 155 665–675 [DOI] [PubMed] [Google Scholar]
- Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta UM, Battchikova N, Aro EM (2006) Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol Genomics 25 142–152 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-A and chlorophyll-B extracted with 4 different solvents—verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A (2006) Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224 556–568 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG (2005) Towards an understanding of photosynthetic acclimation. J Exp Bot 56 435–447 [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P (1995. a) Acclimation of Arabidopsis thaliana to the light environment—changes in photosynthetic function. Planta 197 306–312 [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P (1995. b) Acclimation of Arabidopsis thaliana to the light environment—regulation of chloroplast composition. Planta 197 475–481 [DOI] [PubMed] [Google Scholar]
- Yin ZH, Johnson GN (2000) Photosynthetic acclimation of higher plants to growth in fluctuating light environments. Photosynth Res 63 97–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.