Abstract
The cellular functions of Ku70 in repair of DNA double-stranded breaks and telomere regulation have been described in a wide range of organisms. In this study, we identified the rice (Oryza sativa) Ku70 homolog (OsKu70) from the rice genome database. OsKu70 transcript was detected constitutively in every tissue and developmental stage examined and also in undifferentiated callus cells in rice. Yeast two-hybrid and in vitro pull-down experiments revealed that OsKu70 physically interacts with OsKu80. We obtained loss-of-function osku70 T-DNA knockout mutant lines and constructed transgenic rice plants that overexpress the OsKu70 gene in the sense (35S:OsKu70) or antisense (35S:anti-OsKu70) orientation. The homozygous G2 osku70 mutant lines were more sensitive than wild-type plants to a DNA-damaging agent (0.01%–0.05% methyl-methane sulfonate), consistent with the notion that OsKu70 participates in the DNA repair mechanism. Terminal restriction fragment analysis revealed that telomeres in homozygous G2 osku70 mutants were markedly longer (10–20 kb) than those in wild-type plants (5–10 kb), whereas telomere length in heterozygous G2 osku70 mutant and T2 OsKu70-overexpressing transgenic (35S:OsKu70) rice resembled that of the wild-type plant. In contrast to what was observed in Arabidopsis (Arabidopsis thaliana) atku70 mutants, homozygous G2 osku70 rice plants displayed severe developmental defects in both vegetative and reproductive organs under normal growth conditions, resulting in sterile flowers. Analysis of meiotic progression in pollen mother cells demonstrated that up to 11.1% (seven of 63) of G2 mutant anaphase cells displayed one or more chromosomal fusions. These results suggest that OsKu70 is required for the maintenance of chromosome stability and normal developmental growth in rice plants.
Higher plants are continuously exposed to various environmental stresses, including ionizing radiation and genotoxic agents. These stresses cause DNA damage, including double-strand breaks (DSBs). The repair of DSBs is essential for maintaining the integrity and function of the genome. The major DSB repair mechanism in prokaryotes is homologous recombination, whereas nonhomologous end joining (NHEJ) is predominant over homologous recombination in higher eukaryotes during G1 phase (Lieber and Karanjawala, 2004). The NHEJ pathway is modulated by the DNA-dependent protein kinase (DNA-PK) complex, which consists of a Ku70/Ku80 heterodimer and a catalytic subunit (DNA-PKcs), and the DNA ligase IV (Lig4)/Xrcc4 (Lif1) complex in vertebrates (Critchlow and Jackson, 1998). The first step in the NHEJ pathway is recognition of DSB termini and binding of exposed DNA ends by Ku70/Ku80 heterodimers, which recruit DNA-PKcs and activate its kinase activity (Dynan and Yoo, 1998). The Ku and DNA-PKcs complexes are localized exclusively to the nucleus and have strong affinity for double-stranded DNA ends, independent of DNA sequence and structure (Koike et al., 1998; Wang et al., 1998). In addition, they participate in RNA polymerase I- and II-mediated transcription, in DNA replication, and in regulation of cell cycle progression (Tuteja and Tuteja, 2000). In yeast cells, a Ku70/Ku80 heterodimer is required not only for DNA repair but also for the maintenance of telomere length. The Ku complex binds to telomeric repeats in vivo, and Ku70-deficient cells exhibit shortened telomeres (Boulton and Jackson, 1998; Baumann and Cech, 2000). In human, Ku interacts with hTERT and the telomerase RNA subunit, perhaps to regulate access of telomerase to telomeric DNA ends (Chai et al., 2002; Stellwagen et al., 2003). Ku70 appears to prevent end-to-end chromosome fusion and to bind the telomere repeat-binding protein TRF2 (Hsu et al., 2000; Song et al., 2000). It was recently demonstrated that Ku86 is essential for human telomere integrity and that its inactivation leads to cell death along with massive loss of telomeric DNA via recombination in human somatic cells (Wang et al., 2009).
Although the DNA repair mechanisms in higher plants are less well understood than those in yeast or mammals, components involved in plant DSB repair have been identified and some aspects of their mechanisms have been worked out (Gorbunova and Levy, 1999; Gallego and White, 2005; Riha et al., 2006). For instance, Arabidopsis (Arabidopsis thaliana) AtKu70/AtKu80 heterodimers possess single-stranded DNA-dependent ATPase and ATP-dependent DNA helicase activities in vitro (Tamura et al., 2002). Moreover, the AtKu70 and AtKu80 genes are significantly up-regulated by DNA-damaging drugs in cultured Arabidopsis cells. These findings raise the possibility that AtKu70 and AtKu80 are involved in DSB repair through the NHEJ system (Tamura et al., 2002). Indeed, although AtKu70 T-DNA knockout plants appeared normal under standard growth conditions, their seedlings were highly sensitive to γ-irradiation that induced DSBs (Bundock et al., 2002; Riha et al., 2002). Similarly, disruption of AtKu80 resulted in hypersensitivity to genotoxic agents (West et al., 2002; Gallego et al., 2003a), consistent with the notion that AtKu70/AtKu80 proteins play a role in DSB repair in Arabidopsis. In contrast to what was observed in yeast and some mammalian cells, mutations in AtKu70 and AtKu80 resulted in longer telomeres in Arabidopsis, suggesting their role in telomere regulation as negative factors (Bundock et al., 2002; Riha et al., 2002; Gallego et al., 2003b; Riha and Shippen, 2003; Watson and Shippen, 2007). Interestingly, in Arabidopsis, critically shortened telomeres (<300–400 bp) are treated as DSBs, and the NHEJ pathway functions using both Ku70-dependent and Ku70-independent mechanisms. This indicates that there are multiple systems for chromosome end joining (Riha and Shippen, 2003; Heacock et al., 2004). Most recently, it was reported that AtKu70/AtKu80 heterodimers inhibited the formation of extrachromosomal telomeric circles and alternative telomere lengthening (Zellinger et al., 2007), suggesting that Ku contributes to genome stability by suppressing aberrant homologous recombination.
In this report, we identify a rice (Oryza sativa) homolog (OsKu70) of Arabidopsis Ku70 and describe osku70 T-DNA insertional knockout mutant rice plants. Terminal restriction fragment analysis revealed that homozygous G2 osku70 mutant telomeres are markedly longer (10–20 kb) than wild-type telomeres (5–10 kb). In contrast to Arabidopsis atku70 knockout mutants, homozygous G2 osku70 mutants displayed severe developmental defects in both vegetative and reproductive organs, resulting in sterile flowers. Analysis of meiotic progression in pollen mother cells (PMCs) showed that up to 11.1% (seven of 63) of G2 mutant cells in anaphase displayed one or more chromosomal fusions. These results suggest that OsKu70 is required for the maintenance of chromosomal stability and normal development in rice.
RESULTS
Identification of the Ku70 Homolog in Rice
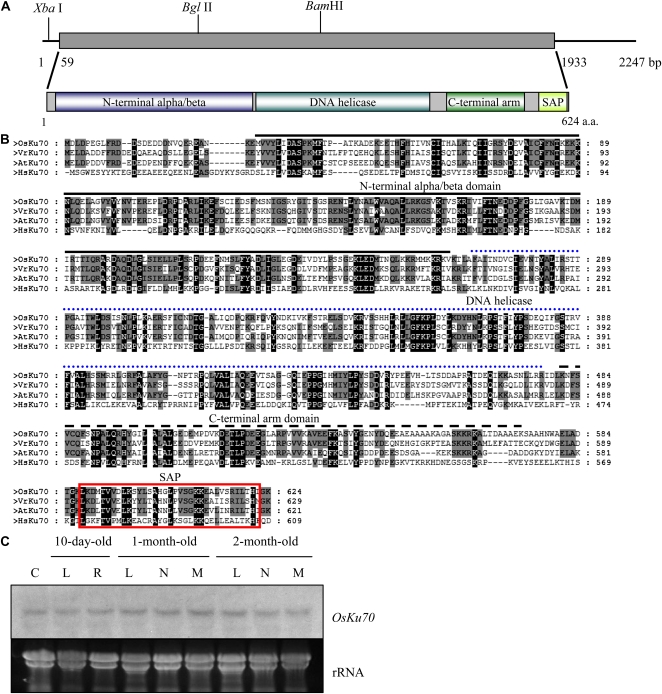
We searched the rice genome database (http://signal.salk.edu/cgi-bin/RiceGE) for sequences similar to that of Arabidopsis AtKu70 (Bundock et al., 2002; Riha et al., 2002; Tamura et al., 2002) and identified the homolog rice Ku70 gene (OsKu70; GenBank accession no. BAC83189). To gain insight into the function of OsKu70, we isolated full-length OsKu70 cDNA by extracting total RNA from leaf tissue of 3-month-old mature rice plants and performing reverse transcription (RT)-PCR using gene-specific primers. The coding region of OsKu70 cDNA is 1,872 bp long and encodes a 624-amino acid protein with a predicted molecular mass of 70.1 kD, similar to human (69.8 kD) and Arabidopsis (70.3 kD) Ku70 proteins (Fig. 1A). As in other Ku70 homologs, the OsKu70 protein contains an N-terminal α/β-domain, a DNA helicase (Ku-type) motif, a C-terminal arm region, and a SAP DNA-binding domain at the extreme C terminus. Multiple alignments of the deduced amino acid sequence of OsKu70 with those of previously characterized Ku70 proteins are shown in Figure 1B. OsKu70 is 62% and 61% identical to the mung bean (Vigna radiata) and Arabidopsis proteins, respectively (Tamura et al., 2002; Liu et al., 2007), while it shares a relatively low degree of sequence identity (29%) with the human protein (Chan et al., 1989).
Figure 1.
Sequence analysis of rice OsKu70. A, Schematic structure of OsKu70 cDNA and its predicted protein. The solid bar represents the coding region, while the solid line indicates 5′ and 3′ untranslated regions. The N-terminal α/β-domain, DNA helicase motif, C-terminal arm domain, and SAP DNA-binding domain are indicated. a.a., Amino acids. B, Comparison of the deduced amino acid sequences of Ku70 homologs from rice, mung bean, Arabidopsis, and human. Amino acid residues conserved in at least three of the four sequences are shaded. Amino acids identical in all four proteins are highlighted in black. The N-terminal α/β-domain, DNA helicase motif, C-terminal arm domain, and SAP DNA-binding domain are indicated by a solid line, dotted line, broken line, and box, respectively. C, RNA gel-blot analysis of OsKu70 mRNA in wild-type rice plants. Total RNA (40 μg per lane) was obtained from various organs at different developmental stages as indicated, separated on an agarose gel, and blotted onto a nylon membrane. The membrane was hybridized to a 32P-labeled OsKu70 cDNA probe under high-stringency conditions. rRNA is shown as a loading control. C, Callus; L, leaf; R, root; N, internode; M, meristem. [See online article for color version of this figure.]
To investigate the spatial and temporal expression patterns of OsKu70, we examined OsKu70 mRNA levels in various rice tissues at different developmental stages by RNA gel-blot analysis. Total RNAs isolated from leaves, roots, internodes, and meristems of 10-d-old, 1-month-old, and 2-month-old wild-type plants were hybridized with 32P-labeled cDNA probe under high-stringency conditions. A steady-state level of OsKu70 transcript was detected in every tissue examined at all developmental stages and was also present in undifferentiated callus cells (Fig. 1C).
OsKu70 Interacts with OsKu80 in Yeast Cells and in Vitro
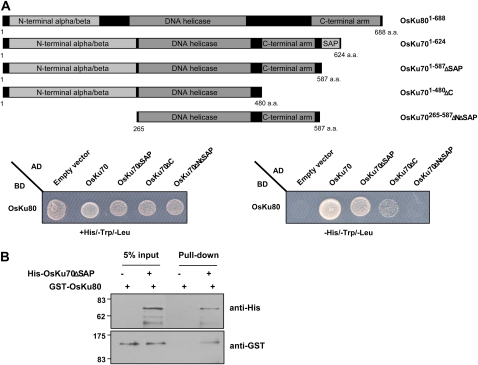
Ku70 is typified by interaction with Ku80 to form a heterodimer. It was shown that Arabidopsis AtKu70 and AtKu80 bind to each other (Riha et al., 2002; Tamura et al., 2002; West et al., 2002). Thus, we examined the interaction between OsKu70 and OsKu80 proteins by yeast two-hybrid and in vitro pull-down assays. For the yeast two-hybrid analysis, full-length or deletion mutant OsKu70 cDNA was cloned into a plasmid (pGADT7) carrying the activation domain of the GAL4 gene, while full-length OsKu80 cDNA (GenBank accession no. AAS01977) was cloned into a plasmid (pGBKT7) containing the DNA-binding domain of GAL4. The resulting constructs were cotransformed into the yeast strain AH109, which cannot grow in minimal medium lacking the amino acid His. As shown in Figure 2A, the cells were able to grow in the absence of His when full-length OsKu701–624 was cotransformed with OsKu80. The intensity of the interaction decreased as the C-terminal region of OsKu70 was serially deleted (OsKu701–587-ΔSAP and OsKu701–480-ΔC). OsKu70265−587-ΔNΔSAP, in which the N-terminal α/β-domain and the C-terminal SAP region were deleted, failed to interact with OsKu80, suggesting that the N-terminal α/β-domain is necessary for binding of OsKu70 to OsKu80 (Fig. 2A).
Figure 2.
Interactions between OsKu70 and OsKu80. A, Yeast two-hybrid assay. Full-length OsKu70 and its deletion mutants (OsKu701–587-ΔSAP, OsKu701–480-ΔC, and OsKu70265–587-ΔNΔSAP) were cloned into pGADT7, while full-length OsKu80 was cloned into pGBKT7. Combinations of the indicated plasmids were cotransformed into yeast AH109 cells. Transformed yeast cells were plated onto SD/+His/−Trp/−Leu or SD/−His/−Trp/−Leu medium containing 10 mm 3-aminotriazole and grown for 4 d at 28°C. The capacity of the yeast cells to grow in the absence of His was used as a readout for the interaction between OsKu70 and OsKu80. a.a., Amino acids; AD, activation domain; BD, binding domain. B, In vitro pull-down assay. The bacterially expressed (His)6-OsKu701–587-ΔSAP fusion protein was incubated with GST-OsKu80 in the presence of Ni-NTA resin. After extensive washing with immunoprecipitation buffer, the bound proteins were eluted from the affinity resin by boiling with SDS sample buffer. The eluted proteins were resolved by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and subsequently immunoblotted with anti-His and anti-GST antibodies. [See online article for color version of this figure.]
To confirm the interaction between OsKu70 and OsKu80, we next carried out an in vitro pull-down assay. Because full-length OsKu70 was insoluble and precipitated in Escherichia coli cells, we used the OsKu701–587-ΔSAP deletion mutant, which is still able to interact with OsKu80 in yeast cells. As shown in Figure 2B, GST-OsKu80 protein was effectively pulled down from the nickel-nitrilotriacetic acid agarose (Ni-NTA) resin in the presence of (His)6-OsKu701–587-ΔSAP but not by GST-OsKu80 alone. Although the stoichiometry of the OsKu70-OsKu80 interaction is not known, these results indicate that the two proteins bind to one another.
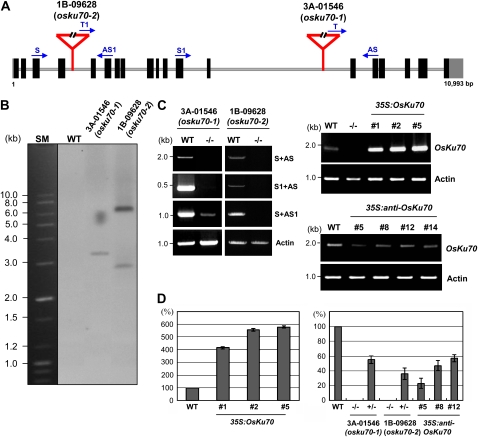
Identification of an OsKu70 T-DNA Insertional Mutant and Construction of 35S:OsKu70 and 35S:anti-OsKu70 Transgenic Rice Plants
To define the physiological roles of OsKu70 in rice plants, we obtained two independent mutant lines containing a T-DNA insertion in the OsKu70 gene from the Rice T-DNA Insertion Sequence Database (http://www.postech.ac.kr/life/pfg/risd). The T-DNA insertions were mapped to the fourth (1B-09268) and 13th (3A-01546) introns in the OsKu70 gene, which is located on chromosome 7 (Fig. 3A). Homozygous mutant plants were identified using multiplex PCR with specific primers: S, S1, AS, AS1, T, and T1. Genomic Southern-blot analysis using the GUS gene as a probe revealed that the 1B-09628 mutant contained two copies of T-DNA, while the 3A-01546 mutant contained one copy (Fig. 3B). Thus, the 3A-01546 and 1B-09268 mutant lines were designated as osku70-1 and osku70-2, respectively. As shown in Figure 3, C and D, full-length OsKu70 transcript is not detectable in both osku70-1 and osku70-2 mutant seedlings. In the osku70-1 line, a low level of partial OsKu70 mRNA transcript was detected with a primer set that amplified a region upstream of the insertion site in OsKu70, indicating that although there are some truncated transcripts still present, osku70-1 lacks full-length, functional OsKu70 mRNA. In contrast, there was no partial OsKu70 mRNA in the osku70-2 mutant. This suggests that osku70-2 is null for the OsKU70 gene (Fig. 3, C and D).
Figure 3.
Molecular characterization of the osku70 T-DNA knockout mutant lines and 35S:OsKu70 and 35S:anti-OsKu70 transgenic rice plants. A, Schematic representation of the osku70 alleles with the positions of the inserted T-DNAs. The triangles represent the integrated T-DNAs. Black bars show coding regions, whereas gray bars represent the 5′ and 3′ untranslated regions. Solid lines indicate introns. Gene-specific (S, S1, AS, and AS1) and T-DNA-specific (T and T1) primers that were used in genotyping and RT-PCR are depicted with arrows. B, DNA gel-blot analysis in rice genomic DNA. Total genomic DNA was isolated from the leaf tissue of wild-type (WT) and 1B-09628 and 3A-01546 mutant plants. Isolated DNA (10 μg per lane) was digested with EcoRI, blotted onto nylon membranes, and hybridized with the 32P-labeled GUS cDNA probe under high-stringency conditions. SM, Size marker. C, RT-PCR analysis of OsKu70 transcripts in wild-type, homozygous G2 osku70 mutant (3A-01546 and 1B-09628), and T2 35S:OsKu70 (1, 2, and 5) and 35S:anti-OsKu70 (5, 8, 12, and 14) transgenic rice plants. The constitutively expressed Actin gene was used as a loading control. −/− indicates homozygous mutant plants. D, Quantitative real-time RT-PCR. Expression of the OsKu70 gene was examined by real-time RT-PCR in wild-type, 35S:OsKu70, 35S:anti-OsKu70, and osku70 mutant (3A-01546 and 1B-09628) plants as indicated. The level of OsKu70 mRNA was normalized to that of the rice Actin gene. −/− indicates homozygous mutant plants; +/− indicates heterozygous plants. Results are means ± sd from three independent experiments. [See online article for color version of this figure.]
We also constructed transgenic rice plants that express OsKu70 in the sense (35S:OsKu70) and antisense (35S:anti-OsKu70) orientations under the control of the cauliflower mosaic virus 35S promoter. A quantitative real-time RT-PCR analysis verified that 35S:OsKu70 plants (lines 1, 2, and 5) contained markedly enhanced levels (4.1- to 5.8-fold increase) of mRNA, while 35S:anti-OsKu70 plants (lines 5, 8, 12, and 14) had significantly reduced levels (42%–73% decrease) of transcript (Fig. 3, C and D).
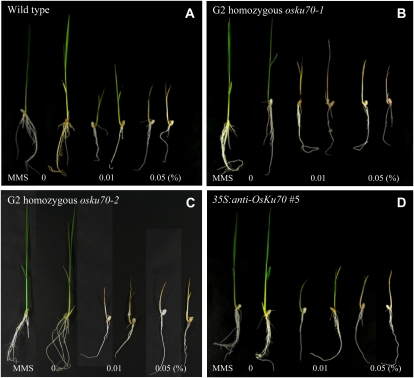
Sensitivity of the osku70 Mutant Lines and 35S:OsKu70 and 35S:anti-OsKu70 Transgenic Rice Plants to DNA-Damaging Agents
The Arabidopsis atku70 and atku80 mutants are hypersensitive to genotoxic agents, such as methyl-methane sulfonate (MMS), x-rays, menadione, and bleomycin (Bundock et al., 2002; Riha et al., 2002; West et al., 2002). To investigate the sensitivity of rice plants to DNA-damaging drugs, light-reared seedlings were transferred to growth medium with different concentrations (0%–0.05%) of MMS and then allowed to grow further for 10 d. Five-day-old wild-type seedlings exhibited significantly retarded growth in the presence of 0.01% MMS and stopped growing and died in medium containing 0.05% MMS (Fig. 4A). Because homozygous G2 osku70 mutant lines grow much more slowly than wild-type plants (see Fig. 6 below), 10-d-old mutant seedlings were used for the MMS-sensitivity assay. As shown in Figure 4, B and C, both G2 homozygous osku70-1 and osku70-2 mutant seedlings were completely bleached and their growth was delayed in the presence of 0.01% MMS for 10 d. These data indicate that homozygous osku70 mutants are more susceptible to DNA-damaging agent than are wild-type plants, consistent with a role for OsKu70 in DNA repair in rice. On the other hand, the sensitivity of 5-d-old T2 35S:anti-OsKu70 transgenic seedlings to 0.01% to 0.05% MMS was indistinguishable from that of wild-type plants (Fig. 4D). We also used hydrogen peroxide, mitomycin C, and zeocin for sensitivity assays, but at given concentrations of these agents, we did not observe different sensitivity between wild-type and mutant plants (Supplemental Fig. S1), suggesting that these agents are not good to observe the differences between the wild-type and mutant rice plants.
Figure 4.
Sensitivity of wild-type (A), homozygous G2 osku70-1 mutant (B), homozygous G2 osku70-2 mutant (C), and T2 35S:anti-OsKu70 transgenic (D) rice plants to MMS. Light-grown 5-d-old (A and D) or 10-d-old (B and C) seedlings were transferred to growth medium containing different concentrations (0%–0.05%) of MMS and then allowed to grow for another 10 d. [See online article for color version of this figure.]
Figure 6.
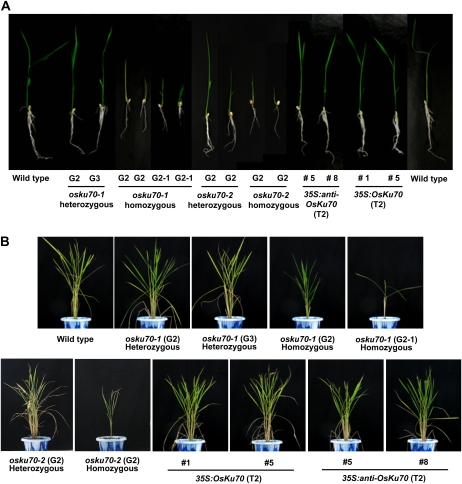
Phenotypic comparisons of vegetative organs between wild-type, osku70 mutant, and OsKu70 transgenic plants. A, Growth retardation of osku70 homozygous mutant seedlings. Wild-type, heterozygous (G2 and G3) and homozygous (G2 and G2-1) osku70-1 and osku70-2 mutant, and 35S:anti-OsKu70 (T2; lines 5 and 8) and 35S:OsKu70 (T2; lines 1 and 5) transgenic seedlings were germinated and grown in half-strength Murashige and Skoog medium for 10 d under normal-light growth conditions. B, Gross morphology of 3-month-old wild-type, heterozygous (G2 and G3) and homozygous (G2 and G2-1) osku70-1 and osku70-2 mutant, and 35S:OsKu70 (T2; lines 1 and 5) and 35S:anti-OsKu70 (T2; lines 5 and 8) transgenic rice plants, which were grown under greenhouse conditions. [See online article for color version of this figure.]
Lengthened Telomeres in osku70 Homozygous Mutant Rice
To examine whether OsKu70 plays a functional role in telomere regulation in rice, we measured telomere length in wild-type, osku70 heterozygous and homozygous mutants, and 35S:OsKu70 and 35S:anti-OsKu70 transgenic plants. Terminal restriction fragments were generated by digestion of leaf genomic DNA with the restriction enzyme TaqI and analyzed by Southern hybridization with a telomere repeat probe (TTTAGGG)70 (Fig. 5). Telomere length in wild-type and heterozygous osku70 plants ranged from about 5 to 10 kb (Fig. 5A), in agreement with the previously identified telomere length in rice plants (Hong et al., 2007). However, telomeres in homozygous osku70-1 and osku70-2 plants (G1 and G2) were markedly longer (approximately 10–20 kb) than those in wild-type plants (Fig. 5). These telomere elongation patterns are consistent with those observed in Arabidopsis atku70 mutants (Bundock et al., 2002; Riha et al., 2002; Riha and Shippen, 2003). On the other hand, independent T2 35S:anti-OsKu70 transgenic lines had varied telomere lengths. In line 5, which had reduced OsKu70 mRNA levels (Fig. 3C), telomere length was intermediate between that of wild-type and osku70-1 homozygous plants (Fig. 5B). Other antisense lines (8, 12, and 14) with only slightly decreased OsKu70 transcript levels had normal-length telomeres (Fig. 5B). This suggests that telomere length is dependent on the level of OsKu70 transcript. However, telomere length in most 35S:OsKu70 plants (lines 1, 2, and 5), which overexpress OsKu70, was similar to that of wild-type plants (Fig. 5B). Since overexpression of OsKu70 has undetectable effects on telomere length, it is likely that the level of OsKu70 in wild-type rice plants is at saturation. Taken together, our data indicate that OsKu70 acts as a negative factor in telomere homeostasis. This contrasts with the role of Ku70 in yeast but is consistent with results obtained in Arabidopsis. Thus, Ku70 homologs in monocot (rice) and dicot (Arabidopsis) plants may have similar roles in telomere length regulation.
Figure 5.
Terminal restriction fragment analysis in wild-type (WT), osku70 mutant, and 35S:OsKu70 and 35S:anti-OsKu70 transgenic rice plants by pulse-field gel electrophoresis. A, Telomere length in wild-type rice and heterozygous (G1) and homozygous (G1) osku70-1 and osku70-2 mutant lines. Telomere length of rtbp1 plants is shown as a positive control. +/− indicates heterozygous mutants; −/− indicates homozygous mutants. B, Telomere length in wild-type, heterozygous (G2) and homozygous (G2 and G2-1) osku70-1 mutant, and 35S:anti-OsKu70 (T2; lines 5, 8, 12, and 14) and 35S:OsKu70 (T2; lines 1, 2, and 5) transgenic rice plants. The experiments were conducted four times independently using randomly chosen individual rice plants.
OsKu70 Is Required for Normal Development of Vegetative Organs in Rice
To examine the physiological outcome of loss of OsKu70 function (Fig. 3) and of alterations in telomere length in rice (Fig. 5), we monitored the phenotypic differences between wild-type, 35S:anti-OsKu70 and 35S:OsKu70 transgenic, and osku70 knockout plants. We first investigated the pattern of vegetative growth. As shown in Figure 6A, 10-d-old light-grown heterozygous G2 and G3 osku70, T2 35S:anti-OsKu70, and T2 35S:OsKu70 seedlings had normal growth. In contrast, homozygous G2 osku70-1 and osku70-2 mutants displayed severely retarded root and shoot growth. Consequently, the height and number of leaves of 3-month-old homozygous G2 plants were reduced to approximately 50% to 70% of the wild-type level under greenhouse growth conditions (Fig. 6B). Because homozygous G2 mutant rice failed to produce seeds (see below), we obtained subsequent homozygous progeny by crossing heterozygous G2 osku70-1 mutants and referred to them as the G2-1 generation. Figure 6 shows that homozygous G2-1 osku70-1 mutants are even more severely growth retarded and are unable to produce healthy leaves and shoots. These results suggest that OsKu70 is required for postgermination growth of vegetative organs in rice plants.
osku70 Mutants Display Impaired Development of Reproductive Organs
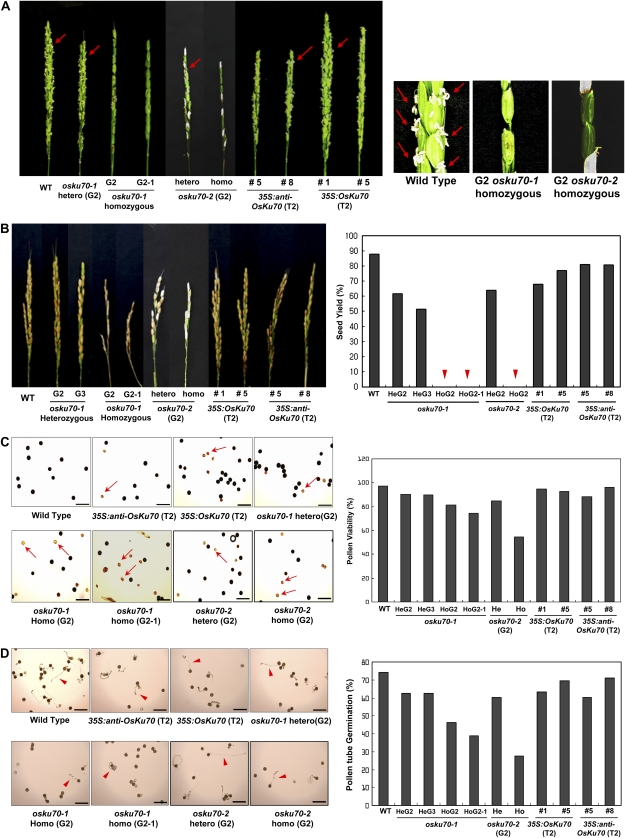
We next examined whether OsKu70 is involved in the development of the reproductive organs. As shown in Figure 7A, the panicles of wild-type, T2 35S:OsKu70 and T2 35S:anti-OsKu70 transgenic, and heterozygous G2 osku70 plants developed normally and contained numerous flowers and pollen grains. In contrast, the panicles of homozygous G2 osku70 mutants appeared to contain fewer flowers. In addition, we rarely observed pollen that had moved out from the G2 osku70-1 and osku70-2 flowers, indicating that elongation of the filaments was suppressed (Fig. 7A). We then measured the seed yield of flowers. Under our experimental conditions, the seed yields of wild-type, T2 35S:OsKu70, T2 35S:anti-OsKu70, and G2 osku70 heterozygous flowers were approximately 90%, 70%, 80%, and 60%, respectively. In contrast, G2 and G2-1 osku70-1 and G2 osku70-2 homozygous flowers were unable to produce seeds (Fig. 7B).
Figure 7.
Phenotypic analysis of floral organs in osku70 mutant rice plants. A, Structure of panicles in wild-type (WT), heterozygous (G2) and homozygous (G2 and G2-1) osku70-1 and osku70-2 mutant, and 35S:OsKu70 (T2; lines 1 and 5) and 35S:anti-OsKu70 (T2; lines 5 and 8) transgenic plants. Arrows indicate pollen that moved out from the flowers. B, Seed yield of wild-type, heterozygous (G2 and G3) and homozygous (G2 and G2-1) osku70-1 and osku70-2 mutant, and 35S:OsKu70 (T2; lines 1 and 5) and 35S:anti-OsKu70 (T2; lines 5 and 8) transgenic plants. He, Heterozygous; Ho, homozygous. C, Pollen viability. Pollen grains in wild-type, heterozygous (G2) and homozygous (G2 and G2-1) osku70-1 and osku70-2 mutant, and 35S:OsKu70 (T2) and 35S:anti-OsKu70 (T2) transgenic plants were stained with I2-KI solution. Pollen grains with black staining were considered to be viable, and those with yellow staining were considered to be infertile. Arrows indicate infertile pollen grains. Bars = 100 μm. D, In vitro germination assay of pollen grains in wild-type, osku70-1 and osku70-2 mutant, and OsKu70 transgenic plants. Arrowheads show elongating pollen tubes. Bars = 100 μm. [See online article for color version of this figure.]
To determine whether the infertility of osku70 homozygous plants resulted from a developmental defect of pollen in the anther, we examined the pollen viability and performed an in vitro pollen germination assay. Pollen grains stained black by I2-KI solution were considered viable, and those stained yellow were considered sterile (Chhun et al., 2007). Pollen viability in wild-type, heterozygous osku70 mutant (G2 and G3), and T2 35S:OsKu70 and T2 35S:anti-OsKu70 transgenic plants was approximately 90% to 95% (Fig. 7C). On the other hand, 20%, 26%, and 47% of pollen grains from homozygous G2 osku70-1, G2-1 osku70-1, and G2 osku70-2 flowers, respectively, were sterile (Fig. 7C). Moreover, under conditions optimized for pollen tube germination and elongation (Han et al., 2006), 60% to 75% of the wild-type, transgenic, and heterozygous mutant grains germinated in vitro (Fig. 7D). However, only about 43%, 37%, and 28% of pollen grains from homozygous G2 and G2-1 osku70-1 and G2 osku70-2 mutant plants were able to germinate (Fig. 7D). These results indicate that pollen from the osku70 mutant is severely impaired.
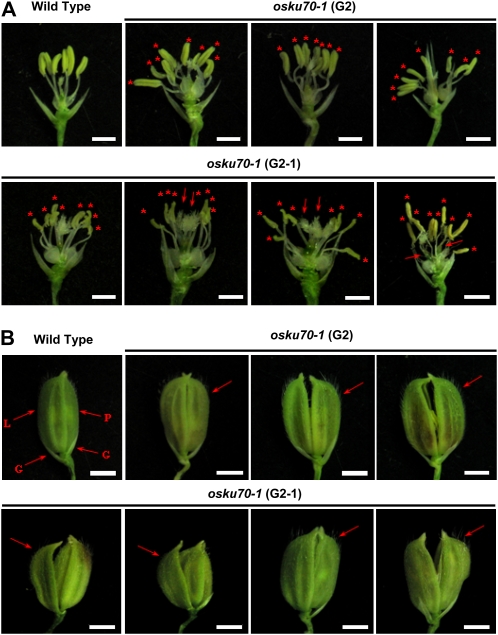
We also compared the morphology of the floral organs between wild-type and osku70-1 mutant plants. The flowers of wild-type rice are characterized by one pair of glumes, one lemma, one palea, and six stamens (Jeon et al., 2000; Hong et al., 2007). However, the flowers of the osku70-1 mutant exhibited an aberrant number of stamens. Figure 8 and Table I show that 0% (none of 370), 0.6% (three of 487), and 3.2% (18 of 561) of the flowers from wild-type and G2 and G3 heterozygous osku70-1 flowers, respectively, contained abnormal stamen numbers. In contrast, the percentages of flowers with abnormal stamen numbers in G2 and G2-1 homozygous osku70-1 mutant flowers were 7.6% (16 of 210) and 14.3% (17 of 119), respectively (Fig. 8A; Table I). Furthermore, impaired development of the palea and lemma was often detected in the G2-1 mutant, resulting in abnormal spikelet structure (Fig. 8B). These overall defects in reproductive organs may explain the infertility of G2 and G2-1 osku70-1 mutant plants. On the other hand, we could not examine the detailed phenotypes of G2 osku70-2 mutant floral organs because we failed to obtain statistically enough flowers due to the severe phenotypic defects in G2 osku70-2 mutant flowers. Taken together, these results indicate that osku70 mutants displayed impaired development not only of vegetative organs but also of reproductive organs.
Figure 8.
Aberrant structure of floral organs in osku70-1 mutant lines. A, Close-up inspection of the stamens of wild-type and G2 and G2-1 osku70-1 flowers. Wild-type filaments contain six stamens, while mutant filaments contain abnormal numbers of stamens. Asterisks depict multiple anthers in one filament; arrows indicate stigmas. Bars = 0.5 cm. B, A spikelet from wild-type and G2 and G2-1 osku70-1 lines. G, Glume; L, lemma; P, palea. Bars = 0.5 cm. [See online article for color version of this figure.]
Table I.
Frequency of abnormal stamen and anaphase bridges
ND, Not determined.
| Genotype | Abnormal Stamen | Anaphase Bridge |
|---|---|---|
| Wild type | 0/370 (0%) | 0/97 (0%) |
| Heterozygous G2 osku70-1 | 3/487 (0.6%) | 1/82 (1.2%) |
| Heterozygous G3 osku70-1 | 18/561 (3.2%) | 4/86 (4.7%) |
| Homozygous G2 osku70-1 | 16/210 (7.6%) | 7/63 (11.1%) |
| Homozygous G2-1 osku70-1 |
17/119 (14.3%) |
ND |
osku70 Mutants Exhibit Abnormal Anaphase Bridges during Meiosis in PMCs
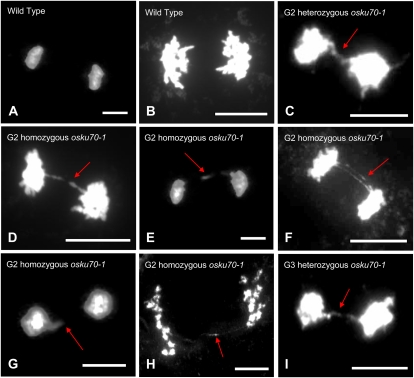
To test whether impaired growth of the osku70 mutant line is associated with telomeric and chromosomal instability in rice, we examined wild-type and mutant chromosomes by 4′,6-diamino-phenylindole (DAPI) staining during meiotic progression of PMCs. As shown in Figure 9, Table I, and Supplemental Figure S2, the percentage of abnormal anaphase bridges was 0% (none of 97), 1.2% (one of 82), and 4.7% (four of 86) in wild-type and heterozygous G2 and G3 osku70-1 PMCs, respectively. However, in homozygous G2 osku70-1 mutant PMCs, the frequency of abnormal anaphase bridges was 11.1% (seven of 63), indicating that loss-of-function osku70 mutants suffer from severe chromosomal instability. We were unable to examine the frequency of abnormal anaphase bridges in PMCs of homozygous G2-1 osku70-1 and G2 osku70-2 lines, because it was extremely difficult to obtain intact PMCs. Collectively, our data suggest that OsKu70 is necessary not only for developmental growth but also for chromosomal stability in rice plants.
Figure 9.
Cytogenetic analysis of PMCs from wild-type (A and B) and heterozygous (G2 and G3; C and I) and homozygous (G2; D–H) osku70-1 mutant rice plants. Anaphase chromosomes were stained by DAPI. Arrows show abnormal anaphase bridges. No anaphase bridges were detected in wild-type PMCs. Bars = 10 μm. [See online article for color version of this figure.]
DISCUSSION
Several previous studies have demonstrated that Ku70 plays important roles in the repair of DNA DSBs and telomere length regulation in Arabidopsis, a dicot model plant (Bundock et al., 2002; Riha et al., 2002; Tamura et al., 2002; Riha and Shippen, 2003; Zellinger et al., 2007). However, nothing is known about its function or regulation in monocots. In this report, we identified the rice Ku70 homolog, OsKu70 (Fig. 1, A and B). Its mRNA was constitutively expressed in all tissues and developmental stages examined (Fig. 1C). As in yeast and Arabidopsis Ku70, OsKu70 contains several distinct motifs, including an N-terminal α/β-domain, a DNA helicase (Ku-type) motif, a C-terminal arm region, and a SAP DNA-binding domain at the extreme C terminus. In addition, OsKu70 binds to OsKu80 in yeast cells and in vitro (Fig. 2), suggesting that they form a heterodimer. These structural conservations suggest that OsKu70 is indeed a homolog of yeast and Arabidopsis Ku70.
We next investigated the effects of knockout, knockdown, and overexpression of OsKu70 (Fig. 3) on developmental growth and genome stability in rice plants. We found that G2 homozygous osku70 mutant alleles were more sensitive to MMS compared with wild-type plants at the postgermination stage (Fig. 4). MMS is known to induce the production of 7-methylguanine and sister chromatid exchanges. Thus, as is the case for Arabidopsis, rice OsKu70 may participate in the repair of DNA DSBs.
Inactivation of Ku70 or Ku80 in yeast cells resulted in shorter telomeres and extensive rearrangement of subtelomeric regions (Boulton and Jackson, 1998; Baumann and Cech, 2000). In mammalian cells, Ku proteins have both positive and negative effects on telomere length. Mouse ku mutants have both longer and shorter telomeres (d'Addadi Fagagna et al., 2001; Espejel et al., 2002), while silencing of human Ku86 leads to telomere shortening (Jaco et al., 2004; Myung et al., 2004). The complete loss of Ku86 results in massive telomere loss in the form of T-circles in human somatic cells (Wang et al., 2009). There was no change in telomere length in Ku70 mutant chicken cells (Wei et al., 2002). In contrast, knockout mutations of AtKu70 and AtKu80 always resulted in longer telomeres in Arabidopsis. Similarly, the homozygous osku70 lines possess markedly longer telomeres than do wild-type rice plants (Fig. 5). These findings suggest that in higher plants, Ku70 works exclusively as a negative regulator of telomere homeostasis. We speculate that Ku70 homologs in monocot (rice) and dicot (Arabidopsis) plants share similar roles with regard to the repair of DSBs and some aspects of telomere metabolism.
Ku-deficient mice exhibited a dwarf phenotype, early onset of senescence, and severe immunological problems (Gu et al., 1997; Ouyang et al., 1997; Vogel et al., 1999). In the absence of functional Ku, there is an elevated level of end-to-end chromosomal fusions in mouse cells (Samper et al., 2000; d'Adda di Fagagna et al., 2001). In contrast, Arabidopsis ku70 mutant plants appeared grossly normal under standard growth conditions, and no chromosomal fusions were detected (Bundock et al., 2002; Riha et al., 2002). Based on these results, it was proposed that Arabidopsis AtKu70 might not be required for normal growth (Bundock et al., 2002) and that animal and plant Ku70 may have differing roles in telomere protection (Riha et al., 2002). However, a more detailed analysis of the cellular function of AtKu70 in Arabidopsis growth and telomere stability was performed using compound mutants. The ku70 tert double mutant exhibited an enhanced shortening of telomeres and a 2- to 3-fold accelerated onset of growth defects compared with single tert knockout plants (Riha and Shippen, 2003). Moreover, analysis of ku70 tert mre11 triple Arabidopsis mutants led to the proposal that together with Mre11, Ku70 functions to fuse critically shortened telomeres via the NHEJ pathway (Heacock et al., 2004).
In contrast, we have shown that single OsKu70 mutants display clearly detectable phenotypes in rice. The homozygous G2 osku70-1 and osku70-2 mutant lines suffer from severe developmental abnormalities in the postgermination growth of vegetative organs (Fig. 6). Furthermore, their reproductive organs are impaired, with aberrant pollen grains, stamens, palea, and lemma, resulting in the lack of functional seeds (Figs. 7 and 8). Up to 11.1% of anaphase events observed in G2 osku70-1 mutant PMCs display unusual anaphase bridges, suggesting that deficiency of functional OsKu70 is accompanied by genome instability in rice (Fig. 9; Table I). These results are different from those observed in Arabidopsis but are in at least partial agreement with what is observed in mammalian cells. Thus, this leads to the tantalizing possibility that unlike its dicot Arabidopsis AtKu70 homolog, monocot rice OsKu70 possesses cellular functions that are necessary for both normal developmental growth and protection of telomeres. At present, however, we do not know the molecular and cellular bases for this difference between OsKu70 and AtKu70 function.
Because homozygous G2 mutant plants were sterile, we obtained subsequent homozygous progeny (G2-1) by crossing heterozygous G2 mutants. As shown in Figures 6 to 8 and Table I, the severity of the defects in the G2-1 osku70-1 allele was worse than in G2 plants. Since the G2 heterozygous plants are relatively normal, these results are intriguing. Although the reason for this severity is not clear, we consider the possibility that the phenotype caused by the suppression of OsKu70 becomes more serious during subsequent generations.
The phenotype of osku70 is somewhat reminiscent of mutants in which the rice double-stranded specific telomere-binding protein 1 (RTBP1) is inactivated (Hong et al., 2007). The rtbp1 mutant was previously shown to exhibit progressive and severe developmental defects in both the vegetative and reproductive organs over four generations (G1–G4). Its telomeres were much longer (approximately 10–30 kb) than wild-type telomeres (approximately 5–10 kb), and genomic anomalies were frequently observed, including abnormal anaphase bridges in rtbp1 PMCs (Hong et al., 2007). The major differences between osku70 and rtbp1 are that, under our experimental conditions, growth retardation of G2 osku70 is more severe than that of G2 rtbp1 and that G4 rtbp1 plants still produce a small number of seeds whereas G2 osku70 flowers are infertile (Fig. 7B). This suggests that the impact of OsKu70 inactivation is greater than that of RTBP1 inactivation in rice plants. One possibility is that OsKu70 and RTBP1 function cooperatively to stabilize the structure and function of telomeres in rice. Thus, it would be interesting to test whether OsKu70 interacts with RTBP1 and/or OsTRBF, another class of rice telomere-binding proteins (Byun et al., 2008), in the presence or absence of OsKu80.
It is worth noting that despite a clear deregulation of telomere length, inactivation of RTBP1 did not cause detectable phenotypes in Arabidopsis, possibly due to a functional redundancy of homologous genes for telomere-binding proteins (Karamysheva et al., 2004; Hwang and Cho, 2007). Similarly, Arabidopsis atku70 mutants are phenotypically normal under standard growth conditions. Given that rice plants are highly susceptible to mutations in either RTBP1 or Ku70, it would be conceivable that there are subtle differences between the dicot Arabidopsis and the monocot rice in the control of the structure and function of telomeres.
Loss of AtMre11 and AtRad50, which are components of the Mre11/Rad50/Nbs1 complex involved in the NHEJ pathway, is intimately tied with developmental defects and chromosomal instability in Arabidopsis (Gallego and White, 2001; Bundock and Hooykaas, 2002; Puizina et al., 2004; Vannier et al., 2006). Thus, we are currently carrying out experiments to examine the phenotypic properties of rice osmre11 and osrad50 knockout mutants. This work will tell us more about the function of the NHEJ mechanism in rice plants. In conclusion, our results suggest that OsKu70 is required for the maintenance of chromosomal stability and normal developmental growth in rice, a monocot model plant.
MATERIALS AND METHODS
Plant Growth and Genotoxic Treatment
Dry rice (Oryza sativa var japonica ‘Dongjin’) seeds were soaked in 70% ethanol for 10 min and washed extensively with sterilized water. Seeds of wild-type, osku70 mutant, and 35S:OsKu70 and 35S:anti-OsKu70 transgenic plants were germinated on Murashige and Skoog medium containing Murashige and Skoog basal salt (Wako Pure Chemical), 3% Suc, 0.2% phytogel, and 0.55 mm myoinositol (Hong et al., 2007). The seedlings were grown for 10 d at 27°C under continuous light in the growth chamber, transplanted to soil, and raised to maturity in the greenhouse. To test the sensitivity to genotoxic agents, 5- or 10-d-old wild-type, heterozygous and homozygous G2 osku70 mutant, and T2 35S:anti-OsKu70 transgenic seedlings were transferred to growth medium containing different concentrations (0%–0.05%) of MMS and then incubated further for 10 d in greenhouse conditions.
Identification of rice OsKu70 and OsKu80 Genes
Homologs of Arabidopsis (Arabidopsis thaliana) AtKu70 and AtKu80 genes were found in the rice genome database (http://signal.salk.edu/cgi-bin/RiceGE) using the BLASTN program, with the E-value set to be lower than 10−14. OsKu70 (GenBank accession no. BAC83189) and OsKu80 (GenBank accession no. AAS01977) were identified as homologs of the AtKu70 and AtKu80 genes, respectively. Full-length cDNA for OsKu70 and OsKu80 were obtained by RT-PCR as described below.
Total RNA Isolation, RT-PCR, and Quantitative Real-Time RT-PCR
Total RNAs were isolated from 3-month-old mature leaf tissue of wild-type, osku70 mutant, 35S:OsKu70, and 35S:anti-OsKu70 rice plants by a method described previously (Jang et al., 2009). First-strand cDNA was synthesized from 10 μg of total RNA and then amplified by PCR using OsKu70-specific primers (5′-ATGGATCTGGACCCCGAGGGC-3′ and 5′-TCACTTGCCTAGATGAGTCAA-3′) and OsKu80-specific primers (5′-ATGGCTCGCAACAAGGAAGCA-3′ and 5′-CTACTGAGAGGACGGCTCGGG-3′) using high-fidelity Ex-Tag polymerase (Takara) as described by Kim et al. (2008). Real-time PCR was conducted with the IQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) using 2× SYBR Premix Ex Taq II (Takara) and a gene-specific primer set (5′-TAGGTGAAGATGAGATGCCAG-3′ and 5′-ATGTGCTGCACTTTTCTCTGC-3′). To evaluate transcript levels, cycle threshold values for OsKu70 in wild-type, 35S:OsKu70, 35S:anti-OsKu70, and osku70 plants were normalized to that of the rice Actin gene. The primers for Actin were 5′-GACCTTTATGGCAACATTGTG-3′ and 5′-TCCACATCTGTTCCAAAGTCG-3′.
RNA Gel-Blot Analysis
Rice total RNAs were obtained from the leaves, roots, internodes, and meristems of 10-d-old, 1-month-old, and 3-month-old plants and from undifferentiated callus cells. Total RNA (40 μg per lane) was separated by 1.0% agarose gel electrophoresis and blotted onto a nylon membrane filter (Amersham). The filter was hybridized to a 32P-labeled OsKu70 cDNA probe under high-stringency hybridization and washing conditions, as described by Lee et al. (2009). The blot was visualized by autoradiography at −80°C using Kodak XAR-5 film with an intensifying screen.
Yeast Two-Hybrid and in Vitro Pull-Down Assays
For the yeast two-hybrid experiment, full-length and deletion mutant OsKu70 cDNA (OsKu701–587-ΔSAP, OsKu701–480-ΔC, and OsKu70265–587-ΔNΔSAP) and OsKu80 cDNA were inserted into pGADT7 and pGBKT7 (Clontech; Matchmaker3) vectors, respectively. These plasmid constructs and an empty vector control were introduced into the yeast strain AH109. After transformation, yeast cells were grown on SD/−His/−Trp/−Leu medium including 10 mm 3-aminotriazole for 4 d at 28°C. The capacity of the yeast cells to grow in the absence of His was used as a readout for the interaction between OsKu70 and OsKu80.
For the in vitro pull-down assay, (His)6-OsKu701–587-ΔSAP and GST-OsKu80 fusion proteins were expressed in Escherichia coli. The purified (His)6-OsKu701–587-ΔSAP protein was coincubated with GST-OsKu80 in the presence of Ni-NTA resin. After extensive washing with immunoprecipitation buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm MgCl2, and 1× complete protease inhibitor cocktail), the bound proteins were eluted from the affinity resin by boiling with SDS sample buffer. The eluted proteins were resolved on a 10% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane. The blot was subjected to immunoblot analysis with anti-His and anti-GST antibodies.
Genotyping of osku70 T-DNA Knockout Mutant Lines
Total genomic DNA was isolated from 1B-09628 and 3A-01546 mutant lines as described below. To obtain homozygous osku70 mutant lines, PCR was performed in a 50-μL total volume, using genomic DNA as a template, 10× ExTaq buffer, 0.2 mm deoxyribonucleotide triphosphate, 0.5 units of ExTaq polymerase (Takara), and 1 μm of each primer. The S (5′-AACAAGGAGATGGTCGTCTAC-3′), AS1 (5′-TGTTGACGGTCTTAAGTTGTG-3′), and T1 (5′-TTGGGGTTTCTACAGGACGTAAC-3′) primers were used for the 1B-09628 line, while the S1 (5′-TTGGAAGCACTCGTGTTTTCG-3′), AS (5′-TTTTCTCTGCAGCTGCATCAG-3′), and T (5′-TTCCCCTTTCTACAGGACGTTTC-3′) primers were used for the 3A-01546 line.
Genomic DNA Isolation and Pulse-Field Gel Electrophoresis
Mature leaves were excised from 2-month-old rice plants and immediately frozen with liquid nitrogen. Frozen leaf tissue was pulverized with a mortar and pestle in liquid nitrogen, and total genomic DNA was isolated as described previously (Hong et al., 2005). Leaf DNA was digested with TaqI restriction enzyme, and digested DNA fragments (10 μg per lane) were separated by pulse-field gel electrophoresis using the CHEF-DRIII system (Bio-Rad) according to the methods described by Yang et al. (2004). The gel was blotted onto a nylon membrane filter (Amersham) and hybridized to a 32P-labeled (TTTAGGG)70 telomeric fragment under high-stringency conditions (Yang et al., 2004).
Genomic Southern-Blot Analysis
Leaf genomic DNA (10 μg per lane) was obtained from wild-type and osku70 knockout mutants and digested with EcoRI restriction enzyme. Digested DNA fragments were separated by electrophoresis on a 0.8% agarose gel, blotted onto a nylon membrane filter (Amersham), and incubated with 32P-labeled GUS cDNA probe under high-stringency hybridization and washing conditions, according to the method described by Hong et al. (2005). The blot was visualized by autoradiography using Kodak XAR-5 film.
Generation of 35S:OsKu70 and 35S:anti-OsKu70 Transgenic Rice
Transgenic rice plants were generated by the method described by Jeon et al. (2000) with the following modifications. Full-length pOsKu70 cDNA was inserted into the binary vector pMBP2 in the sense and antisense orientations. The constructed plasmids were electroporated into Agrobacterium tumefaciens strain AGL1, and transfected cells and rice leaf discs were cocultivated as described previously (Jeon et al., 2000). Transgenic rice plants were selected on a 40 mg L−1 hygromycin B-containing medium. The regenerated plants were transferred and grown in a greenhouse at 28°C to 30°C during the day and 20°C to 22°C at night. The light/dark cycle in the greenhouse was 14/10 h (Hong et al., 2007).
I2-KI Staining of Pollen and in Vitro Pollen Tube Germination Assay
To estimate the pollen viability, pollen grains from wild-type, osku70 mutant, 35S:OsKu70, and 35S:anti-OsKu70 plants were placed on a glass slide. Pollen grains were stained with I2-KI solution for 1 to 2 min and observed using a light microscope (Olympus) under bright-field conditions. Pollen grains that stained black were considered to be viable and living, whereas those with yellow staining were considered to be sterile or dead (Chhun et al., 2007).
For the in vitro pollen tube germination assay, pollen grains of wild-type, osku70 mutant, 35S:OsKu70, and 35S:anti-OsKu70 plants were placed on glass slides for 2 to 3 h at 35°C in pollen tube germination medium containing 1 mm CaCl2, 1 mm KCl, 0.8 mm MgSO4, 1.6 mm H3BO3, 30 μm CaSO4, 0.03% casein, 0.3% MES, 10% Suc, and 10% polyethylene glycol (molecular weight 8,000; Han et al., 2006). The pollen grains were observed with a light microscope (Olympus) under bright-field conditions.
Cytogenetic Studies
Anaphase chromosomes were obtained from actively dividing PMCs of wild-type plants as well as heterozygous and homozygous osku70 mutant plants and stained with DAPI by previously described methods (Hong et al., 2007). Anthers from wild-type and mutant plants were fixed in 1:3 (v/v) acetic acid:ethanol for 2 h, gently rinsed with distilled water, and treated with cell wall digestion enzyme mixture (2% cellulose, 1.5% macerozyme, 0.3% pectolyase, and 1 mm EDTA [pH 4.2] in distilled water) for 1 h. After digestion, anaphase chromosomes were gently squashed on a glass slide, stained with DAPI, and observed by fluorescence microscopy (Axio imager; Zeiss).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number BAC83189 (OsKu70).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Response of osku70 homozygous plants to genotoxic agents.
Supplemental Figure S2. Meiotic stages of PMCs in wild-type rice plants.
Supplementary Material
This work was supported by the BioGreen 21 Program (funded by the Rural Development Administration) and the Plant Diversity Research Center (21st Century Frontier Research Program funded by the Ministry of Education, Science, and Technology). M.Y.B. was the recipient of a Brain Korea 21 graduate student scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Woo Taek Kim (wtkim@yonsei.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baumann P, Cech TR (2000) Protection of telomeres by the Ku protein in fission yeast. Mol Biol Cell 11 3265–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P (2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, van Attikum H, Hooykaas P (2002) Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res 30 3395–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun MY, Hong JP, Kim WT (2008) Identification and characterization of three telomere repeat-binding factors in rice. Biochem Biophys Res Commun 372 85–90 [DOI] [PubMed] [Google Scholar]
- Chai W, Ford LP, Lenertz L, Wright WE, Shay JW (2002) Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem 277 47242–47247 [DOI] [PubMed] [Google Scholar]
- Chan JY, Lerman MI, Prabhakar BS, Isozaki O, Santisteban P, Kuppers RC, Oates EL, Notkins AL, Kohn LD (1989) Cloning and characterization of a cDNA that encodes a 70-kDa novel human thyroid autoantigen. J Biol Chem 264 3651–3654 [PubMed] [Google Scholar]
- Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, Kitano H, Ashikari M, Matsuoka M, Ueguchi-Tanaka M (2007) Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19 3876–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23 394–398 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP (2001) Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol 11 1192–1196 [DOI] [PubMed] [Google Scholar]
- Dynan WS, Yoo S (1998) Interaction of Ku protein and DNA dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res 26 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA (2002) Mammalian Ku86 mediates chromosomal functions and apoptosis caused by critically short telomeres. EMBO J 21 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego ME, Bleuyard JY, Daoudal-Cotterell S, Jallut N, White CI (2003. a) Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J 35 557–565 [DOI] [PubMed] [Google Scholar]
- Gallego ME, Jalut N, White CI (2003. b) Telomerase dependence of telomere lengthening in ku80 mutant Arabidopsis. Plant Cell 15 782–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego ME, White CI (2001) RAD50 function is essential for telomere maintenance in Arabidopsis. Proc Natl Acad Sci USA 98 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego ME, White CI (2005) DNA repair and recombination functions in Arabidopsis telomere maintenance. Chromosome Res 13 481–491 [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA (1999) How plants make ends meet: DNA double strand break repair. Trends Plant Sci 4 263–269 [DOI] [PubMed] [Google Scholar]
- Gu Y, Siedl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng H-L, Sekiguchi JM, Frank K, et al (1997) Growth retardation and leaky SCID phenotype of Ku70 deficient mice. Immunity 7 653–665 [DOI] [PubMed] [Google Scholar]
- Han MJ, Jung KH, Yi G, Lee DY, An G (2006) Rice immature pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol 47 1457–1472 [DOI] [PubMed] [Google Scholar]
- Heacock M, Spangler E, Riha K, Puizina J, Shippen DE (2004) Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J 23 2304–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JP, Byun MY, Koo D, An K, Bang J, Chung IK, An G, Kim WT (2007) Suppression of Rice Telomere Binding Protein1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell 19 1770–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JP, Kim SM, Ryu MY, Choe S, Park PB, An G, Kim WT (2005) Structure and expression of OsMRE11 in rice. J Plant Biol 48 229–236 [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev 14 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang MG, Cho MH (2007) Arabidopsis thaliana telomeric DNA-binding protein 1 is requited for telomere length homeostasis and its Myb-extension domain stabilizes plant telomeric DNA binding. Nucleic Acids Res 35 1333–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I, Munoz P, Blasco MA (2004) Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res 64 7271–7278 [DOI] [PubMed] [Google Scholar]
- Jang S, Cho K, Shibato J, Han O, Iwahashi H, Tamogami S, Zargar SM, Kubo A, Masuo Y, Agrawal GK, et al (2009) Rice OsOPRs: transcriptional profiling responses to diverse environmental stimuli and biochemical analysis of OsOPR1. J Plant Biol 52 229–243 [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jand S, Yang K, Nam J, et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22 561–570 [DOI] [PubMed] [Google Scholar]
- Karamysheva ZN, Surovtseva YV, Vespa L, Shakirov EV, Shippen DE (2004) A C-terminal Myb-extension domain defines a novel family of double-strand telomeric DNA binding proteins in Arabidopsis. J Biol Chem 279 47799–47807 [DOI] [PubMed] [Google Scholar]
- Kim JH, Cheon YM, Kim BG, Ahn JH (2008) Analysis of flavonoids and characterization of the OsFNS gene involved in flavone biosynthesis in rice. J Plant Biol 51 97–101 [Google Scholar]
- Koike M, Miyasaka T, Mimori T, Shiomi T (1998) Subcellular localization and protein-protein interaction regions of Ku proteins. Biochem Biophys Res Commun 252 679–685 [DOI] [PubMed] [Google Scholar]
- Lee SC, Kwon SY, Kim SR (2009) Ectopic expression of a cold-responsive CuZn superoxide dismutase gene, SodCc1, in transgenic rice (Oryza sativa L.). J Plant Biol 52 154–160 [Google Scholar]
- Lieber MR, Karanjawala ZE (2004) Ageing, repetitive genomes and DNA damage. Nat Rev Mol Cell Biol 5 69–75 [DOI] [PubMed] [Google Scholar]
- Liu PF, Chang WC, Wang YK, Munisamy SB, Hsu SH, Chang HY, Wu SH, Pan RL (2007) Differential regulation of Ku gene expression in etiolated mung bean hypocotyls by auxins. Biochim Biophys Acta 1769 443–454 [DOI] [PubMed] [Google Scholar]
- Myung K, Ghosh G, Fattah FJ, Li G, Kim H, Dutia A, Pak E, Smith S, Hendrickson EA (2004) Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol Cell Biol 24 5050–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H, Nussenzwieg A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Iliakis G, et al (1997) Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J Exp Med 186 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K (2004) Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE (2006) The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet 40 237–277 [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE (2003) Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci USA 100 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE (2002) Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in KU70. EMBO J 21 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep 1 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Jung D, Jung Y, Lee SG, Lee I (2000) Interaction of human Ku70 with TRF2. FEBS Lett 481 81–85 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H (2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J 29 771–781 [DOI] [PubMed] [Google Scholar]
- Tuteja R, Tuteja N (2000) Ku autoantigen: a multifunctional DNA-binding protein. Crit Rev Biochem Mol Biol 35 1–33 [DOI] [PubMed] [Google Scholar]
- Vannier JB, Depeiges A, White C, Gallego ME (2006) Two roles for Rad50 in telomere maintenance. EMBO J 25 4577–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P (1999) Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA 96 10770–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dong X, Reeves WH (1998) A model for Ku heterodimer assembly and interaction with DNA. J Biol Chem 273 31068–31074 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA (2009) Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA 106 12430–12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Shippen DE (2007) Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol Cell Biol 27 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Skopp R, Takata M, Takeda S, Price CM (2002) Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res 30 2862–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, Bray CM (2002) Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J 31 517–528 [DOI] [PubMed] [Google Scholar]
- Yang SW, Kim SK, Kim WT (2004) Perturbation of NgTRF1 expression induces apoptosis-like cell death in tobacco BY-2 cells and implicates NgTRF1 in the control of telomere length and stability. Plant Cell 16 3370–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K (2007) Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell 27 163–169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.