Abstract
Epicardial epithelial-mesenchymal transition (EMT) is hypothesized to generate cardiovascular progenitor cells that differentiate into various cell types, including coronary smooth muscle and endothelial cells, perivascular and cardiac interstitial fibroblasts and cardiomyocytes. Here we show that an epicardial-specific knockout of Wt1 leads to a reduction of mesenchymal progenitor cells and their derivatives. We demonstrate that Wt1 is essential for repression of the epithelial phenotype in epicardial cells and during Embryonic Stem (ES) cell differentiation, through direct transcriptional regulation of Snail (Snai1) and E-cadherin (Cdh1), two of the major mediators of EMT. Some mesodermal lineages fail to form in Wt1 null embryoid bodies but this effect is rescued by the expression of Snai1, underlining the importance of EMT in generating these differentiated cells. These new insights into the molecular mechanisms regulating cardiovascular progenitor cells and EMT will shed light on the pathogenesis of heart diseases and may help the development of cell based therapies.
Keywords: EMT, Wt1, epicardium, progenitor cells, Snail, stem cells
In the developing heart, very little is known about the molecular and cellular mechanisms controlling epicardial EMT and the formation of cardiovascular progenitor cells1. Wt1 encodes a zinc finger protein, which plays a critical role in the normal development of several organs such as kidney, gonads, spleen and heart2. Coronary vascular defects in Wt1 mutant mice are conjectured to arise through defective EMT3. However, until now it has not been possible to test if Wt1 in the epicardium is directly involved in EMT or whether EMT is essential for the formation of cardiovascular progenitor cells.
To investigate the role of Wt1 in the epicardium we generated Wt1 conditional knockout mice (Supplementary Fig. 1) that were crossed with Wt1-GFP knockin mice4 and transgenic Gata5-Cre 5 mice (See Online Methods for details of breeding). The resulting Wt1 mutants die between embryonic day 16.5 (E16.5) and embryonic day 18.5 (E18.5) due to cardiovascular failure. Embryos at E16.5 display edema and accumulation of blood in the systemic veins (Fig. 1a, b). Efficient deletion of Wt1 in epicardial cells was confirmed using immunohistochemistry on heart sections and real time-PCR analysis on FACS-sorted GFP+ epicardial cells isolated from Gata5-Cre+/Wt1 loxP/gfp (Cre+) mice (Supplementary Fig. 2a-c). The gross morphology of Cre+ embryos is normal. However the right ventricle (RV) of some mutant embryos (Fig. 1d) showed a thinner free wall as compared to the control (Fig. 1c) (arrows), while the left ventricle (LV) was apparently normal. Mutant embryos also display pericardial haemorrhaging (* in Fig. 1d).
Figure 1.
Heart defects in epicardial-specific Wt1 mutant embryos. (b) Gata5-Cre+/Wt1 loxP/gfp (Cre+) embryos display edema and accumulation of blood in the systemic veins; a littermate control (Cre−) is shown in panel (a). Scale bars represent 100 μm. (c, d) Haematoxylin and eosin-stained sections of Cre− and Cre + E16.5 embryos. The right ventricle (RV) of some the mutant embryos (d) shows a thinner wall (arrows) as compared to the control (c), while the left ventricle (LV) is apparently normal. Mutant embryos show pericardial haemorrhage (* in d). Scale bars represent 50 μm. (e, f) OPT image of control and mutant hearts at E16.5. Scale bars represent 50 μm. (g, h ) Immunofluorescence staining for the indicated blood vessel markers. Only control embryos show arteries with a well differentiated smooth muscle layer (g). Analysis of EMT markers with antibodies against: Snail (i, j), E-cadherin (k, l), Vimentin and Cytokeratin (m, n). Abnormal E-cadherin (l) and decreased Snail (j) and Vimentin (n) expression is observed in epicardial cells from Cre+ embryos. Scale bars represent 50 μm.
We confirmed the embryonic expression of the Gata5-Cre transgene in the heart. At E10.5 we detected cells expressing Cre in the epicardium, while at E12.5 Gata5-Cre derived cells were abundant within the heart (Supplementary Fig. 3 and data not shown) 5.
Optical projection tomography (OPT) showed that the coronary arteries failed to form in Wt1 mutant mice (Fig. 1e, f and Supplementary movies 1, 2). This result was confirmed by staining for platelet/endothelial cell adhesion molecule-1 (PECAM-1) and α-smooth muscle actin (α-SMA), markers for endothelial and smooth muscle cells respectively (Fig. 1g, h).
The presence of GFP+ epicardial cells covering the surface of the myocardium in Cre+ mice demonstrated the integrity of this structure in mutant mice (Supplementary Fig. 4b). Since epicardial EMT plays a very important role in the formation of coronary vascular precursor cells, we studied the expression patterns of the major markers of EMT in the Wt1 mutant epicardium. We found that E-cadherin and Cytokeratin (CK), epithelial markers, were upregulated in mutant epicardial cells in comparison with control hearts (Fig. 1k-n). Conversely, the expression of mesenchymal markers, Snail (Fig. 1i, j) and Vimentin was reduced (Fig. 1m, n). We also compared the levels of Slug (Snai2) in Cre+ and Cre− FACS-sorted epicardial cells by real time-PCR and confirmed that similar to Snail (Snai1), Snai2 levels were reduced by 70% in Cre+ in comparison with Cre− cells (data not shown).
To determine whether Wt1 plays a direct and cell autonomous role in epicardial EMT we generated tamoxifen inducible Wt1 KO immortalised epicardial cells (Cre+ CoMEEC) Fig. 2c-f (See Online Methods). Loss of Wt1 after tamoxifen treatment leads to a robust increase in E-cadherin expression in a dose dependent manner (Fig. 2g) which correlates with a downregulation in the levels of N-cadherin and α-SMA (Fig 2g). RT-PCR analysis also showed that there was a striking downregulation of Snai1 expression after Wt1 deletion (Fig. 2h). Not only does treatment of the cells with tamoxifen lead to changes in the EMT marker pattern, but the cells also display decreasing cell migration (Fig. 2i). We have not observed any difference in the markers that were analysed or in the migration properties of Cre−CoMEEC after tamoxifen treatment (Supplementary Fig. 5 a, b).
Figure 2.
Wt1 expression is necessary to maintain a mesenchymal phenotype in immortalised epicardial cells. (a) Heart of Wt1-GFP knockin mouse. Scale bar represents 50 μm. (b) Direct GFP expression on heart cryosection of Wt1-GFP knockin embryos shows GFP expression in epicardial cells. Scale bar represents 50 μm. (c) Phase-contrast micrograph of Wt1 conditional KO immortalised mouse epicardial cells (CoMEEC). Scale bar represents 100 μm. (d-f) GFP, ZO-1 and Wt1 expression in CoMEEC. Scale bars represent 50 μm. CoMEEC displayed a cobblestone monolayer typical of epicardial cells (c, e). (g) Western blot analysis was conducted in Cre+ tamoxifen inducible CoMEEC cultured in the presence of different concentrations of tamoxifen. (h) RT-PCR analyses of Snai1 and Cdh1 expression in Cre+ CoMEEC in presence of tamoxifen. (i) The migratory behaviour of Cre+ CoMEEC in presence of tamoxifen was analyzed in an in vitro wound model. Scale bar represents 100 μm.
We examined whether or not the Snai1 gene, one of the master regulators of EMT 6, could be directly regulated by Wt1. We identified three conserved potential Wt1 binding sites in the Snai1 genomic sequence schematically represented in Fig. 3a. The −KTS Wt1 isoform, which functions as a transcription factor 2, is able to activate the Snai1 fragment containing the promoter binding site in a dose dependent manner (Fig. 3b), while the rest of the fragments were insensitive to Wt1 activation (data not shown). The transcriptional activation was abolished when the functional binding site was mutated (Fig. 3c). In addition, Chromatin immunoprecipitation (ChIP) demonstrated the in vivo binding of Wt1 to the endogenous Snai1 promoter but not to the intronic and 3′ UTR regions in epicardial cells (Fig. 3d). The endogenous Snai1 promoter of epicardial cells is enriched in acetylated histones H3 and trimethylated K4 of histone H3, but depleted in trimethylated K27, which is compatible with the activated state of the Snai1 gene in these cells (Fig. 3d).
Figure 3.
Wt1 is an activator of Snai1 and a repressor of Cdh1 in epicardial cells. (a, e) Schematic representation of the putative conserved Wt1 binding sites ( , □) in the Snai1 and Cdh1 loci,
, □) in the Snai1 and Cdh1 loci,  (functional binding site), □ (putative but non functional binding site) and ■ (exons). (b) Luciferase activity of reporter construct carrying mouse Snai1 promoter in epicardial cells in the presence of indicated amounts of −KTS Wt1 expression vector. (c) Luciferase activity of wild-type (Control) or mutated Snai1 promoters in the presence of -KTS Wt1 isoform. (d, f) Cell extracts from epicardial cells were chromatin immunoprecipitated (ChIP), using antibodies against Wt1, anti-acetyl-histone H3 (AcH3), anti-H3 tri methyl K4 (K4Me) and anti- H3 tri methyl K27 (K27Me) or an irrelevant antibody (IgG). The input was used as a positive control for PCR of the Snai1 (d) and Cdh1 (f) promoters, intronic regions and 3′UTRs. (g, h) Luciferase activity of constructs carrying DNA fragments identified by the Cdh1 ChIP assay in epicardial cells, together with different concentrations of −KTS Wt1 isoform. (i) Luciferase activity of Control or mutated Cdh1 constructs in the presence of -KTS Wt1 isoform (100ng). (j) ChIP assays of Snail and Wt1 at the endogenous Cdh1 promoter in epicardial cells. The graphs represent the mean values ±s.e.m from three independent experiments.
(functional binding site), □ (putative but non functional binding site) and ■ (exons). (b) Luciferase activity of reporter construct carrying mouse Snai1 promoter in epicardial cells in the presence of indicated amounts of −KTS Wt1 expression vector. (c) Luciferase activity of wild-type (Control) or mutated Snai1 promoters in the presence of -KTS Wt1 isoform. (d, f) Cell extracts from epicardial cells were chromatin immunoprecipitated (ChIP), using antibodies against Wt1, anti-acetyl-histone H3 (AcH3), anti-H3 tri methyl K4 (K4Me) and anti- H3 tri methyl K27 (K27Me) or an irrelevant antibody (IgG). The input was used as a positive control for PCR of the Snai1 (d) and Cdh1 (f) promoters, intronic regions and 3′UTRs. (g, h) Luciferase activity of constructs carrying DNA fragments identified by the Cdh1 ChIP assay in epicardial cells, together with different concentrations of −KTS Wt1 isoform. (i) Luciferase activity of Control or mutated Cdh1 constructs in the presence of -KTS Wt1 isoform (100ng). (j) ChIP assays of Snail and Wt1 at the endogenous Cdh1 promoter in epicardial cells. The graphs represent the mean values ±s.e.m from three independent experiments.
Wt1 has been shown to transcriptionally activate the E-cadherin (Cdh1) promoter in NIH3T3 cells 7. However, we also decided to explore whether Wt1 might directly repress Cdh1 in epicardial cells. ChIP with primers flanking the previously identified Wt1 binding sequence in the Cdh1 promoter confirmed that Wt1 binds directly to the endogenous Cdh1 promoter in epicardial cells (Fig. 3f). Consistent with a repressed state, the Cdh1 promoter is depleted in trimethyl-K4 of histone H3 and enriched for trimethylated K27 (Fig. 3f). An extended analysis of the Cdh1 genomic sequence revealed two new conserved potential Wt1 binding sites in the intron and in the 3′UTR (Fig. 3e). ChIP with primers flanking these regions demonstrated the in vivo binding of Wt1 in epicardial cells to the 3′UTR but not to the intronic region (Fig. 3f). In epicardial cells the −KTS Wt1 isoform does exert a repressive effect on the Cdh1 promoter fragment containing the Wt1 binding site shown previously to be activated in NIH3T3 cells (Fig. 3g and data not shown) and on the 3′-UTR construct (Fig. 3h). The transcriptional repression was abolished when the functional binding sites were mutated (Fig. 3i). The +KTS isoform was able to regulate the Snai1 and Cdh1 constructs but to a much lesser degree (data not shown).
Our finding that Wt1 can directly repress the Cdh1 gene raises the question of whether Snail is also a repressor of Cdh1 in this system. ChIP analysis showed that Snail does interact with the Cdh1 promoter in epicardial cells (Fig. 3j). Furthermore the knockdown of Snai1 in epicardial cells with a shRNA-Snai1 leads to an increase in Cdh1 promoter activity (Supplementary Fig. 6). These results lead us to propose a model in which Wt1 can promote downregulation of Cdh1 directly and indirectly through increasing the levels of Snail.
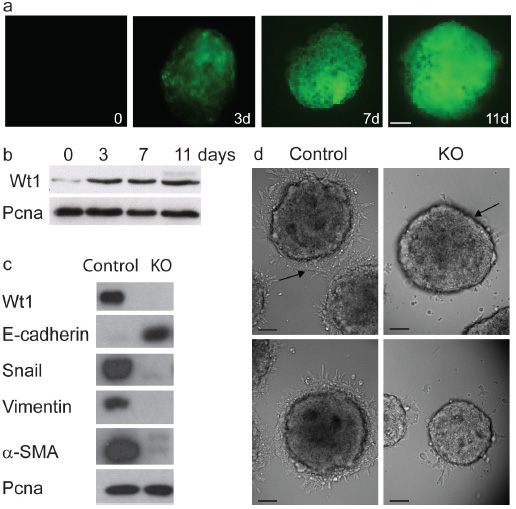
We wanted to determine whether Wt1 might regulate EMT and cardiovascular differentiation in another cellular system, so we chose to investigate ES cells 8. Wt1 expression analysis, detected by either fluorescence (Wt1-GFP Knockin ES cells) or Western blots, showed that Wt1 was almost undetectable in undifferentiated ES cells (day 0), while expression was first detectable at 3 days and peaked between days 9 and 11 during embryoid bodies (EB) differentiation (Fig. 4a, b) (a proliferative period for undifferentiated mesenchymal cells). Thereafter, Wt1 expression declined, as various terminally-differentiated cell types started appearing (data not shown).
Figure 4.
Wt1 is required for EMT in ES cells. (a) Wt1-GFP expression during EB differentiation of Wt1-GFP knockin ES cells. Scale bar represents 50 μm (b) Western Blot (WB) analysis of the endogenous Wt1 protein in EB. (c) WB of epithelial and mesenchymal protein markers in wild type EB (control) and Wt1 KO EB (KO). (d) Phase-contrast microscopy of control and Wt1 KO EB migrating cells. Scale bars represent 50 μm.
To determine whether Wt1 is required for EMT in ES cell we examined the expression of the principal epithelial and mesenchymal markers in wild type (control EB) versus Wt1 KO (KO EB) EB (Fig. 4c). Control EB express high levels of Vimentin, α-SMA and Snail but not E-cadherin, indicating that these cells have undergone EMT. By contrast, Wt1 KO EB maintain high levels of E-cadherin expression and fail to induce the mesenchymal markers, reflecting their failure to undergo EMT (Fig. 4c). Additionally, control EB derived cells possess the capacity to migrate; whereas Wt1 KO EB derived cells have a clear impairment in their migration properties (Fig. 4d).
Since Wt1 KO EB failed to undergo EMT, we asked whether Wt1 is required for the generation of mesodermal lineages from ES cells. We analysed the expression patterns of the primitive streak genes Brachyury (T) and Goosecoid (Gsc). Both of these genes were normally expressed in Wt1 KO EB (Fig. 5a). The analysis of the more mature mesodermal markers, haematopoietic (Flk1 (Kdr) and Hbb-b1), endothelial (Kdr, Tie2 (Tek) and Ve-cadherin (Cdh5)) and cardiac markers (Kdr, Nkx2.5, Hand1 and Isl1) demonstrated a clear reduction in their expression levels in Wt1 KO EB (Fig. 5a). The ectodermal (Sox2, Nestin (Nes)) and endodermal markers (Fgf5, Afp and Hnf4 were also analysed and no differences were found (Fig. 5b).
Figure 5.
Wt1 is required for the formation of some mesodermal lineage in EB differentiation. RT-PCR analysis for expression of mesodermal (a), endodermal and ectodermal markers (b) in wild type EB (control) and Wt1 KO EB (KO). (c) RT-PCR analysis for expression of Snai1 and Cdh1 in Wt1 KO Snai1 expressing clones (snailc1 and snailc2) versus Wt1 KO GFP expressing clones (c1 and c2). (d) RT-PCR analysis for expression of mesodermal markers in Snai1 clones (Snai1c1 and Snai1c2) versus GFP clones (c1 and c2). (e) Quantification of the number of beating cardiomyocytes in control, and Wt1 KO clones (c1, c2 and Snai1c1, Snai1c2). The graph represents the mean values ±s.e.m from three independent experiments.
To test whether the EMT defect in Wt1 KO EB could be the principal cause of impairment in the formation of mesoderm precursors, we expressed Snai1 in Wt1 KO ES cells. Snail positive clones (Snai1c1 and Snai1c2) expressed reduced levels of Cdh1 and higher levels of Snail2 and Twist1, relative to control GFP clones (c1 and c2) (Fig. 5c, d), suggesting a rescue of the EMT process. We also observed that the levels of expression of the endothelial and cardiac mesodermal markers were rescued after Snai1 expression (Fig. 5d). To measure the presence of mature functional mesoderm, we analysed the formation of beating cardiomyocytes. These were observed in control EB and Snai1 rescue clones but were absent in the control GFP clones (c1 and c2) (Fig. 5e).
Our new findings support a crucial role for Wt1 in the generation of mesenchymal cardiovascular progenitor formation in the epicardium and during ES cell differentiation, through the direct regulation of the Snail transcription factor and E-cadherin. A model built on our principal findings is shown in Supplementary Fig. 7. It has been shown previously that Wt1 expression is reactivated in hearts following ischaemia9. It will be interesting to determine whether Wt1 is required for the regeneration and repair of damaged hearts through the pathways described here.
The ES cell/EB model has been particularly useful in elucidating the molecular events involved in the specification of cell lineage differentiation10, 11. Our data suggest that in Wt1 null ES cells, disruptions during EMT causes defects in the differentiation of some mesodermal lineages. This conclusion is based on the finding that Snail, the master regulator of EMT, rescued the Wt1 null phenotype. The role of Wt1 in coronary blood vessel cell formation has been demonstrated previously 3, 12 and in the present study. However, this is the first evidence that Wt1 is also required for the formation of cardiomyocytes, through the regulation of EMT.
It is not clear whether the role of Wt1 in EB recapitulates the transitions in the epicardium that are required for the formation of cardiovascular progenitor cells, or whether the role of Wt1 in EB reflects an earlier function of the Wt1 gene in mesoderm formation. Interestingly, Mitiku and Baker have recently developed a dataset that samples the mouse transcriptome from gastrulation through organogenesis 13. Surprisingly, there was a massive increase in the levels of Wt1 expression during gastrulation, implicating this gene in the extensive morphological changes that occur within the embryo throughout this stage. Despite early embryonic expression of Wt1, the deficient embryo displays no overt phenotype just after gastrulation, although the formation of several mesodermal tissues is impaired. More extensive analysis will be required to evaluate whether Wt1 may have an earlier role in mesoderm formation.
Intriguingly in recent unpublished studies we have consolidated the evidence that Wt1 regulates the reverse process, mesenchymal-epithelial transition (MET), in the kidney and started to dissect the molecular mechanisms involved. Together, all of our findings suggest that Wt1 plays a major role regulating the balance between two fundamental cell states in several mesodermal tissues. In this context it is fascinating to consider the pattern of expression of Wt1 during development. The highest sites of expression are in the mesothelium, podocytes of the kidneys and Sertoli cells of the testis. All of these cells display dual epithelial/mesenchymal properties. The other major sites of expression are mesenchymal cell populations whose fates have not been determined. Further studies will be necessary to elucidate whether these cells are progenitors for particular cell types in the developing fetus.
Our findings may have a much broader relevance than for the cardiovascular field alone. Wt1 and Snail are both expressed at high levels in a range of adult cancers. Expression of both genes is correlated with poor prognosis in breast cancer 14, 15. Very recently it was shown that introduction of Snai1 into mammary epithelial cells converts them into cells with properties of mammary cancer stem cells 16. We speculate that Wt1 may regulate Snai1 and EMT, which may be involved in establishing cancer stem cells and tumour cells with invasive properties.
Methods
Generation of the loxP flanked Wt1 targeting construct and Wt1 loxP/loxP mice
A genomic fragment 5.8kb in length surrounding exon 1 of the murine Wt1 locus (representing nucleotides 104963917 to 104969768 of Chromosome 2 in Ensembl release 50) was subcloned into PolyIII. An oligonucleotide containing a loxP site marked with a PstI restriction enzyme recognition site was cloned in the unique AatII restriction site at position 2270 within this fragment. A neomycin cassette, flanked by Frt sites and carrying in addition a single loxP site (from vector p451) was targeted by bacterial recombination 17 to position 3955. This produced a targeting vector with two external homology arms of 2.2kb and 1.9 kb and an internal homology arm of 1.7kb; the completed targeting vector is shown in Supplementary Fig. 1a-e. The insert was removed from the vector backbone by digestion with NotI and purified using an Elutrap (Schleicher and Schuell). After homologous recombination in E14Tg2AIV ES cells (a kind gift from Austin Smith) using standard techniques 18, 307 clones were screened by Southern blotting of PstI digested genomic DNA using a 3′ probe external to the targeting vector. Of the 9 clones correctly targeted at the 3′end, subsequent screening using an internal probe demonstrated that 3 correctly carried the lone 5′ loxP site (Supplementary Fig. 1c). Two of these lines were electroporated with a vector expressing FLPe recombinase to remove the neomycin cassette (Supplementary Fig. 1d), and subsequently with a vector expressing Cre recombinase to verify accurate excision of exon 1 (Supplementary Fig. 1e). Clones were re-analysed by Southern blotting (Supplementary Fig. 1f), karyotyped and injected into mouse blastocysts. Initially, germ-line transmission of the mutant allele was demonstrated by Southern blotting but subsequent genotyping has been done by PCR using the primers Wt1 loxP/loxP (Supplementary. Table I).
Generation of epicardial specific Wt1 mutant mice
Epicardial Wt1 mutant mice (Gata5-Cre+/Wt1loxP/gfp) were generated by cross breeding Wt1 loxP/loxP with mice that are heterozygous for Wt1-GFP knockin 4 and Gata5-Cre 5. Transgenic mice were genotyped by PCR using the primers: Gata5cre, Wt1-GFP and Wt1 loxP/loxP (Supplementary Table 1).
Histology and Immunohistochemistry
For histological analysis, wax sections of 6-μm thickness were stained with H&E. For immunostaining, antigens were retrieved by boiling samples in a pressure cooker for 4 minutes in TEG-buffer pH 9 (10 mM Tris, 0.5mM EGTA, pH 9.0). Slides were then incubated in 50 mM NH4Cl/PBS pH 7.4 for 30 min and blocked in 1% BSA, 0.2% gelatine, and 0.05% saponin (3 times, 10 min each). Samples were then incubated overnight at 4°C with the following antibodies: Wt1 1:800 (C19, Santa Cruz); GFP 1:800 (abcam) diluted in 0.1% BSA, 0.3% Triton x-100/ PBS pH7.4; followed by washing with 0.1% BSA, 0.2% gelatine, 0.05% saponin.
The following antibodies were analysed using standard Immunohistochemistry procedure: Anti α-SMA 1:100 (Clone 1A4 Sigma); E-cadherin 1:800 (BD Bioscience); Cytokeratin 1: 200 (Dako); Vimentin 1: 200 (Dako) ; PECAM 1:50 (clone Mec 13.3) and Snail19 1:25. Samples were incubated with the secondary antibodies and counterstained with Vectashield with DAPI or Propidium Iodide (Vector laboratories).
Real-time PCR of FACS-sorted GFP+ epicardial cells
The heart ventricles of Cre+/Wt1loxP/gfp and Cre−/Wt1loxP/gfp mice were trypsinised for 15 minutes at 37°C. RNA isolated from FACS-sorted GFP+ epicardial cells was reverse-transcribed using first strand cDNA kit for RT-PCR (Roche). Analysis of gene expression was carried out by Taqman real-time PCR. The expression levels of Wt1, Snail and Slug were normalized to the housekeeping gene gapdh (Roche). Relative values were shown as the ratio gene/Gapdh levels.
Generation of Wt1loxP/gfp immorto Epicardial cells (CoMEEC)
To generate Wt1 loxP/gfp immortalized epicardial cell lines Wt1gfp/+ mice were crossed with the H-2KbtsA58 “immorto” mice carrying a temperature-sensitive simian virus 40 (SV40) large T antigen 20. Wt1gfp/+ /Immorto +/− were mated with Wt1 loxP/loxP mice. Hearts from E11.5 Wt1 loxP/gfp/Immorto +/− mice were placed on 24 well gelatinized dishes. After 24 hrs, the hearts were removed and the epicardial monolayer of cells attached to the gelatin coated surface was grown until confluent. Cells were propagated at 33°C in DMEM, 10% heat inactivated FCS and 20 ng/mL mouse gamma interferon (Peprotech). We generated tamoxifen inducible Wt1 mutant cells by transfecting Wt1 loxP/gfp immortalized epicardial cells with a CAGGs-CreERT2-IRES-puro R expression construct (kind gift of Dr. Lars Grotewold). Stable clones were generated after selection with 5 μg/ml puromycin.
Migration assays
Immortalised epicardial cells that had been cultured for 6 days in presence or absence of 100nM of tamoxifen were seeded in 6-well culture dishes at a density of 0.5 × 106 cells per well. A wound was incised 24 h later in the central area of the confluent culture, carefully washed to remove detached cells and fresh medium added before being incubated for further 24 h. The images were captured using a live cell imaging system: Zeiss Axiovert 200 fluorescence microscope.
ES cell culture and EB differentiation
Wt1 KO ES cells 21, Snai1 rescue ES cells, Wt1-GFP knockin ES cells and wild-type E14tg2aIV ES cells were used in this study. Briefly, undifferentiated ES cells were cultured on gelatin-coated dishes in BHK-21 medium (Glasgow MEM, GIBCO) supplemented with 10% FCS, 2 mM L-glutamine, 1 mM sodium pyruvate (SIGMA), 0.1 mM non essential amino acids (SIGMA), 0.1mM mercapthoethanol (SIGMA) and leukaemia inhibitory factor (LIF). EB were formed by culturing ES cells (1 × 106 per 9cm bacterial dishes) for the indicated number of days on non-adherent bacterial plates in medium without LIF.
For migration analysis, day 11 EB were transferred to gelatinized plates in DMEM medium and after 6 hours of migration representative images were recorded from each type of EB.
To analyze the differentiation of cardiomyocytes, EB were transferred into gelatinized plates at day 7 of differentiation. EB were monitored for beating from day 1 to 7. Spontaneously contracting EB were counted by visual inspection under a light microscope.
Generation of Snail transfectant cells
The myc-Snail and GFP constructs were made by cloning the coding region of Snai1 and GFP into pcDNA6.2-v5-DEST (Invitrogen). 100 μg of Snai1 and GFP vectors were electroporated into 1 × 107 Wt1 KO ES cells. Stable clones were generated after selection in 10 μg/ml blasticidin. Two independent clones from each construct were analyzed during the rescue experiments: Snai1; Snai1-C1, Snai1-C2; GFP; C1 and C2.
Western Blot (WB) and RT-PCR analysis
For WB, lysates from 7- 11 day EB or six day treated epicardial cells were analyzed using antibodies against E-cadherin (1:2000, Transduction Laboratories), Wt1 (C-19, 1:2000, Santa Cruz Biotechnology), Vimentin (1:1000, ABCAM), α-SMA (1:5000, SIGMA), Pcna (1:10000, Santa Cruz) and Snail (1:100) 19.
For gene expression analysis, total RNA was isolated from six days tamoxifen treated immortalised epicardial cells and EB (3, 7 and 11 days) using an RNA purification kit (Invitrogen). Total RNA was reverse-transcribed as described above. Primers used are listed in Supplementary Table 1.
Luciferase assays and ChIP
ChIP-positive fragments were cloned into a pGL3 plasmid (Supplementary Table 1). These reporter constructs (0.1 μg) were transfected into immortalised epicardial cells, in the presence of indicated amounts of expression construct encoding −KTS Wt1 isoform 22. The total amount of transfected DNA was normalized with lacz plasmid. Renilla plasmid was also cotransfected as a control for efficiency. 24 hours after transfection, firefly luciferase and renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega). To mutate the putative Wt1 binding sequences, a QuikChange Site-Directed Mutagenesis kit (Stratagene) was used.
ChIP assays were performed on immortalised epicardial cells. Immunoprecipitations of the cross-linked chromatin were carried out with Wt1 (rabbit polyclonal antibody, C-19, Santa Cruz Biotechnology) and Snail (rabbit polyclonal antibody, abcam). Rabbit serum served as a negative control and a 1:10 dilution of the ‘input sample’ as positive control. The modification status of histones at the Snai1 and Cdh1 promoter was checked. Immunoprecipitations of the cross-linked chromatin were carried out with the following antibodies anti-acetyl-histone H3 (abcam), anti-H3 (tri methyl K4, abcam) and anti- H3 (tri methyl K27) (upstate). The amplified DNA was separated on 2% agarose gel and visualized with ethidium bromide. Primers used are listed in Table 1.
Snai1 knockdown experiments
Immortalised epicardial cells were cultured in the presence of Snail and control shRNA lentiviral particle (sc-38399-V and sc108080; Santa Cruz Biotehcnology). After three days of culture Snail and control infected cells were transfected with the Cdh1 promoter. Renilla plasmid was also cotransfected as a control for efficiency. The luciferase and renilla activities were measured as described above.
OPT scanning
Embryos were fixed in 4% paraformaldehyde. The samples were then bleached in H2O2 and paraformaldehyde at 4°C. Samples were mounted in 1% Low Melting Point agarose, dehydrated in methanol and then cleared in 1 Benzyl Alcohol: 2 Benzyl Benzoate. The sample was imaged using fluorescence channels 488nm excitation light and imaged using a 520LP filter. The images were reconstructed using in-house software designed as part of the Edinburgh mouse atlas project. Bioptonics 3001 OPT Scanner software was used to generate the 3D embryos image. An MIP (Maximum Image Projection) digital filter was used to increase the over all contrast of the image.
Supplementary Material
Acknowledgments
We thank Dr. Amparo Cano for the kind gift of the Snai1 promoter, Dr. Antonio Garcia de Herreros for Snail antibody and Dr. Pilar Ruiz-Lozano for the Gata5-Cre transgenic mice. We also thank Harris Morrison for the OPT analysis, Anna Thornburn for help maintaining mouse colonies, Craig Nicol for assistance with graphics (Supplementary Fig. 7) and all members of NDH's lab for helpful discussions and comments. This work was supported by a core grant to NDH from the MRC, and by a grant from the Spanish Ministry of Science, MICINN (BFU08-02384) to RMC. A.E. was supported by EuReGene, a FP6 grant by the European Union (05085). O.M.M.E. was supported by an EU Marie Curie (FP6) personal fellowship.
Footnotes
The authors declare no conflict of interest.
Reference List
- 1.Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- 2.Hohenstein P, Hastie ND. The many facets of the Wilms' tumour gene, WT1. Hum. Mol. Genet. 2006;15(Spec No 2):R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 3.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 4.Hosen N, et al. The Wilms' tumor gene WT1-GFP knock-in mouse reveals the dynamic regulation of WT1 expression in normal and leukemic hematopoiesis. Leukemia. 2007;21:1783–1791. doi: 10.1038/sj.leu.2404752. [DOI] [PubMed] [Google Scholar]
- 5.Merki E, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 7.Hosono S, et al. E-cadherin is a WT1 target gene. J. Biol. Chem. 2000;275:10943–10953. doi: 10.1074/jbc.275.15.10943. [DOI] [PubMed] [Google Scholar]
- 8.Spencer HL, et al. E-cadherin inhibits cell surface localization of the pro-migratory 5T4 oncofetal antigen in mouse embryonic stem cells. Mol. Biol. Cell. 2007;18:2838–2851. doi: 10.1091/mbc.E06-09-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner KD, et al. The Wilms' tumor suppressor Wt1 is expressed in the coronary vasculature after myocardial infarction. FASEB J. 2002;16:1117–1119. doi: 10.1096/fj.01-0986fje. [DOI] [PubMed] [Google Scholar]
- 10.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 11.Nishikawa S, Jakt LM, Era T. Embryonic stem-cell culture as a tool for developmental cell biology. Nat. Rev. Mol. Cell Biol. 2007;8:502–507. doi: 10.1038/nrm2189. [DOI] [PubMed] [Google Scholar]
- 12.Wagner N, et al. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms' tumor transcription factor Wt1. Genes Dev. 2005;19:2631–2642. doi: 10.1101/gad.346405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitiku N, Baker JC. Genomic analysis of gastrulation and organogenesis in the mouse. Dev. Cell. 2007;13:897–907. doi: 10.1016/j.devcel.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi Y, et al. High expression of Wilms' tumor suppressor gene predicts poor prognosis in breast cancer patients. Clin. Cancer Res. 2002;8:1167–1171. [PubMed] [Google Scholar]
- 15.Blanco MJ, et al. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21:3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- 16.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lettice LA, et al. The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9695–9700. doi: 10.1073/pnas.96.17.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franci C, et al. Expression of Snail protein in tumor-stroma interface. Oncogene. 2006;25:5134–5144. doi: 10.1038/sj.onc.1209519. [DOI] [PubMed] [Google Scholar]
- 20.Jat PS, et al. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5096–5100. doi: 10.1073/pnas.88.12.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spraggon L, et al. hnRNP-U directly interacts with WT1 and modulates WT1 transcriptional activation. Oncogene. 2007;26:1484–1491. doi: 10.1038/sj.onc.1209922. [DOI] [PubMed] [Google Scholar]
- 22.Niksic M, Slight J, Sanford JR, Caceres JF, Hastie ND. The Wilms' tumour protein (WT1) shuttles between nucleus and cytoplasm and is present in functional polysomes. Hum. Mol. Genet. 2004;13:463–471. doi: 10.1093/hmg/ddh040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.