Abstract
Mycobacteria are a group of obligate aerobes that require oxygen for growth, but paradoxically have the ability to survive and metabolize under hypoxia. The mechanisms responsible for this metabolic plasticity are unknown. Here, we report on the adaptation of Mycobacterium smegmatis to slow growth rate and hypoxia using carbon-limited continuous culture. When M. smegmatis is switched from a 4.6 h to a 69 h doubling time at a constant oxygen saturation of 50%, the cells respond through the down regulation of respiratory chain components and the F1Fo-ATP synthase, consistent with the cells lower demand for energy at a reduced growth rate. This was paralleled by an up regulation of molecular machinery that allowed more efficient energy generation (i.e. Complex I) and the use of alternative electron donors (e.g. hydrogenases and primary dehydrogenases) to maintain the flow of reducing equivalents to the electron transport chain during conditions of severe energy limitation. A hydrogenase mutant showed a 40% reduction in growth yield highlighting the importance of this enzyme in adaptation to low energy supply. Slow growing cells at 50% oxygen saturation subjected to hypoxia (0.6% oxygen saturation) responded by switching on oxygen scavenging cytochrome bd, proton-translocating cytochrome bc1-aa3 supercomplex, another putative hydrogenase, and by substituting NAD+-dependent enzymes with ferredoxin-dependent enzymes thus highlighting a new pattern of mycobacterial adaptation to hypoxia. The expression of ferredoxins and a hydrogenase provides a potential conduit for disposing of and transferring electrons in the absence of exogenous electron acceptors. The use of ferredoxin-dependent enzymes would allow the cell to maintain a high carbon flux through its central carbon metabolism independent of the NAD+/NADH ratio. These data demonstrate the remarkable metabolic plasticity of the mycobacterial cell and provide a new framework for understanding their ability to survive under low energy conditions and hypoxia.
Introduction
Microorganisms show a remarkable metabolic flexibility that allows them to adapt to various environmental changes (e.g. nutrient starvation, oxygen deprivation and various exogenous stress conditions). A crucial feature in the adaptation of any bacterium to alternative energy sources and changing environmental parameters is the balance of oxidative and reductive reactions in the metabolic scheme that are dependent on electron donor and acceptor availability. Mycobacteria are a group of obligate aerobes that require oxygen for growth, but paradoxically have the extraordinary ability to survive and metabolize under hypoxia suggesting a high degree of metabolic plasticity. The mechanisms responsible for this metabolic flexibility are unknown.
The paucity of basic knowledge in this area reflects the limitations of available models to study the adaptation of mycobacteria to slow growth rate and/or hypoxia. A conventional approach has been to put mycobacterial cells into a low metabolic state using the following stimuli; e.g. low oxygen (Wayne model) [1], nutrient starvation [2] and extended stationary phase [3], [4]. These approaches have provided valuable information, but frequently have multiple factors changing throughout the experiment, and the mycobacteria often fail to grow and metabolize.
A powerful method to study gene expression in response to changing environmental conditions, where the growth rate can be maintained at a constant value, is continuous culture. This also allows the researcher to change the growth rate (i.e. dilution rate) while maintaining other factors (e.g. oxygen tension) at a constant value. For example, carbon-limited continuous culture was used to study the effect of growth rate on gene expression in Mycobacterium bovis BCG [5]. Slowing the growth rate of M. bovis BCG from a 23 h to a 69 h doubling time at 70–100% oxygen saturation has been shown to precipitate a transcriptional response more akin to growth of M. tuberculosis in macrophages compared to other in vitro models. However, in the study by Beste et al. [5], the growth rate varied by only 3-fold whereas the change in growth rate during the transition to hypoxia or nutrient limitation is likely to be much greater than this value. Bacon et al. [6] report on the response of M. tuberculosis to lowering oxygen saturation (20% and 1%) at a constant mean generation time of 24 h in glycerol-limited continuous culture. However, no study has yet been performed that dissects both the response to carbon starvation and oxygen limitation simultaneously.
The aim of the current study was to determine the molecular response of M. smegmatis to changing growth rate (15-fold difference) and decreasing oxygen supply (50% to 0.6% oxygen saturation) using carbon-limited continuous culture. We rationalized that this would be a valid model to elucidate the metabolism and energetics of M. smegmatis growth in slowly metabolizing cells and in cells confronted with oxygen limitation.
Results
Establishing Carbon-Limited Continuous Culture of M. smegmatis
M. smegmatis was grown in carbon-limited continuous culture at dilution rates of either 0.15 h−1 (fast) or 0.01 h−1 (slow) that corresponded to doubling times of 4.6 h and 69 h respectively. A doubling time of 69 h has been used in other continuous culture studies with mycobacteria (e.g. M. bovis BCG) to simulate growth rates akin to those in vivo [5]. Oxygen supply was maintained at 50%, 2.5% or 0.6% oxygen saturation and these values were chosen to be comparable with the oxygen tensions observed in the Wayne model [1] i.e. during exponential growth phase (50%), beginning of NRP1 (2.5%) and advanced NRP1 (0.6%), respectively. Oxygen saturation values of 70–100% [7] and 20–50% [6] have been used in other continuous culture studies with mycobacteria. Steady state growth was confirmed in all cultures by monitoring the optical density of the culture as well as oxygen consumption. Carbon limitation was tested by pulsing glycerol into the culture vessel and showing an increase in OD600, as well as respiration rate, and by measuring residual glycerol (Figure 1). The steady-state optical density was significantly lower at low dilution rate (OD600 = 4.2±0.1) compared to high dilution rate (5.1±0.2). The number of viable cells was higher at low dilution rate than at high dilution rate indicating a difference in cell volume (Table 1). This was confirmed by fluorescence microscopy, which revealed a 7-fold difference in cell length between slow and fast growing cells (Figure 1B and 1C). The growth yields on glycerol and the magnitude of the proton-motive force were comparable at both dilution rates suggesting the cells were energetically equivalent (Table 1).
Figure 1. Growth of M. smegmatis in continuous culture.
A: Growth curve of M. smegmatis based on OD600 (solid squares) and residual glycerol concentration (solid diamonds) measured during chemostat cultivation at a dilution rate of 0.15 h−1. B&C: Fluorescence microscopy pictures of Mycobacterium smegmatis grown in continuous cultivation at a dilution rate of (B) 0.15 h−1 (fast) and (C) 0.01 h−1 (slow) and stained with SYBRGreen I.
Table 1. Bulk parameters measured at steady state in continuous culture experiments with M. smegmatis.
| Dilution rate (h−1) | OD600 | CFU (*109)/ml | Yield glycerol (g biomass/g glycerol) | Membrane potential (mV) | ZΔpH (mV)a | ΔµH+ (mV) |
| 0.01 | 4.2±0.1 | 1.5±0.6 | 0.56±0.04 | 75±5 | 55±7 | 130±12 |
| 0.15 | 5.1±0.2 | 0.7±0.3 | 0.62±0.01 | 64±8 | 48±5 | 111±13 |
Extracellular pH was 6.6.
When the oxygen saturation was lowered from 50% to 2.5%, the oxygen demand decreased (stirring rate) and the culture maintained growth at a 69 hours doubling time. At 0.6% oxygen saturation, the doubling time was greater (86 hours) indicating that the chemostat culture was oxygen-limited.
Differential Gene Expression in Response to Slow Growth Rate and Low Oxygen Concentration
A total of 1294 genes of M. smegmatis were differentially expressed (p-value<0.05, expression ratio >2 or<0.5) at low dilution rate compared to high dilution rate. Of these, 691 genes were upregulated and 603 were downregulated. For a complete list of genes, see Dataset S1. At 2.5% oxygen saturation and a dilution rate of 69 h, 28 genes were downregulated and 91 upregulated. For a complete list of genes, see Dataset S1. Most prominently, two gene clusters (msmeg_3932-3955 and msmeg_5241-5246) with homology to DosR-regulated genes in M. tuberculosis were upregulated. Both clusters contain a copy of the gene devR, the homologous gene to dosR in M. tuberculosis. At 0.6% oxygen saturation, the expression ratio of these gene clusters increased and additional genes were differentially regulated. In total, we found 870 genes differentially expressed (p-value<0.05) at 0.6% oxygen saturation compared to 50% oxygen saturation, 429 genes above a 2-fold expression ratio and 441 genes below 0.5-fold threshold level. For a complete list of genes, see Dataset S1.
To check the quality of our microarray data, we used quantitative RT-PCR to validate expression ratios for selected genes (Figure S3). Figure S3 demonstrates that real-time PCR results correlated well with the microarray results.
Energy Metabolism
Energy-limiting conditions
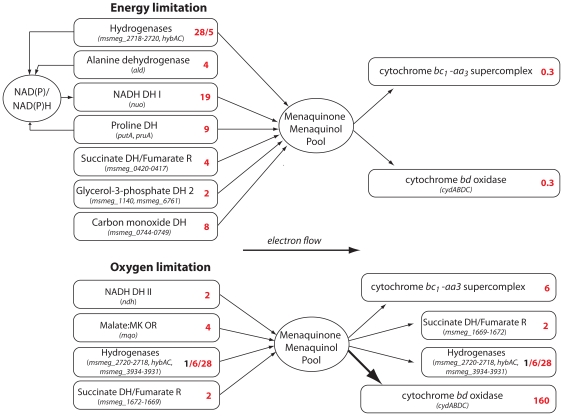
In response to low dilution rate at 50% oxygen saturation, 128 genes involved in energy metabolism were upregulated and 62 downregulated. The majority of the downregulated genes encode components of the oxidative phosphorylation machinery. These included the gene clusters for the F1FO-ATP synthase, succinate dehydrogenase (msmeg_1669-1672), cytochrome bc1-aa3 supercomplex, and cytochrome bd (Table S1). Strikingly, strong upregulation of gene clusters encoding for two Ni-Fe-hydrogenases (msmeg_2262-2264 and msmeg_2720-2713), NADH-quinone oxidoreductase (nuo), propane monoxygenase, carbon-monoxide dehydrogenase and a succinate dehydrogenase (msmeg_0417- 0419) were observed. Furthermore, several primary dehydrogenases proposed to be electron donors to the electron transport chain were upregulated at slow growth rate. These included proline dehydrogenase, alanine dehydrogenase, glycerol-3-phosphate dehydrogenase 2, and a putative pyruvate dehydrogenase (Figure 2). The data for genes with a purported role in energy metabolism is summarized in Table S1 and specifically for hydrogenases in Table 2. Additionally, we found 17 out of 32 cytochrome P450 enzymes, several alcohol and aldehyde dehydrogenases were all upregulated (Dataset S1).
Figure 2. Proposed scheme of enzymes preferentially used in the respiratory chain of M. smegmatis under energy- and oxygen-limiting conditions.
Data shown is based on gene expression ratios (indicated as red numbers next to gene name). See Table 2, Table S1 and Dataset S1 for detailed expression data.
Table 2. Expression ratios of gene clusters encoding for hydrogenases and assembly genes in M. smegmatis at different growth rates and oxygen concentrations.
| Gene ID | Gene name | Slow/Fast1 | Hypoxia2 2.5% O2 | Hypoxia2 0.6% O2 | Description |
| MSMEG_2262 | hybA | 5.4 | 4.1 | 5.7 | hydrogenase-2, small subunit |
| MSMEG_2263 | hybC | 5.4 | 3.7 | 6.2 | hydrogenase-2, large subunit |
| MSMEG_2264 | 2.3 | 3.5 | 4.4 | peptidase M52, hydrogen uptake protein | |
| MSMEG_2271 | hypB | 1.5 | 2.2 | 2.3 | hydrogenase accessory protein |
| MSMEG_2272 | hypA | 1.9 | 2.8 | 1.8 | hydrogenase nickel insertion protein HypA |
| MSMEG_2273 | hypF | 2.0 | 2.0 | 3.0 | [NiFe] hydrogenase maturation protein HypF |
| MSMEG_2274 | hypC | 1.8 | 2.1 | 2.5 | hydrogenase assembly chaperone HypC/HupF |
| MSMEG_2275 | hypD | 1.8 | 3.0 | 5.2 | hydrogenase expression/formation protein HypD |
| MSMEG_2276 | hypE | 2.0 | 2.0 | 2.0 | hydrogenase expression/formation protein HypE |
| MSMEG_2702 | hypD | 3.6 | 1.4 | 2.4 | hydrogenase expression/formation protein HypD |
| MSMEG_2703 | hypC | 7.0 | 1.5 | 3.5 | hydrogenase assembly chaperone HypC/HupF |
| MSMEG_2705 | hypE | 4.9 | 1.2 | 3.7 | hydrogenase expression/formation protein HypE |
| MSMEG_2711 | hypF | 7.0 | 1.6 | 11.2 | [NiFe] hydrogenase maturation protein HypF |
| MSMEG_2712 | hypC | 15.1 | 1.5 | 12.1 | hydrogenase assembly chaperone HypC/HupF |
| MSMEG_2713 | 14.5 | 1.4 | 8.9 | peptidase M52, hydrogen uptake protein | |
| MSMEG_2718 | 15.4 | 0.9 | 3.5 | iron-sulfur cluster-binding protein, Rieske family | |
| MSMEG_2719 | 18.2 | 1.1 | 6.6 | hydrogen:quinone oxidoreductase | |
| MSMEG_2720 | 27.8 | 1.2 | 3.1 | NADH ubiquinone oxidoreductase, 20 kda subunit | |
| MSMEG_2721 | hypB | 7.9 | 1.0 | 1.0 | hydrogenase accessory protein HypB |
| MSMEG_2722 | hypA | 1.5 | 1.2 | 0.8 | hydrogenase nickel insertion protein HypA |
| MSMEG_3927 | 1.4 | 5.2 | 18.6 | peptidase M52, hydrogen uptake protein | |
| MSMEG_3928 | 2.6 | 2.2 | 7.6 | [NiFe] hydrogenase, alpha subunit, putative | |
| MSMEG_3929 | 1.0 | 8.2 | 26.9 | [NiFe] hydrogenase, delta subunit, putative | |
| MSMEG_3930 | 1.2 | 10.0 | 28.0 | [NiFe] hydrogenase, gamma subunit, putative | |
| MSMEG_3931 | 1.0 | 16.7 | 67.3 | [NiFe] hydrogenase, beta subunit, putative |
Expression ratios with a p-value<0.05 are indicated with bold numbers.
Gene expression ratios of slow growing (0.01 h−1) versus fast growing (0.15 h−1) chemostat cultures at 50% oxygen saturation.
Gene expression ratios of slow growing (0.01 h−1, 50% oxygen saturation) versus slow growing cultures at either 2.5% or 0.6% oxygen saturation.
Oxygen-limiting conditions
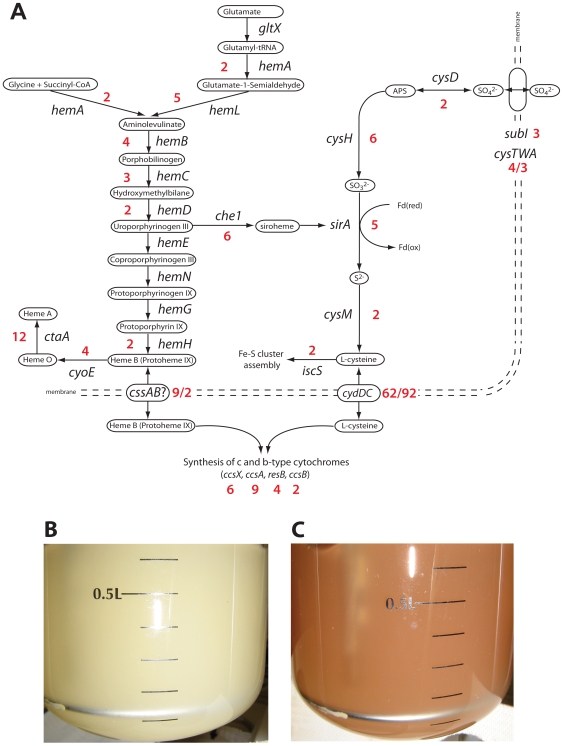
When the oxygen saturation was lowered to 2.5%, 8 genes involved in energy metabolism were upregulated. These included, two Ni-Fe-hydrogenase gene clusters [msmeg_2262-2264 (Hyd1) and msmeg_3931-3927 (Hyd3)], soluble pyridine nucleotide transhydrogenase sthA and a conserved hypothetical protein. In contrast, at an oxygen saturation of 0.6% we observed a distinct gene expression pattern (Table S1). The high-affinity cytochrome bd oxidase was upregulated >50-fold, along with heme and cysteine biosynthesis pathways (Figure 3A), and genes annotated or with similarity to known cytochrome c biogenesis proteins (msmeg_0971-0974). Furthermore, cysteine desulfurase (msmeg_2347), a protein involved in Fe-S cluster assembly [8] was upregulated 2-fold. Unexpectedly, given the low oxygen saturation, cytochrome bc1-aa3 complex was upregulated (5-fold), along with the cytochrome aa3 controlling protein msmeg_3117 (12-fold). Nearly all dehydrogenases that were upregulated during slow growth showed very low mRNA levels under hypoxia (Table S1). The only exceptions were the hydrogenases, which showed a 2- to 5-fold increase in expression (Table 2). This indicates that M. smegmatis makes a distinct transcriptional switch from energy starvation to oxygen limitation and that hydrogenases would appear to play an important role in this adaptation. Furthermore, oxygen-limited growth of M. smegmatis was accompanied by a change in the coloration of the cells from a light yellow pigmentation to a red pigmentation (Figure 3B and C).
Figure 3. Effect of oxygen limitation on M. smegmatis growing at slow growth rate.
(A) Diagram of the upregulated heme, cysteine and cytochrome c biosynthesis pathways in M. smegmatis under oxygen-limited growth. Numbers in red represent gene expression ratios (0.6% oxygen saturation/50% oxygen saturation) above the 2-fold threshold. B&C: Comparison of M. smegmatis mc2155 wild type growing in continuous culture at a dilution rate of 0.01 h−1 (69 h doubling time) with 50% oxygen saturation (B) or 0.6% oxygen saturation (C).
TCA and Glyoxylate Shunt Pathways
Energy limiting conditions
The majority of genes of the TCA cycle and glycolysis/gluconeogenesis did not show a change in gene expression at low growth rate. The two putative succinate dehydrogenase operons [msmeg_0420-0417 (Sdh1) and msmeg_1672-1669 (Sdh2)] showed opposite expression ratios. While Sdh1 was upregulated 4-fold, Sdh2 was downregulated 3-fold like most other genes of the respiratory chain. Succinate dehydrogenase plays an important role in respiration and the TCA cycle. Both enzyme complexes have close homologues in M. tuberculosis (Rv0250-Rv0247, Rv3316-Rv3319). Additionally, three genes annotated as subunits of pyruvate dehydrogenase (msmeg_4710-4712) were strongly upregulated (up to 11-fold) in response to slow growth rate. The genome of M. smegmatis encodes two isocitrate lyase enzymes and both are homologous to those found in M. tuberculosis CDC1551. At low dilution rate, one isocitrate lyase (msmeg_3706) was upregulated while the second copy msmeg_0911 was downregulated (Dataset S1).
Oxygen-limiting conditions
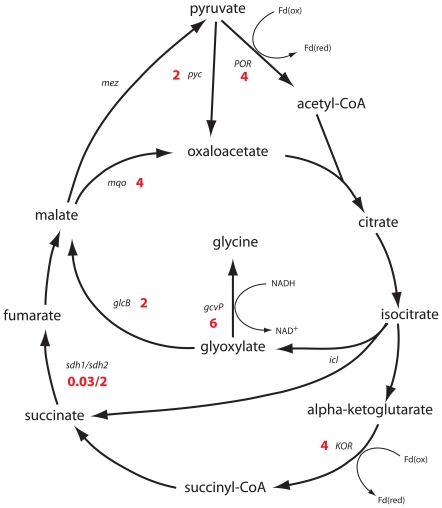
Under oxygen-limiting conditions (0.6% oxygen saturation), several enzymes of the TCA cycle and glyoxylate shunt were differentially regulated compared to cultures grown at 50% oxygen saturation (Figure 4). Genes for malate synthase were upregulated as well as glycine dehydrogenase, which converts glyoxylate to glycine and recycles NADH to NAD+. Furthermore, M. smegmatis does not harbour a NAD+-dependent malate dehydrogenase, but instead expresses a malate:quinone oxidoreductase (MQO), which feeds electrons directly into the menaquinone pool. MQO was upregulated 4-fold under hypoxia (Figure 4). Additionally, a putative pyruvate:ferredoxin oxidoreductase or α-ketoglutarate:ferredoxin oxidoreductase (4-fold p = 0.02), pyruvate carboxylase (2.5-fold, p = 0.02), succinate dehydrogenase (Sdh2) (2-fold p = 0.02) and phosphoenolpyruvate synthase (23-fold, p = 0.08) were upregulated (Dataset S1). Genes downregulated under these conditions included malic enzyme, succinate dehydrogenase (Sdh1), pyruvate kinase, putative pyruvate dehydrogenase and succinic semialdehyde dehydrogenase (Dataset S1).
Figure 4. Expression of genes encoding enzymes of the TCA cycle and glyoxylate shunt during growth of M. smegmatis under microaerobic conditions.
Annotation: sdh1/sdh2 – succinate dehydrogenase (msmeg_0420-0417/msmeg_1672-1669), pyc – pyruvate carboxylase, POR – pyruvate:ferredoxin oxidoreductase, mqo – malate:quinone oxidoreductase, mez – malic enzyme, gcvP - glycine dehydrogenase, glcB – malate synthase, icl – isocitrate lyase, KOR – alpha-ketoglutarate:ferredoxin oxidoreductase.
Regulation and signal transduction
The transcriptional response to 2.5% oxygen saturation at slow growth rate was manifested predominantly in the upregulation of two gene clusters with 31 genes (msmeg_3926-msmeg_3955) and 6 genes (msmeg_5241-msmeg_5246) respectively. Both clusters are near the two-component regulatory system DevR/DevS, of which M. smegmatis harbors two copies (see Dataset S1). Most of these genes have homologues in M. tuberculosis and are part of the Dos regulon [9]. Exceptions to this are the 5 genes encoding a Ni-Fe hydrogenase, a cation-transporting P-type ATPase and the gene encoding for phosphoenolpyruvate synthase. The expression pattern of these genes was similar at 0.6% oxygen saturation although a few genes showed increased expression (Dataset S1). Three two-component regulatory systems (i.e. msmeg_0854/msmeg_0856, msmeg_4968/msmeg_4969 and msmeg_6236/msmeg_6238) were upregulated at slow growth rate while senX3/regX3 and msmeg_5487/msmeg_5488 (MprA) were upregulated under hypoxia.
The M. smegmatis genome is annotated to have 29 sigma factors and 4 anti-sigma factors. At slow growth rate, the expression ratio of 17 sigma factors and 3 anti-sigma factors was significantly different from 1 (p<0.05) (Dataset S1). All but one sigma factor (sigF) and one anti-sigma factor were upregulated. The genes sigG, sigH, sigL, sigJ, sigE (all with homologues in M. tuberculosis) and msmeg_1970 were upregulated more than 2-fold (Dataset S1). Sigma factor genes sigB, sigE, sigD, sigH and sigJ showed increased mRNA levels under oxygen limiting conditions, while sigG and sigL levels were decreased. One out of 6 WhiB transcription factors, i.e. msmeg_6199, showed a >4-fold induction in response to slow growth. The homologue in M. tuberculosis is WhiB4 and is believed to be involved in redox signaling [10]. When the oxygen saturation was lowered to 0.6%, the same gene was downregulated 20-fold. Additionally, msmeg_1919 (WhiB1) responded to hypoxia with an 8-fold upregulation (Dataset S1).
Hydrogenase Gene Clusters in M. smegmatis and Characterization of a Hydrogenase Mutant
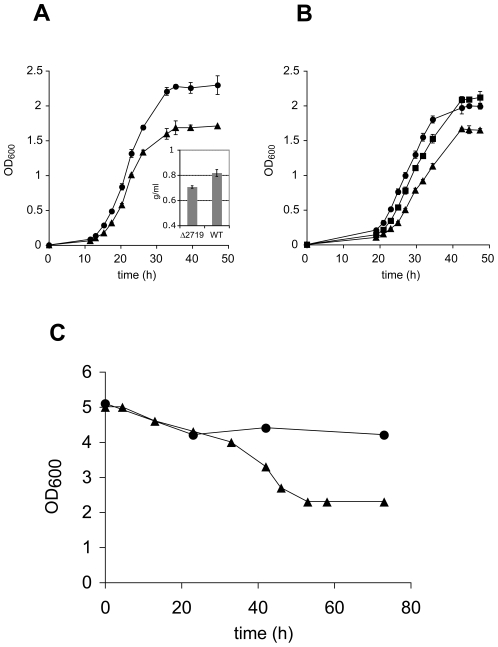
There are three different putative hydrogenase gene clusters (complexes) in the genome of M. smegmatis, two of which showed strong upregulation under energy limiting conditions and all three were upregulated under hypoxia (Table 2). All three enzyme complexes are predicted to have a Ni-Fe center (or Ni-Fe-Se) and to be soluble proteins (PFAM and THMMH analysis). According to these predictions, their cofactors could be either NADH, ferredoxin or coenzyme F420. A phylogenetic analysis on the catalytic subunit of each of the three enzymes revealed that they share homology with hydrogenases found in cyanobacteria (msmseg_2263), other soil microorganisms (msmeg_2719), and pathogenic microorganisms (msmeg_3928) (Figure S1). To elucidate the role of these hydrogenases in response to slow growth rate (i.e. energy limitation), we constructed a markerless mutation in msmeg_2719, a gene encoding a putative hydrogen:quinone oxidoreductase that was the most strongly upregulated of the three hydrogenases in response to growth rate (see Table 2). The gene disruption in msmeg_2719 was confirmed via Southern hybridization (Figure S2). When grown in LBT medium (batch growth), the mutant exhibited a 20% reduction in biomass compared to the isogenic wild-type parent strain (Figure 5A). Complementation of msmeg_2719 with the operon msmeg_2720-msmeg_2718, under control of its native promoter, restored growth behavior to wild type levels (Figure 5B). The difference in growth yield was significantly greater (40%) when grown in continuous culture at a dilution rate of 69 h (50% oxygen saturation) demonstrating the essential role of this hydrogenase in the adaptation of M. smegmatis to energy limitation. No H2 gas was detected in the culture headspace from both batch and continuous culture using gas chromatography suggesting that M. smegmatis did not produce detectable levels of H2 using the culture conditions employed in this study.
Figure 5. Comparative analysis of wild-type M. smegmatis mc2155 with an isogenic hydrogenase mutant (Δ2719).
A. Growth curve of M. smegmatis mc2155 wild-type (solid circles) versus strain Δ2719 (solid triangles) in LB Tween80. Insert: Biomass of the two strains based on dry weight measurements. Error bars represent standard deviations from 3 biological replicates. B. Complementation experiment: Growth curve of M. smegmatis mc2155 mutant strain Δ2719_pOLYG (solid triangles), strain wt_pOLYG (solid circles) and the complemented mutant strain Δ2719_pOLYGHyd2 (solid squares) in LB Tween80. C. Optical density of wild-type strain (solid circles) and mutant strain Δ2719 (solid triangles) measured during chemostat runs at a dilution rate of 0.01 h−1. Time zero represents start of chemostat mode.
Discussion
Adaptation of M. smegmatis to Low Growth Rate Activates the Expression of Alternative Primary Dehydrogenases and Hydrogenases for Energy Generation and Unique Pathways for Scavenging Carbon
The results of this study demonstrate that M. smegmatis represents an important model to dissect the physiological response mycobacteria exhibit during adaptation to low energy conditions and hypoxia. As mycobacteria make the transition from fast to slow growth rate, the cell adjusts its metabolic machinery to match physiological demands for energy to drive catabolic and anabolic reactions in the cell. This effect was manifested by a downregulation of respiratory chain components commensurate with decreased rates of oxygen consumption. Despite the lower rates of respiration, the membrane potential was maintained at the same magnitude as fast growing cells. This suggests that energy-limited cells switch to more energy-efficient mechanisms for membrane potential generation and this was supported by the upregulation of the proton-translocating NADH:menaquinone dehydrogenase complex I. To power the respiratory chain under energy-limited conditions, mycobacteria switch to alternative primary dehydrogenases to provide reducing equivalents for respiration. For example, succinate dehydrogenase, alanine dehydrogenase, Ni-Fe-hydrogenases and proline dehydrogenase were all upregulated in response to slow growth rate (Figure 2).
The physiological adaptation of M. smegmatis to energy limitation included the induction of enzymes to transport sugars and degrade a wide array of natural and xenobiotic compounds. However, apart from glycerol, Tween80 and sodium-glutamate (0.05%), there were no additional carbon sources present in the growth medium. The observed derepression of alternative catabolic functions in the absence of the respective inducers (substrates) presumably is advantageous in many natural growth situations [11]. Two putative succinate dehydrogenases were the only TCA cycle enzymes that showed induction/repression under energy limiting conditions. The role of succinate dehydrogenase and the reason for the presence of two putative Sdh enzymes with different membrane-spanning regions in mycobacterial species is still unclear [12]. M. tuberculosis contains close homologues of both Sdh enzymes and expresses an additional fumarate reductase. Succinate dehydrogenase (Sdh) and fumarate reductase (Frd) are enzymatically and structurally similar proteins. In mycobacteria the membrane-bound electron acceptor for Sdh is menaquinone, indicating that the conversion of fumarate to succinate is a thermodynamically favourable reaction [12]. The M. smegmatis genome does not encode a designated fumarate reductase, thus we hypothesize that at least one of the annotated Sdh enzymes could function as a fumarate reductase suggesting that fumarate could be used as an electron acceptor in the absence of oxygen.
Adaptation to Hypoxia by M. smegmatis Results in the Upregulation of Oxygen Scavenging Oxidases, Low Redox Potential Electron Carriers and Hydrogenases
All mycobacteria species are obligate aerobes and are therefore faced with a number of energetic challenges as the cells encounter hypoxic conditions (e.g. recycling of reducing equivalents). Our data indicates that M. smegmatis adopts three strategies to metabolize under hypoxia. Firstly, the cell induces the high-affinity (oxygen scavenging) cytochrome bd oxidase and machinery needed for its assembly and synthesis (i.e. porphyrin/heme and cysteine biosynthesis). Secondly, it switches to NAD+/NADH-independent enzymes, especially those with ferredoxins as a redox partner. Thirdly, soluble hydrogenases are upregulated to either oxidize hydrogen or produce hydrogen, or both activities operate in concert with other energetic partners.
The high-affinity cytochrome bd oxidase is able to scavenge oxygen under microaerobic conditions and by operating in concert with menaquinone oxidoreductases (ndh2) and the proton-translocating cytochrome bc1-aa3 supercomplex would result in the formation of an electrical potential at low oxygen tensions. During M. tuberculosis infection of macrophages, the cytochrome bc1-aa3 complex is downregulated, but this is based on a relative comparison with broth cultures [13] and therefore this most likely reflects the change in growth rate and not oxygen per se. Our results also show a downregulation of the terminal oxidases in response to slow growth rate, but an upregulation (relative to aerobic slow growth) when switched to hypoxia. Upregulation of cytochrome bd in mycobacteria has been observed in vitro [14] and in vivo [15], however the designated role and the regulator(s) of this enzyme are still unknown. In Streptomyces coelicor, a transcriptional repressor of cytochrome bd was identified and named Rex [16]. This NADH/NAD+ redox sensor regulates the expression of cydABDC as well as the hemACDB operon [16]. No homologue of this protein has been found in mycobacterial species, however the simultaneous upregulation of both operons observed in our study could point to a common regulatory protein.
A novel, but yet unexplained observation is the red pigmentation observed in hypoxic cultures (Figure 3A&B). This could be due to the increased expression of heme containing cytochromes, which is supported by our expression data showing strong upregulation of heme/porphyrin, cytochromes and cysteine biosynthesis pathways as well as a potential heme transporter (CssAB) [17] (Figure 3A). It has been suggested that the presence of porphyrins is the main reason for the slightly red color in Mycobacteria, because the absence of porphyrins in iron-deficient grown cells has been reported to account for their very pale appearance [18].
An unanswered question in the physiology and metabolism of mycobacteria under hypoxia is how cells recycle reducing equivalents (e.g. NADH) in the lack of exogenous electron acceptors and maintain metabolic flux through the TCA cycle. NADH-producing enzymes commonly found in the TCA cycle of bacteria (e.g. α-ketoglutarate dehydrogenase and malate dehydrogenase) are absent in the genome of M. smegmatis. Instead, these enzymes are replaced with a malate:quinone oxidoreductase (MQO) and a putative α-ketoglutarate:ferredoxin oxidoreductase (KOR) and/or pyruvate:ferredoxin oxidoreductase (POR), which reduce menaquinone or ferredoxin, respectively. It has been suggested that MQO in C. glutamicum allows cells to attain a higher TCA cycle flux independently of the NADH/NAD+ ratio [19]. In E. coli MQO is regulated by the oxygen limitation sensor ArcBA [20]. The gene msmeg_4645 is annotated as KOR and its neighbouring gene, msmeg_4646, encodes a protein with a POR family domain. This raises the possibility that this gene pair encodes for an enzyme that is able to metabolize both α-ketoglutarate and pyruvate under hypoxia. Support for this idea comes from a recent report demonstrating that M. tuberculosis does indeed express an “anaerobic type” α-ketoglutarate:ferredoxin oxidoreductase (KOR) (Rv2454c/Rv2455c) and a mutant defective in this enzyme is impaired for aerobic growth in the absence of CO2 [21]. The strategy of substituting NAD+-dependent with NAD+-independent enzymes ensures functionality of the TCA cycle under hypoxia and this appears to apply to other metabolic pathways. We observed the induction of several ferredoxin-reducing and -oxidizing enzymes: Ni-Fe hydrogenases, sulfite reductase, ferredoxin–NADP+ reductase, several cytochrome P450s (CYP) and for ferredoxin itself. This indicates that under hypoxic conditions, M. smegmatis is dependent on the alternative electron carrier ferredoxin, a feature only reported for anaerobic bacteria. Recently, it was shown that a ferredoxin-dependent CYP is essential in M. tuberculosis [22]. Notwithstanding this, NADH recycling remains important as demonstrated by the induction of glycine dehydrogenase, an enzyme that produces NAD+ [23], [24].
M. smegmatis has assembled a large arsenal of tools to degrade complex substrates and to adapt to large fluctuations in energy and oxygen supply. This metabolic plasticity allows the bacterium to survive in diverse environmental compartments, such as different soils, water distribution systems [25] and under human skin [26]. M. smegmatis harbors gene clusters encoding for three putative soluble hydrogenases that are differentially regulated in response to growth rate and hypoxia. Hydrogenases catalyze the reversible reaction: 2H+ + 2e− ⇔ H2 and represent the ideal mechanism to deal with fluctuations in energy and oxygen supply [27]. Under energy limiting conditions, hydrogenases can scavenge hydrogen as an energy source and under hypoxia, hydrogen can be produced as a redox sink and ultimately re-utilized by hydrogenases when the intracellular carriers become less reduced as observed for many fermentative organisms [28]. To validate this proposal, we disrupted msmeg_2719 and this mutant exhibited a significantly reduced growth yield compared to the wild type, indicating an essential role of hydrogenases in the adaptation of Mycobacteria to low energy environments. It has been shown that M. smegmatis, among other mycobacterial species, can oxidize molecular hydrogen in the presence of carbon monoxide implying that they indeed express a functional hydrogenase [29]. However, in our experiment no additional hydrogen was added so the only hydrogen available could be scavenged from air or produced by the cells themselves. We could not detect H2 gas production in either batch or continuous growth and this data would support the notion that hydrogen is rapidly recycled in M. smegmatis, as it has been observed in Salmonella typhimurium [30].
M. tuberculosis harbours a single gene cluster for a putative membrane-bound hydrogenase complex and this complex has been shown to be upregulated during M. tuberculosis infection of human macrophage-like cells [31]. Moreover, the transcription of the first genes of the hycP/hycQ - containing operon was shown to be upregulated during anaerobic adaptation in several studies and is thought to be part of the DevR/DevS regulon [4], [6], [9], [32], [33]. Boshoff & Barry (2005) hypothesize that the hyc operon in M. tuberculosis is regulated by other signals, in addition to low oxygen tension, because it was upregulated in resting as well as activated murine bone marrow macrophages [13]. Both hycP and hycQ (encoding for a possible hydrogenase components) were shown to be essential for optimal growth [34]. The putative hydrogenase gene cluster of M. tuberculosis shows homology to components of hydrogenase 4 and 3 complex of Escherichia coli, the latter of which can catalyze hydrogen evolution at acidic pH [35]. It is tempting to argue that the acidic, hypoxic and lipid-rich environment in the macrophages might require the expression of a hydrogenase to help with the recycling of reducing equivalents under these conditions. Alternatively, hydrogen could serve as an additional means of energy in the host, a much better energy source than host lipids. The use of hydrogen as an electron donor for respiration in the host by pathogenic bacteria is not unprecedented [36], [37], [38], but has been overlooked in the pathogenesis of mycobacteria.
In summary, we propose that mycobacteria adapt to slow growth rate and hypoxia using a battery of strategies that involve the use of alternative primary dehydrogenases and hydrogenases to maintain the flow of reducing equivalents to the electron transport chain during conditions of energy limitation. When the cell is additionally confronted with hypoxia, mycobacteria adapt further by switching to hydrogenases as a potential conduit for disposing of electrons in the absence of exogenous electron acceptors, and unique electron carriers (ferredoxins) that are more akin to those employed by anaerobic bacteria. These data demonstrate the incredible metabolic diversity that mycobacteria possess and provides a framework for understanding their ability to survive for prolonged periods of time under low energy conditions and hypoxia. The regulators and signaling pathways required for this extraordinary adaptive ability remains to be uncovered, but are clearly unique.
Materials and Methods
Strains and Media
Mycobacterium smegmatis strain mc2155 [39] was used in all experiments. For batch culture experiments, either Middlebrook 7H9 medium (Difco) or Luria-Bertani broth (diluted 3×) both supplemented with 0.05% (w/v) TWEEN 80 was used. For continuous cultivation, Middlebrook 7H9 was supplemented with Na2HPO4 (1.25 gL−1) and KH2PO4 (0.5 gL−1) to buffer the growth medium at pH 6.6±0.2 for high bacterial concentrations (OD600≈5). The medium contained 27 mM glycerol, 0.1% antifoam and 0.4% TWEEN 80.
DNA Manipulation and Cloning of Constructs
All molecular biology techniques were carried out according to standard procedures [40]. Restriction or DNA modifying enzymes and other molecular biology reagents were obtained from Roche Diagnostics or New England Biolabs. Genomic DNA of M. smegmatis was isolated as described previously [41]. All primer sequences are listed in Table S2. To create a markerless deletion of msmeg_2719, a 1058 bp fragment flanking msmeg_2719 on the left, including 208 bp coding sequence, was amplified with primers Hyd2.1 and Hyd2.2 and a 985 bp fragment flanking msmeg_2719 on the right, including 136 bp coding sequence, was amplified with primers Hyd2.3 and Hyd2.4. The two products were fused by PCR-overlap extension [42], cloned into the SpeI site of the pPR23-derived [43] vector pX33 [41], creating pHyd2KO, and transformed into M. smegmatis mc2155. Deletion of msmeg_2719 was carried out using the two-step method for integration and excision of the plasmid as described previously [44]. Correct integration and excision were confirmed by Southern hybridization analysis (Figure S2) as described previously [41]. The deletion resulted in loss of 82% of the msmeg_2719 coding sequence, creating strain Δ2719. For complementation of the msmeg_2719 deletion, the operon msmeg_2720-2718, including 72 bp downstream and 413 bp upstream DNA, was PCR amplified with primers cHyd2f and cHyd2r and cloned as a HindIII fragment into the integrative E. coli/mycobacteria shuttle vector pOLYG [45], creating plasmid pOLYHyd2. This construct was transformed into the Δ2719 strain to yield strain Δ2719_pOLYHyd2. Empty pOLYG plasmid was transformed into the wild type as well as the Δ2719 strain to yield strain wt_pOLYG and Δ2719_pOLYG respectively.
Batch and Continuous Culture
Prior to each experiment, bacteria were inoculated from a cryo-culture (−80°C) onto a LBT plate and incubated for 3 days at 37°C. A single colony was picked from the plate and inoculated into the 5 ml batch medium (LBT or 7H9) in a 30 ml glass universal and incubated at 37°C on a rotary shaker at 200 rpm. At an OD600 between 0.3 − 0.5, a sample was withdrawn to inoculate the bioreactor (700 ml) or batch culture (500 ml Erlenmeyer flask containing 50 LBT medium) to a starting OD600 of 0.005. The flasks were incubated at 37°C on a rotary shaker at 200 rpm. Before inoculation, the culture medium in the bioreactor was stirred at 320 rpm at 37°C for at least 10 minutes to fully saturate the water with oxygen. This corresponded approximately to an O2 concentration of 6.9 mg/L. The oxygen probe was then set to 100%. After inoculation, the culture was left in batch mode maintaining 50% oxygen saturation until OD600 reached around 80% of steady-state OD600 and then switched to chemostat mode either at a dilution rate of 0.15 h−1 or 0.01 h−1 corresponding to doubling times of 4.6 h and 69 h respectively. The chemostat was then left running for at least two volume changes before cell harvest. Glycerol concentration was measured according to Garland & Randle [46].
Cells were harvested into cold glycerol saline [47]. In short, 50 ml of culture was extracted with a syringe. Four centrifugation tubes filled with 20 ml of cold glycerol saline (3:2) (−20°C) were then mixed with 10 ml of culture by vigorous shaking and immediately placed into the centrifuge. Samples were centrifuged for 20 min at 27,200 g at −20°C in a Beckman J2-21M/E with a JA-20 rotor. After centrifugation, the supernatant was removed, the cell pellet resuspended in glycerol saline (1∶1) (−20°C) transferred to a 1.5 ml Eppendorf tube and snap frozen in a dry ice ethanol bath. The samples were stored at −80°C until further use.
For the analysis of gene expression at low oxygen concentration, five individual chemostats were performed at the low dilution rate (D = 0.01 h−1) with 2.5% oxygen saturation and another five with 0.6% oxygen saturation. These chemostats were first brought into steady state at 50% oxygen saturation (as described above) before lowering the oxygen concentration slowly over two consecutive days. All other parameters were left unchanged. At an oxygen saturation of 0.6% the chemostat was truly oxygen-limited. The growth rate of the culture was 0.008±0.0005 h−1.
Fluorescence Microscopy
Bacterial cell samples were withdrawn from the chemostat and 100 µl stained with 1 µl of SYBRGreen I stain. After 15 min incubation the sample was analyzed by epifluorescence microscopy (Olympus BX51) equipped with a 100× magnification lens and a bandpass filter at 520±30 nm.
RNA Extraction
Total RNA was extracted using TRIZOL® reagent (Invitrogen) according to the manufacturer's instructions. Cell lysis was achieved by three cycles of bead-beating in a Mini-Beadbeater (Biospec) at 5,000 rpm for 30 sec. DNA was removed from the RNA preparation by treatment with 2 U RNase-free DNase using the TURBO DNA-free kit (Ambion), according to the manufacturer's instructions. RNA concentrations were determined using a NanoDrop® ND-1000 spectrophotometer and the quality of RNA checked on a 1.2% agarose gel.
Quantitative Real-Time PCR
cDNA was synthesized from 1 µg of RNA for each sample with the SuperScript III reverse transcriptase kit (Invitrogen). After cDNA synthesis, real-time PCR was conducted according to the manufacturer with Platinum®SYBR®Green qPCR SuperMix-UDG with ROX (Invitrogen). Primers (Invitrogen) for 10 genes (Table S3) were designed with the publicly available Primer 3 software. Primer optimization was performed. The real-time PCR reactions were conducted in ABI Prism®7500 (Applied Biosystems) and the results were normalized to the gene sigA (msmeg_2758) as endogenous control (Figure S3).
Microarray Analysis
Microarray analysis was performed using arrays provided by the Pathogen Functional Genomics Research Center (PFGRC) funded by the National Institute of Allergy and Infectious Diseases using protocols SOP# M007 and M008 from The Institute of Genomic Research (TIGR) [48]. Ten µg of total RNA was used to create cDNA labeled with aminoallyl dUTP (Sigma). Fluorescent Cy3 and Cy5 dyes (GE Healthcare) were then covalently attached to the aminoallyl tags. Each pair of differentially labeled probes was resuspended in 60 µl of hybridization buffer (500 µl formamide, 250 µl 20× SSC, 5 µl 10% SDS, 245 µl ultrapure water) and hybridized to the microarray slide overnight in a 42°C water bath. Slides were then washed in increasingly stringent wash conditions (3 times 5 min 1× SSC 0.1% SDS, 3 times 5 min 0.1× SSC 0.1% SDS, 3 times 5 min 0.1× SSC and a final dip in 0.22 µm-filtered MQ water). Arrays were scanned in a Genepix 4000A scanner and spots quantified with TIGR Spotfinder. The data was then processed in TIGR MIDAS. Two color tiff images of the microarrays were visually inspected for defects and obvious spatial biases. Prior to normalization pin-tip (block) intensity box plots were used to detect slides exhibiting spatial and/or pin biases, with post-normalization intensity box plots and MA plots used to assess the effectiveness of normalization in correcting these biases. RI plots were also used to identify outlier slides and for assessing the effectiveness of the normalization process, and Z-score histograms were used to look for slides with abnormal intensity distributions. Slides that did not pass the quality control process were rejected, and repeat hybridizations were performed. Post normalization (Total Intensity and LOWESS) the in-slide replicate spots were averaged before expression ratios were calculated. The results from 5 biological replicates including two dye swaps were then subjected to a t-test without false discovery correction in TIGR MeV software. The analysis was used as a ranking method. For a general overview, genes with expression ratios of >2 and<0.5 and a p-value<0.05 were used for data interpretation. In a second step data was extracted for all genes involved in energy metabolism. In all tables, genes with a p-value of<0.05 are marked with a grey background. Expression ratios and p-values for all genes are listed in Dataset S1. All data has been deposited at Gene Expression Omnibus (GEO, NCBI) with the accession number GSE17520.
Gas Chromatography
Gaseous end products were analyzed by gas chromatography by using a HP 5890 Series II gas chromatograph (Hewlett Packard Co., California, USA) equipped with a thermal conductivity detector and Carbosphere 80/100 column (Alltech, Illinois, USA). Nitrogen was used as the carrier gas at a pressure of 50 psi and a flow rate of 30 ml/min. Oven and detector temperature were 150°C and 180°C respectively.
Phylogenetic Analysis
For phylogenetic analysis, the amino acid sequences of the proteins of interest were compared to the database of MicrobesOnline (http://www.microbesonline.org/) using FastBLAST. For proteins with more than 40% identity, the amino acid sequence was downloaded and all the sequences of interest were aligned using the Clustal W method in MegAlign (DNASTAR, Lasergene) software. The alignment was then transferred to TreeView X software and depicted as a Phylogram.
Supporting Information
(0.96 MB PDF)
Expression ratios of genes involved in energy metabolism. Expression ratios with a p-value<0.05 are indicated with bold numbers.
(0.22 MB DOC)
Primer sequences used for quantitative RT-PCR.
(0.05 MB DOC)
Primer sequences used for deletion of msmeg_2719 and complementation.
(0.03 MB DOC)
Phylogenetic trees of the large subunits of the three hydrogenases found in the genome of M. smegmatis mc2155. Proteins with >40% identity are depicted.
(3.47 MB TIF)
Construction of an unmarked msmeg_2719 deletion mutant of M. smegmatis mc2155. A. Schematic diagram of the two-step approach for deletion of msmeg_2719. The knockout construct consisted of two fragments flanking msmeg_2719 on the left (LF) and right (RF) in PX33. Integration of the vector (thick black line) into the chromosome (thin black line) via the left flank (Int LF) or right flank (Int RF) and subsequent deletion of msmeg_2719 (KO) are shown. Restriction sites of SmaI (S) and fragment sizes as detected in Southern hybridization are indicated. Drawing not to scale. WT, wild-type. B. Southern hybridization analysis of the integration event. Left panel, 1. crossover: SmaI-digests of genomic DNA of wild-type mc2155 (lane 1) and a candidate colony (lane 2) were probed with radiolabeled left flank PCR product of the deletion construct. Right panel, 2. crossover: Southern hybridization of msmeg_2719 deletion. Analysis of msmeg_2719 deletion strain (line 1) and wild-type mc2155 (lane 2) was performed as in the left panel. Molecular masses are indicated in kb. M, marker.
(3.30 MB TIF)
Validation of M. smegmatis gene expression ratios (slow growth rate at 50% oxygen saturation versus fast growth rate at 50% oxygen saturation) by quantitative RT-PCR (qPCR) (grey bars) compared to microarray results (black bars). For qPCR the expression of each gene was normalized to the expression of sigA. Error bars represent standard deviations of gene expression ratios from three biological replicates for each condition.
(5.48 MB TIF)
Acknowledgments
We thank Les McNoe from Otago Genomics Facility for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: New Zealand Lottery Health, Health Research Council NZ, The Maurice Wilkins Centre for Molecular Biodiscovery, Swiss National Science Foundation and National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 3.Hampshire T, Soneji S, Bacon J, James BW, Hinds J, et al. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinb) 2004;84:228–238. doi: 10.1016/j.tube.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voskuil MI, Visconti KC Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Beste DJ, Laing E, Bonde B, Avignone-Rossa C, Bushell ME, et al. Transcriptomic analysis identifies growth rate modulation as a component of the adaptation of mycobacteria to survival inside the macrophage. J Bacteriol. 2007;189:3969–3976. doi: 10.1128/JB.01787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon J, James BW, Wernisch L, Williams A, Morley KA, et al. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84:205–217. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Beste DJ, Espasa M, Bonde B, Kierzek AM, Stewart GR, et al. The genetic requirements for fast and slow growth in mycobacteria. PLoS One. 2009;4:e5349. doi: 10.1371/journal.pone.0005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam MS, Garg SK, Agrawal P. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- 11.Ihssen J, Egli T. Global physiological analysis of carbon- and energy-limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environ Microbiol. 2005;7:1568–1581. doi: 10.1111/j.1462-2920.2005.00846.x. [DOI] [PubMed] [Google Scholar]
- 12.Kana BD, Machowski EE, Schechter N, Teh J-S, Rubin H, et al. Parish T, Brown A, editors. Electron transport and respiration in mycobacteria. 2009. pp. 35–64. Mycobacterium: Genomics and molecular biology: Horizon Scientific Press.
- 13.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, et al. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J Bacteriol. 2001;183:7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci U S A. 2005;102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brekasis D, Paget MS. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). Embo J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A. 2009;106:10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratledge C. Iron metabolism. In: Ratledge C, Dale J, editors. Mycobacteria Molecular Biology and Virulence. Oxford: Blackwell Science Ltd; 1999. pp. 260–286. [Google Scholar]
- 19.Molenaar D, van der Rest ME, Drysch A, Yucel R. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J Bacteriol. 2000;182:6884–6891. doi: 10.1128/jb.182.24.6884-6891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Rest ME, Frank C, Molenaar D. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J Bacteriol. 2000;182:6892–6899. doi: 10.1128/jb.182.24.6892-6899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baughn AD, Garforth SJ, Vilcheze C, Jacobs WR. An Anaerobic-Type alpha-Ketoglutarate Ferredoxin Oxidoreductase Completes the Oxidative Tricarboxylic Acid Cycle of Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000662. doi: 10.1371/journal.ppat.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean KJ, Carroll P, Lewis DG, Dunford AJ, Seward HE, et al. Characterization of active site structure in CYP121. A cytochrome P450 essential for viability of Mycobacterium tuberculosis H37Rv. J Biol Chem. 2008;283:33406–33416. doi: 10.1074/jbc.M802115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang YJ, Shui W, Myers S, Feng X, Bertozzi C, et al. Central metabolism in Mycobacterium smegmatis during the transition from O2-rich to O2-poor conditions as studied by isotopomer-assisted metabolite analysis. Biotechnol Lett. 2009;31:1233–1240. doi: 10.1007/s10529-009-9991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 25.Vaerewijck MJ, Huys G, Palomino JC, Swings J, Portaels F. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol Rev. 2005;29:911–934. doi: 10.1016/j.femsre.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Best C, Best T. Mycobacterium smegmatis infection of the hand. Hand. 2009;4:165–166. doi: 10.1007/s11552-008-9147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz E, Friedrich B. The H2-metabolizing prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes A handbook on the biology of bacteria: Ecophysiology and biochemistry. 3 ed. New York: Springer; 2006. pp. 496–563. [Google Scholar]
- 29.King GM. Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl Environ Microbiol. 2003;69:7266–7272. doi: 10.1128/AEM.69.12.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zbell AL, Maier RJ. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl Environ Microbiol. 2009;75:1456–1459. doi: 10.1128/AEM.02064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fontan P, Aris V, Ghanny S, Soteropoulos P, Smith I. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect Immun. 2008;76:717–725. doi: 10.1128/IAI.00974-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman DR, Guinn KM, Hickey MJ, Mathur SK, Zakel KL, et al. Mycobacterium tuberculosis H37Rv: Delta RD1 is more virulent than M. bovis bacille Calmette-Guerin in long-term murine infection. J Infect Dis. 2004;190:123–126. doi: 10.1086/421472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mnatsakanyan N, Bagramyan K, Trchounian A. Hydrogenase 3 But Not Hydrogenase 4 is Major in Hydrogen Gas Production by Escherichia coli Formate Hydrogenlyase at Acidic pH and in the Presence of External Formate. Cell Biochem Biophys. 2004;41:357–366. doi: 10.1385/CBB:41:3:357. [DOI] [PubMed] [Google Scholar]
- 36.Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 37.da Silva SM, Venceslau SS, Fernandes CL, Valente FM, Pereira IA. Hydrogen as an energy source for the human pathogen Bilophila wadsworthia. Antonie Van Leeuwenhoek. 2008;93:381–390. doi: 10.1007/s10482-007-9215-x. [DOI] [PubMed] [Google Scholar]
- 38.Zbell AL, Benoit SL, Maier RJ. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology. 2007;153:3508–3516. doi: 10.1099/mic.0.2007/009027-0. [DOI] [PubMed] [Google Scholar]
- 39.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- 41.Gebhard S, Tran SL, Cook GM. The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology. 2006;152:3453–3465. doi: 10.1099/mic.0.29201-0. [DOI] [PubMed] [Google Scholar]
- 42.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 43.Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, et al. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran SL, Cook GM. The F1Fo-ATP synthase of Mycobacterium smegmatis is essential for growth. J Bacteriol. 2005;187:5023–5028. doi: 10.1128/JB.187.14.5023-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garbe TR, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, et al. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 46.Garland PB, Randle PJ. A rapid enzymatic assay for glycerol. Nature. 1962;196:987–988. doi: 10.1038/196987a0. [DOI] [PubMed] [Google Scholar]
- 47.Villas-Boas SG, Bruheim P. Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal Biochem. 2007;370:87–97. doi: 10.1016/j.ab.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 48.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, et al. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–550, 552-544, 556 passim. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.96 MB PDF)
Expression ratios of genes involved in energy metabolism. Expression ratios with a p-value<0.05 are indicated with bold numbers.
(0.22 MB DOC)
Primer sequences used for quantitative RT-PCR.
(0.05 MB DOC)
Primer sequences used for deletion of msmeg_2719 and complementation.
(0.03 MB DOC)
Phylogenetic trees of the large subunits of the three hydrogenases found in the genome of M. smegmatis mc2155. Proteins with >40% identity are depicted.
(3.47 MB TIF)
Construction of an unmarked msmeg_2719 deletion mutant of M. smegmatis mc2155. A. Schematic diagram of the two-step approach for deletion of msmeg_2719. The knockout construct consisted of two fragments flanking msmeg_2719 on the left (LF) and right (RF) in PX33. Integration of the vector (thick black line) into the chromosome (thin black line) via the left flank (Int LF) or right flank (Int RF) and subsequent deletion of msmeg_2719 (KO) are shown. Restriction sites of SmaI (S) and fragment sizes as detected in Southern hybridization are indicated. Drawing not to scale. WT, wild-type. B. Southern hybridization analysis of the integration event. Left panel, 1. crossover: SmaI-digests of genomic DNA of wild-type mc2155 (lane 1) and a candidate colony (lane 2) were probed with radiolabeled left flank PCR product of the deletion construct. Right panel, 2. crossover: Southern hybridization of msmeg_2719 deletion. Analysis of msmeg_2719 deletion strain (line 1) and wild-type mc2155 (lane 2) was performed as in the left panel. Molecular masses are indicated in kb. M, marker.
(3.30 MB TIF)
Validation of M. smegmatis gene expression ratios (slow growth rate at 50% oxygen saturation versus fast growth rate at 50% oxygen saturation) by quantitative RT-PCR (qPCR) (grey bars) compared to microarray results (black bars). For qPCR the expression of each gene was normalized to the expression of sigA. Error bars represent standard deviations of gene expression ratios from three biological replicates for each condition.
(5.48 MB TIF)