Abstract
Protein glycosylation regulates protein function and cellular distribution. Additionally, aberrant protein glycosylations have been recognized to play major roles in human disorders, including neurodegenerative diseases. Glycoproteomics, a branch of proteomics that catalogs and quantifies glycoproteins, provides a powerful means to systematically profile the glycopeptides or glycoproteins of a complex mixture that are highly enriched in body fluids, and therefore, carry great potential to be diagnostic and/or prognostic markers. Application of this mass spectrometry-based technology to the study of neurodegenerative disorders (e.g., Alzheimer's disease and Parkinson's disease) is relatively new, and is expected to provide insight into the biochemical pathogenesis of neurodegeneration, as well as biomarker discovery. In this review, we have summarized the current understanding of glycoproteins in biology and neurodegenerative disease, and have discussed existing proteomic technologies that are utilized to characterize glycoproteins. Some of the ongoing studies, where glycoproteins isolated from cerebrospinal fluid and human brain are being characterized in Parkinson's disease at different stages versus controls, are presented, along with future applications of targeted validation of brain specific glycoproteins in body fluids.
Keywords: glycoproteomics, mass spectrometry, Alzheimer's diseases, Parkinson's disease, biomarkers, cerebrospinal fluids
I. Introduction
Advances in proteomic concepts and technologies, particularly unbiased techniques, have stimulated a great interest in application of mass spectrometry (MS) to explore neurodegenerative disorders, e.g., Alzheimer's disease (AD), Parkinson's disease (PD), and amyotrophic lateral sclerosis (ALS) - one of the most important groups of diseases in our rapidly aging population in developing and industrialized countries (Chiang et al., 2008). Application of these techniques to neurodegenerative disorders is especially advantageous because, despite decades of “mechanism”- or “pathway”-based pursuits, the pathogenesis of most of these diseases remains largely unknown (Arakawa et al., 2008; Cookson, 2005; Moore et al., 2005; Siddique & Siddique, 2008; Thomas & Beal, 2007). Indeed, in the past several years, proteomic investigations that use different platforms with samples collected from AD and PD patients have already revealed quite a few novel proteins that are potentially critical, not only to the understanding of the mechanisms of the diseases but also to new avenues to diagnose these diseases and to monitor disease progression (Butterfield et al., 2003; Castegna et al., 2002; Finehout et al., 2007; Jin et al., 2006; Leverenz et al., 2007; Osorio et al., 2007; Simonsen et al., 2007).
Defining protein biomarkers unique to a disease diagnosis or progression in body fluids, particularly cerebrospinal fluid (CSF), is currently one of the most exciting areas of research in neurodegenerative disorders (Rite et al., 2007; Simonsen et al., 2007; Tumani et al., 2008; Yang et al., 2008; Yuan & Desiderio, 2003,2005). CSF, which originates within the ventricles and surrounds the brain and spinal cord, is an ideal source for biomarker discovery for diseases of the central nervous system (CNS) like AD and PD. The reasons include (Abdi et al., 2006; Srivastava et al., 2008; Zhang, 2007): 1) CSF is the only body fluid that directly interchanges with the extracellular fluid of the CNS, and therefore reflects pathological changes in the CNS most directly, and 2) multiple CSF taps can be obtained with minimal risk to make possible a longitudinal analysis of biomarkers in a given cohort. That said, among thousands of proteins identified by proteomics in human CSF thus far (Pan et al., 2007b; Zougman et al., 2008), only a small portion are related to the CNS structurally or functionally. This deficit in identifying CNS-specific proteins is mainly due to the fact that most of the CNS-specific proteins are low in abundance, and all current proteomic techniques are biased towards abundant proteins in a sample with a large dynamic range (Gulcicek et al., 2005). One of the approaches to get around this difficulty is to focus on a subproteome(s) that can be isolated readily (e.g., proteins with glycosylation, phosphorylation, or oxidation) before proteomic profiling, thereby effectively reducing the dynamic range of a given complex sample (Bahl et al., 2008; Korolainen et al., 2002; Kubota et al., 2008). To this end, characterizing glycoproteins is especially appealing because they are intimately related to the health of cells, and in addition, are relatively enriched in body fluids like CSF and plasma (Ohtsubo & Marth, 2006).
In this report, we will begin by summarizing the current understanding of glycoproteins in biology and neurodegenerative disease, followed by an introduction of existing proteomic technologies used to characterize glycoproteins. Next, we will present some of the ongoing studies where glycoproteins isolated from human CSF and brain tissue are characterized in PD at different stages and in controls. Future applications of targeted proteomics - to identify unique proteins in the CNS first, followed by confirmation/validation of known proteins in CSF or plasma – also will be addressed briefly.
II. Glycoproteins in heath and neurodegenerative disease
A. Glycosylation in health and disease
Post-translational modifications (PTMs) play a key role to modulate the activities and functions of most proteins in biological systems (Hann, 2006). Among various PTMs, glycosylation represents the most common and complicated form. It is estimated that 50–60% of proteins in the human body are modified by glycosylation (Apweiler et al., 1999; Hagglund et al., 2004; Kameyama et al., 2006). A glycoprotein often contains more than one oligosaccharide attachment site, and each glycosylation site can be modified with multiple oligosaccharide chains. Additionally, on a single glycoprotein, the structure of oligosaccharides at each site can be significantly different. Various glycosylated proteins are synthesized mainly in the endoplasmic reticulum and Golgi via reactions that involve sugar nucleotide synthases, transporters, glycosyltransferases, glycosidases, and other sugar-modifying enzymes. In addition, the structures of glycans can be easily altered by changes of the physiological condition of the cells (Haltiwanger & Lowe, 2004; Lowe & Marth, 2003). It should be noted that, although it is beyond a review focused on glycoproteins, a mass spectrometric study of glycans is itself an active area of current research (Morelle & Michalski, 2005; Zaia, 2008).
The amino acids known to be involved in glycosylation are asparagine, arginine, serine, threonine, proline, hydroxyproline, tryptophan and tyrosine (Spiro, 2002). Typically, protein glycosylation is categorized as either O-linked or N-linked. The N-linked glycosylation, characterized by the attachment of the glycan to an asparagine side chain of the protein, is by far the most common (Nalivaeva & Turner, 2001). The consensus sequence for N-glycosylation is Asn-Xaa-Ser/Thr, where Xaa is any amino acid other than proline (Johansen et al., 1961). The asparagine is linked to N-acetylglucosamine (GlcNAc) residues. Additional sugar residues in the glycan depend on whether the glycosylation is the high-mannose hybrid or complex type (Suzuki et al., 1995). In O-linked glycosylation, on the other hand, the glycan is attached to the serine/threonine side chain (Spiro, 1973). O-linked glycosylation usually starts with an N-acetylgalactosamine (GalNAc) linked to serine/threonine and, unlike N-linked glycosylation, no consensus sequence that defines an O-linked glycosylation site exists (Spiro, 1964,1973,2002; Tanaka et al., 1964). This type of glycosylation is observed most abundantly in mucin-like glycoproteins that form part of epithelial secretions in, for example, the gut, cervix, and lungs (Gendler & Spicer, 1995; Hanisch, 2001). Another variation of O-linked glycans is the Ser/Thr-O-GlcNAc sequence, which is abundant in nucleocytosolic proteins that aid in signal transduction (Spiro, 2002).
One of the initial functions of glycosylation of a given protein is to direct the protein to the appropriate subcellular location; for example, many lysosomal proteins contain a mannose-6-phosphate moiety, a signaling molecule for lysosome (Kaplan et al., 1977; Varki & Kornfeld, 1980). Additionally, glycosylation has been implicated in numerous biological processes, including cell growth and developmental biology, immune response, tumor growth, metastasis, anticoagulation, cell-to-cell communication, and microbial pathogenesis (Casu et al., 2004; Collins & Paulson, 2004; Dube & Bertozzi, 2005; Guo et al., 2004; Hwang et al., 2003; Inatani et al., 2003; Kinjo et al., 2005; Lin, 2004; Liu et al., 2002; Lowe & Marth, 2003; Miller et al., 2005; Sasisekharan et al., 2002). Aberrant protein glycosylations could also contribute to human disorders, including neurodegenerative diseases (Liu et al., 2002; Saez-Valero et al., 2003).
B. Glycosylation alterations in human neurodegenerative disorders
Alterations in protein glycosylation have been related to human neurodegenerative disease states, such as Creutzfeldt-Jakob disease (CJD), AD, and PD (Saez-Valero et al., 2003; Silveyra et al., 2006). Although the structural elucidation of glycoproteins is a challenge because of their inherent complexity and heterogeneity in biological systems, advances have been made to identify a few proteins where glycosylation appears to be important in the disease processes of AD and PD (Sihlbom et al., 2004). A few key proteins involved in AD and PD pathogenesis are discussed below.
Acetylcholinesterase (AChE), one of the critical enzymes targeted in the current clinical management of AD, hydrolyzes the neurotransmitter acetylcholine at cholinergic synapses, and is widely distributed in brain regions. The glycosylation of AChE is altered in the post-mortem brain and CSF of AD patients (Saez-Valero et al., 2000; Saez-Valero et al., 1999). Additionally, the change in glycosylation of AChE appears to be specific for AD because it is not seen in other neurological diseases. More recently, the glycosylation of a related enzyme, butyrylcholinesterase (BuChE), also appears to be altered in AD CSF (Saez-Valero & Small, 2001). Unfortunately, the sensitivity of diagnosing AD with AChE and BuChE in the CSF is lower than that considered necessary for a satisfactory biomarker (Saez-Valero et al., 2003).
Microtubule-associated protein (MAP) tau, another essential protein involved in AD pathogenesis and related tauopathies, undergoes several PTMs, and aggregates into paired helical filaments. Known modifications of tau include hyperphosphorylation, glycosylation, ubiquitination, glycation, polyamination, nitration, and proteolysis. Glycosylation of tau is an early abnormality that might facilitate the hyperphosphorylation of tau, a pathological hallmark, in an AD brain (Liu et al., 2002). Robertson et al. (Robertson et al., 2004) observed a significant decrease in the glycosylated tau (O-linked) in AD post-mortem brain samples compared with control; that decrease suggested an inverse relationship between the two PTMs (i.e., glycosylation vs. hyperphosphorylation). Furthermore, cells transfected with the cDNA coding for O-GlcNAc transferase displayed altered tau phosphorylation patterns as compared with control cells; these alterations again suggested that changes in tau glycosylation might influence its phosphorylation state. However, glycosylation of tau as a biomarker for AD has not been reported.
Until recently, very little has been known about the role of glycosylated proteins in PD. Farrer and colleagues noted a potential connection between the dysfunction of parkin, an E3 ubiquitin ligase involved in the ubiquitination of protein substrates that targets them for degradation by the proteasomal complex, and the formation of α-synuclein inclusions (Farrer et al., 2001). It turned out that the mechanism that underlies this process could be the parkin-mediated ubiquitination of an O-linked glycosylated form of α-synuclein (Shimura et al., 2001). It should be emphasized that mutations of parkin and α-synuclein result in the development of autosomal recessive and dominant familial PD, respectively (Tan & Skipper, 2007; Wakabayashi et al., 2007), and that changes in the total amount of α-synuclein in CSF have been tested as potential biomarkers of PD (also see later discussion).
From what has been discussed above, it is obvious that glycosylation and glycoproteins play critical roles not only in normal physiological conditions but perhaps also in neurodegenerative disorders like in AD and PD. On the other hand, aside from two earlier reports of CSF glycoproteins (Pan et al., 2006; Sihlbom et al., 2004), there is no systematic analysis of glycoproteins in human tissue or CSF for any disease or even in control subjects. Thus, in this report, we will present the glycoproteins identified in human brain in addition to CSF after an introduction of the current proteomic techniques used for characterization of glycoproteins.
III. Characterization of glycoproteins by mass spectrometry-based proteomics
A. Enrichment of glycoproteins
As discussed above, the glycoproteome represents one of the most important sub-proteomes in tissues and body fluids. However, many glycoproteins might be low in abundance in their glycosylated forms, even though the parent proteins are abundant in CSF or plasma. Consequently, numerous attempts have been made to develop methods to enrich glycoproteins present in complex biological samples prior to mass spectrometric analysis.
1. Enrichment by lectin column
Lectins are widely distributed in nature and can recognize carbohydrates on the surface of proteins. To isolate glycoproteins or glycopeptides by affinity chromatography, various lectins can be used (Cummings & Kornfeld, 1982; Hirabayashi, 2004). Concanavalin A (ConA) is a lectin that binds mannosyl and glucosyl residues that contain unmodified hydroxyl groups at positions C3, C4, and C6, and can be utilized for the targeted binding of certain oligosaccharide structures of N-glycosylated proteins (Goldstein et al., 1965; Kamra & Gupta, 1987; Yahara & Edelman, 1972). The use of wheat germ agglutinin (WGA) isolates glycostructures with N-acetylglucosamine and sialic acids (Nagata & Burger, 1974). Arachis hypogaea agglutinin (PNA) is specific to glycans that contain β-Gal, whereas Datura stramonium agglutinin (DSA) is specific to glycans that contain GlcNAc residues (Novogrodsky et al., 1975; Yamashita et al., 1987). Due to their ability to specifically recognize distinct oligosaccharide epitopes (Sharon & Lis, 1989), lectins bound to appropriate matrices like agarose, membranes, or magnetic beads, can be used to isolate, fractionate, and characterize glycoproteins on the basis of their different glycan structures (Bundy & Fenselau, 2001; Wiener & van Hoek, 1996). In this regard, affinity chromatography with lectins is a useful and powerful technique to fractionate and isolate glycans and glycopeptides. The combination of lectin chromatography and MS analysis provides high-sensitive detection and useful information on glycan structures, and enables further biological approaches. However, because individual lectins display unique binding specificities, separation with a particular lectin will isolate only a fraction of glycoproteins or glycopeptides that bind to that lectin with high affinity (Bunkenborg et al., 2004; Ghosh et al., 2004; Xiong et al., 2003). To overcome the limitation of selective capture of a subset of glycoproteins for a given lectin, a technique has been introduced for glycoprotein/peptide isolation and enrichment from complex mixtures that involves double lectin chromatography prior to identification with liquid chromatograph (LC)-electrospray ionization (ESI) MS (Bunkenborg et al., 2004). Recently, a more elegant method has been established with a multi-lectin column, which allows for an almost complete enrichment of glycoproteins from biological fluids (Wang et al., 2006; Yang & Hancock, 2004). In a similar manner, lectin arrays have been developed that contain more than 35 different lectins that allow a qualitative and quantitative profiling of glycoprotein glycan patterns in a rapid and sensitive high-throughput manner (Kuno et al., 2005). Finally, lectin microcolumns have also been generated that are applicable to high-pressure analytical schemes, and thus, can be directly coupled on-line to ESI-MS to enable a highly sensitive semi-automated profiling of glycoproteins (Madera et al., 2006,2007; Madera et al., 2005).
2. Enrichment with hydrazide
Hydrazide chemistry has been used to selectively isolate, identify, and quantify N-linked glycopeptides in a much more specific and efficient manner (Zhang et al., 2003). This method is based on the conjugation of glycoproteins to a solid support with hydrazide chemistry after periodate-mediated oxidation of the carbohydrate. Peptide moieties of the covalently captured glycopeptides are released with PNGase F treatment to allow the peptide and glycosylation site to be identified. Recently, Sun and colleagues (Sun et al., 2007) reported a novel chemical capture approach that focuses on a more efficient glycopeptide enrichment. In this approach, glycopeptides derived from glycoproteins are enriched by selective capture onto a solid support with hydrazide chemistry followed by enzymatic release of the peptides and subsequent analysis by tandem MS. Digestion of proteins into peptides improves the solubility of large membrane proteins, and exposes all of the glycosylation sites (at least in theory) to ensure an equal accessibility to external capture reagents. Notably, whereas the specificity has been increased by capturing N-linked glycopeptides/glycoproteins with the hydrazide chemistry, this method is restricted to N-glycopeptides and, in addition, information on the carbohydrate structures is lost due to the destruction and removal of the glycan moieties.
3. Other methods for enriching glycoproteins
Besides lectins and hydrazide, a few other techniques, including treatment with boronic acids, have also been employed to facilitate enrichment of glycoproteins. Because boronic acids enhance the capture of the more heterogeneous group of O-linked oligosaccharides, this method has been incorporated into lectin methodology; e.g., a boronic acid-lectin affinity chromatography column has been used to isolate glycoproteins with selective and/or combined elution (Monzo et al., 2007).
B. Mass spectrometric analysis of glycoproteins/peptides
Modern MS has greatly facilitated the characterization of glycoproteins because it provides glycosylation site-specific information by conducting glycopeptide-based analysis, wherein the glycan and its attachment site to the protein can be elucidated in the same experiment; at least in theory. This glycosylation site-specific information is useful to elucidate functional properties of the glycoprotein. Typically, glycopeptide-based MS analysis entails an enzymatic cleavage of glycoproteins with an endoprotease, followed by a separation technique and mass analysis.
1. Desalting
When analyzing glycopeptides and glycoproteins, it is necessary to desalt the sample and remove organic contaminants in order to avoid the formation of salt adducts, thereby obtaining more-informative MS spectra. Cation and anion exchange materials have been used commonly for desalting (Lattard et al., 2006). One of the efficient methods is to use a microcolumn in a GELoader tip (Eppendorf) into which a mixed bed resin column of AG-3 (to remove anions), AG-50 (to remove cations), and C18 (to remove organic materials) are packed (Kussmann et al., 1997). Hydrophilic interaction liquid chromatography (HILIC) (Hagglund et al., 2004) and graphite columns (Larsen et al., 2002) are also useful for desalting.
2. Identification of glycoproteins by mass spectrometric technologies
Most of the large-scale glycoprotein identification studies have used a shotgun proteomics approach, in which glycoproteins are typically trypsin-digested and deglycosylated so that glycosylated peptides can be sequenced in their deglycosylated forms with MS/MS. For glycopeptides with N-linked glycosylation site(s), most of the glycans can be removed with PNGase F. The enzyme cleavage of a glycan group converts asparagine to aspartic acid in a peptide, to introduce a mass difference of 0.984 Da and a negative charge. This phenomenon was used to map the N-linked glycosylation site(s) using MS (Zhou et al., 2007). In the past few years, several studies have used MS to profile N-linked glycoproteins in human body fluids. Liu et al. applied immunoaffinity subtraction and hydrazide chemistry to enrich glycoproteins from human plasma (Liu et al., 2005). The captured plasma glycoproteins were subjected to two-dimensional (2D) LC separation (strong cation exchange [SCX] and reverse-phase capillary LC) followed by tandem MS or MS/MS analysis with a Fourier transform ion cyclotron resonance mass spectrometer. A detection sensitivity at low ng/ml was achieved. A total of 2,053 different N-glycopeptides, representing 303 nonredundant glycoproteins, were identified, including many low-abundance glycoproteins. Other studies applied a lectin affinity-based approach to characterize serum and plasma N-linked glycoproteins, and have added significant numbers of glycoproteins to the blood glycoproteome database (Yang & Hancock, 2004; Zhang et al., 2003). Related to the study of neurodegenerative diseases, the CSF glycoproteome has been investigated in an experiment, where lectin affinity and hydrazide chemistry enrichment methods were both applied to reveal 216 glycoproteins (Pan et al., 2006).
Different approaches have characterized O-glycosylation with tandem mass spectrometry. A very sensitive technique to identify O-glycosylated sites employs the use of ammonia or ethylamine for the specific release of O-linked glycan chains. The integrity of the peptide backbone was retained and ammonia or ethylamine was incorporated into the amino acid residue(s) to which the glycan(s) had been attached. Thus, the former glycosylation site was labeled, and thus, can be identified by the mass alteration of −1 Da and +27 Da for ammonia and ethylamine, respectively (Hanisch et al., 2001; Rademaker et al., 1998). The limitations of collision-induced dissociation (CID) ESI-MS/MS for glycosylation site analysis (i.e., the dominating fragmentation of the glycan chains) can be overcome with different tandem MS techniques. Haynes et al. demonstrated a technique that provided simultaneous detection and identification of O-GlcNAc-modified peptides with low-energy collisions in tandem MS (Haynes & Aebersold, 2000). The differential between the energy required to remove the O-GlcNAc group versus the energy required to fragment the peptide chain allows the O-GlcNAc group to be detected and the peptide sequence, and therefore the protein, to be identified. More recently, ‘soft’ collision techniques, such as electron capture dissociation (ECD) and electron transfer dissociation (ETD) (Catalina et al., 2007; Mormann et al., 2005), have led to a preferential cleavage of the peptide backbone and to leaving glycan structures intact, to thus allow an unambiguous assignment of the glycosylation site in N- and O-glycopeptides (Hakansson et al., 2001; Hogan et al., 2005). To enhance the specificity of O-glycosylation analysis, Durham et al. applied a serial lectin affinity chromatography that combine ConA and Jacalin to enhance the identification of O-glycosylated sites on proteins from the human blood proteome (Durham & Regnier, 2006). The enriched O-glycopeptides were deglycosylated with oxidative elimination and analyzed with ESI and MALDI (matrix-assisted laser desorption/ionization) tandem MS to identify over thirty O-glycosylated glycoproteins from human serum.
MALDI-based mass spectrometric analysis usually produces singly charged glycopeptide ions that can be analyzed off-line with high sensitivity after deposition of nano-LC-derived glycopeptide fractions onto the MALDI-target (Lochnit & Geyer, 2004). This technique is complementary to ESI technology, because ESI mass spectra are sometimes too complicated to fully assign oligosaccharide structures due to the formation of many multiply charged ions. With a MALDI-TOF/TOF-instrument, glycopeptides can be further analyzed via characteristic fragment ions that can sequence the glycan and the peptide simultaneously (Krokhin et al., 2004; Kurogochi & Nishimura, 2004; Stephens et al., 2004; Wuhrer et al., 2004). Nonetheless, a more systematic assessment of O- or N-glycosylation sites on glycoproteins might require the use of mass spectrometers with higher mass accuracies; for example, ESI or MALDI with Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR) (Irungu et al., 2008). FT-ICR MS provides the high mass accuracy needed to improve the specificity for protein database search results, and enhances the prediction of glycoforms. Sihlbom's group applied FT-ICR MS and infrared multi-photon dissociation (IRMPD) to determine the glycosylation states of isoforms of CSF proteins from individual AD patients compared to controls (Sihlbom et al., 2004). In that study, they reported that the sub-femtomole sensitivities of FT-ICR MS analyzed 2D gel-separated complex human protein mixtures. An additional advantage to IRMPD is that it selectively dissociates the glycosidic bonds of N-linked glycans (amino acid consensus sequence N–X–S/T/C, in which X cannot be P).
3. Isotope labeling for quantification of glycoproteins
Characterizing glycoproteins as extensively as possible is just the first step to define biomarkers unique to a disease or disease progression. A more important process is to quantify the changes associated with a disease or a disease stage. Additionally, quantitative glycoproteomics can help to characterize the regulatory pathways and complex system networks by providing protein concentration information that corresponds to different cellular states. Although label-free techniques have been developed by numerous investigators (Levin et al., 2007), most published studies with human samples largely rely on various isotope-labeling techniques for quantification, particularly when large-scale profiling is the main focus. Examples include the use of chemical reactions to introduce isotopic tags at specific functional groups on peptides or proteins, such as ICAT (isotope-coded affinity tags) (Gygi et al., 1999; Haqqani et al., 2008) and iTRAQ (isobaric tags for relative and absolute quantitation) (Aggarwal et al., 2006) as well as the methods that introduce stable-isotope tags via enzymatic reaction, such as enzymatic 18O incorporation (Kaji et al., 2003; Kaji et al., 2006; Zhang et al., 2003).
ICAT labels the side chains of cysteinyl residues in two reduced protein samples with the isotopic light or heavy reagent, respectively, and generates the mass signatures that identify sample origin and serve as the basis for accurate quantification, to thus afford simultaneous comparison of two proteomes. However, ICAT selectively targets cysteine residues, and therefore approximately 3% of mammalian proteins that lack cysteine residues cannot be analyzed (Colangelo & Williams, 2006). In addition, some cysteines are blocked or are inaccessible to the labeling reagent. More recently, iTRAQ technology, which labels lysines and N-termini, has been used for quantitative proteomics in human body fluid and tissue (Martin et al., 2008; Song et al., 2008). The iTRAQ technology has a significant advantage over other methods due to its capability to multiplex up to eight samples in a single experiment (D'Ascenzo et al., 2008). Another positive aspect includes unbiased peptide labeling, because iTRAQ isobaric tags theoretically label lysine side groups and all free amino-terminal groups of the peptides present in a sample. The iTRAQ tags consist of a reporter group, a balance group, and a peptide reactive group that covalently binds to the peptides. The tandem mass spectra include contributions from each sample, and the individual contributions of each sample can be measured by the intensity of the reporter ion peaks. Moreover, a chemical approach for the N-glycosylation identification (i.e., hydrazide chemistry capture) can be incorporated with the iTRAQ quantification method because an iTRAQ-labeled peptide is chemically stable in other buffer systems.
Notably, in addition to quantification, isotope-labeling methods might also increase the certainty of glycoproteome assignments and enable quantitative comparisons of glycosylated samples. For example, several groups have used isotope-coded glycosylation-site-specific tagging (IGOT) for the large-scale identification of N-glycosylated proteins from a complex biological sample (Kaji et al., 2003; Kaji et al., 2006). The IGOT approach is based on the lectin column-mediated affinity capture of a set of glycopeptides generated by tryptic digestion of protein mixtures, followed by peptide-N-glycosidase-mediated incorporation of a stable isotope tag, 18O, specifically into the N-glycosylation site. The 18O-tagged peptides are identified with multi-dimensional LC-MS-based technology. The application of this method to characterize N-linked high-mannose and/or hybrid-type glycoproteins from an extract of Caenorhabditis elegans proteins identified 250 glycoproteins, including 83 putative transmembrane proteins, with the simultaneous determination of 400 unique N-glycosylation sites. A similar approach was later used to identify and quantify N-linked glycoproteins in serum (Zhang et al., 2003). In this study, the N-linked glycopeptides were oxidized and captured directly on a hydrazide column; the quantitation was achieved by comparing two samples that were tagged differentially with 18O- or 16O- labeled water.
IV. Characterization of glycoproteins associated with Parkinson's disease and disease progression
In the next few sections, we will use one of the ongoing projects focused on PD to illustrate a strategy that identifies and quantifies CNS-specific glycoproteins at the same time. However, only identification data will be shown in this report.
A. Parkinson's disease and its progression
PD is traditionally considered a movement disorder that results from a relatively selective loss of neurons in the brainstem, including dopaminergic (DAergic) neurons in the substantia nigra pars compacta (SNpc), with subsequent loss of striatal dopamine and accompanied by the formation of intraneuronal inclusions called Lewy bodies that contain α-synuclein as one of the major proteins (Jankovic, 2001; Lowe et al., 1997). More recently, however, it has become increasingly clear that neurodegeneration in PD is widespread with associated presentation of multiple “non-motor” symptoms, including cognitive impairment, particularly as the disease advances. Cognitive impairment in PD, ranging from mild dysfunction to severe dementia, has major clinical consequences, because it has been associated with a reduced quality of life (Schrag et al., 2000), shortened survival (Nussbaum et al., 1998), and increased caregiver distress compared to PD without cognitive impairment (Aarsland et al., 1999). It should be emphasized that the risk of developing dementia in PD patients is several-fold higher than for community-dwelling controls (Aarsland et al., 2003; Aarsland et al., 2001; Marder et al., 1995). Furthermore, in more recent studies, when PD patients are tested more rigorously, it has been estimated that 36% of patients newly diagnosed with PD had mild cognitive impairment (MCI) - a prodrome of PD dementia (Foltynie et al., 2004; Levin & Katzen, 2005), and that 57% of patients with newly diagnosed PD will develop MCI within three to five years (Williams-Gray et al., 2007).
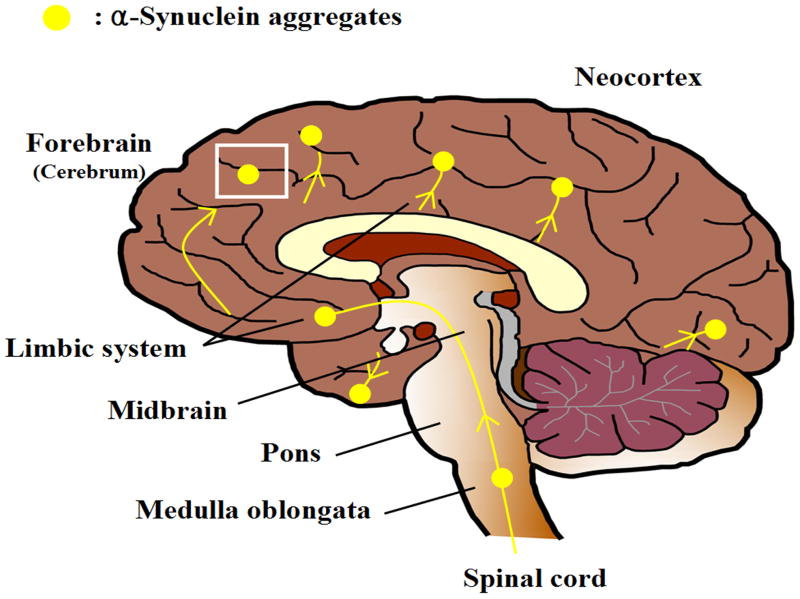
Numerous clinicopathological studies have sought to identify the structural basis of cognitive impairment in patients with PD dementia (PDD). Though remaining to be investigated, it appears that, in a significant portion of PD patients, PD progression is characterized pathologically by the spreading of aggregated α-synuclein deposits from the brainstem to other parts of the brain (Braak et al., 2002; Braak et al., 2003). A staging procedure for the PD-related inclusion body pathology (i.e., Lewy neurites and Lewy bodies) in the brain proposes that the pathological process begins at two sites (the medulla oblongata and olfactory bulb) and progresses in a topographically predictable sequence in six stages. During stages 1-2, the inclusion body pathology remains confined to the medulla oblongata, pontine tegmentum, and anterior olfactory structures. In stages 3-4, the basal midbrain, including SNpc, and forebrain become the foci of the pathology, and the illness reaches its symptomatic phase (motor symptoms). In the final stages 5-6, the pathological process is seen in the association areas and primary fields of the neocortex. The basic concept is diagramed in Figure 1.
Figure 1.
Cognitive impairment associated with PD progression is characterized pathologically by the spreading of α-synuclein aggregates, the main component of Lewy bodies, from brainstem to limbic system and eventually to the neocortex (Braak et al., 2003). The boxed area, the middle frontal gyrus, is the tissue source for a recent nonbiased profiling (Pan et al., 2007a; Shi et al., 2008) as well as characterization of glycoproteins to reveal proteins unique to PD and/or PD progression, particularly development of dementia.
B. Biomarkers for Parkinson's disease and Parkinson's disease progression
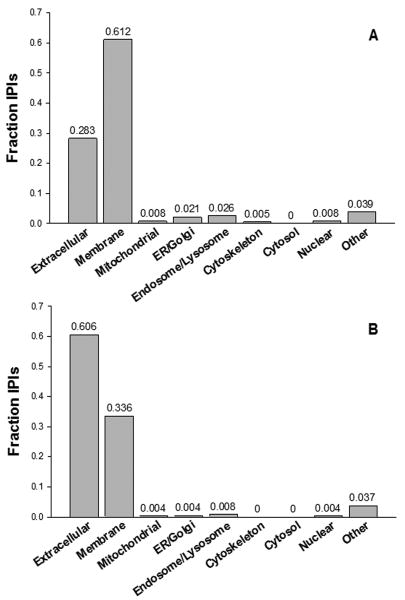
Two approaches, protein-specific, e.g., α-synuclein (Borghi et al., 2000; El-Agnaf et al., 2006; Jakowec et al., 1998; Tokuda et al., 2006; Verbeek et al., 2003) and DJ-1 (Hirotani et al., 2008; Waragai et al., 2006), and unbiased profiling (Abdi et al., 2006) have been undertaken to define protein biomarkers unique to PD diagnosis. Profiling is advantageous because it provides an unbiased view of a disease or stage of a disease whose pathogenesis is largely unknown. In addition, multiple markers can be generated for a given disease when a profiling approach is taken, and generally speaking, a combination of multiple markers offers better sensitivity and specificity than a single protein alone for disease diagnosis (Zhang et al., 2008). There are no known markers that can predict PD progression, whether related to motor symptoms or cognitive impairment; that concept has been emphasized more recently. To resolve this issue, in the last few years, with unbiased proteomics, we have compared the proteome of brain tissue associated with Lewy body progression as PD advances, with the goal of identifying proteins before Lewy body formation in the neocortex (Figure 1). However, in an earlier analysis, among ∼1,500 proteins identified in CSF only 9% were present in the proteins identified in human brain tissue (Pan et al., 2007a; Pan et al., 2007b). It has been hypothesized that there are at least two limitations associated with the previous approaches: 1) the cellular fractionation technique is biased against extracellular proteins, because most of them are discarded along with cell debris (Jin et al., 2006; Pan et al., 2007a), and 2) the large dynamic range of the CSF proteome makes it very challenging to identify proteins of low abundance [albumin and immunoglobulins constitute more than 75% of CSF proteins (Srivastava et al., 2008)]. Indeed, both limitations are the major problems that must be dealt with not only in diseases related to the CNS but also in the biomarker discovery field in general (Aebersold et al., 2005; Qian et al., 2006). To increase the likelihood of identifying proteins that are accessible clinically, most investigators have turned their attention either to removing high-abundance proteins before profiling or to a specific sub-proteome with a unique PTM; e.g. proteins with glycosylation. As mentioned earlier, protein glycosylation, and in particular N-linked glycosylation, is prevalent in proteins destined for extracellular environments (Roth, 2002).
C. Glycoprotein/peptide in human cerebrospinal fluid and brain tissue
1. Glycoproteins in human cerebrospinal fluid
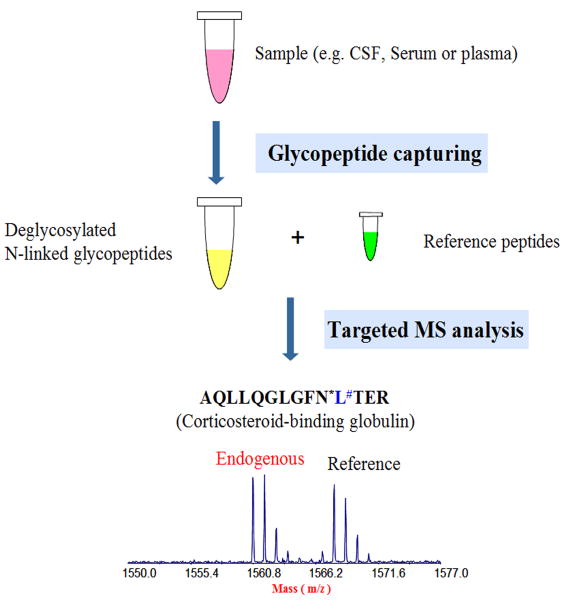
This investigation consisted of four groups of control subjects, AD and PD patients at two different stages. More specifically, the control group consisted of 29 individuals aged 70±6 years, 18 men and 11 women, with no history, symptoms, or signs of psychiatric or neurological disease. The AD group consisted of 51 patients aged 69±9 years, 28 men and 23 women, all of whom underwent a comprehensive clinical examination, and were diagnosed with AD according to NINCDS ADRDA criteria (Jobst et al., 1997). The early-stage PD group consisted of 11 patients aged 61±8 years, 9 men and 2 women, all of whom underwent a comprehensive clinical examination and were diagnosed with PD at a Hoehn and Yahr stage of 1.5 or less. The late stage PD group consisted of 11 patients aged 66±7 years, 7 men and 4 women, all of whom underwent a comprehensive clinical examination and were diagnosed with PD at a Hoehn and Yahr stage of 3 or greater. All CSF samples have been controlled for blood contamination before pooling samples into four groups (Abdi et al., 2006). The pooled CSF samples were mixed with a protease inhibitor cocktail, and stored at -80°C before use. To perform quantitative analysis of glycoproteins unique to PD and PD progression, samples were digested with trypsin, followed with iTRAQ labeling, before hydrazide bead capture. Of note, quantitative data are still being evaluated currently and will be published separately at a later time. The glycopeptides derived from glycoproteins in human CSF were enriched by hydrazide bead capture followed by enzymatic release of the N-linked glycosylated peptides. Peptides from each sample were dissolved in 0.5% trifluoroacetic acid (TFA), and separated with reverse phase (RP) chromatography. MS/MS analysis used the 4800 Proteomics Analyzer with TOF/TOF Optics™ (Applied Biosystems). The MS/MS spectra were extracted and searched against the International Protein Index (IPI) human protein database (version 3.42 from the European Bioinformatics Institute [EBI]) with ProteinPilot™ software (version 2.0.1, revision 33087, Applied Biosystems) with the Paragon™ method. The raw peptide identification results from the Paragon™ Algorithm (Applied Biosystems) searches were further processed with the Pro Group™ Algorithm (Applied Biosystems) within the ProteinPilot™ software before final display. The Pro Group Algorithm uses the peptide identification results to determine the minimal set of proteins that can be reported for a given protein confidence threshold. For each protein, Pro Group Algorithm reports two types of scores for each protein: unused ProtScore and total ProtScore. The total ProtScore is a measurement of all the peptide evidence for a protein, and is analogous to protein scores reported by other protein identification software. The unused ProtScore, however, is a measurement of all the peptide evidence for a protein that is not better explained by a higher ranking protein. In other words, the unused ProtScore is calculated with the unique peptides (peptides that are not used by the higher ranking protein), and it is a clearer indicator of protein evidence and assists in singling out members of a multiprotein family. All reported data were based on 95% confidence for protein identification as determined by ProteinPilot (ProtScore ≥ 1.3). Identified glycoproteins were checked against the UniProtKB/Swiss-Prot database and the Institute for Systems Biology (ISB) database as glycoproteins with known glycosylation sites or probable/potential glycosylation sites.
The MALDI-TOF-TOF analysis revealed a total of 283 non-redundant glycoproteins in human CSF (Appendix I). In comparison with the existing publicly accessible database, 243 of these proteins were annotated in UniProtKB/Swiss-Prot and the ISB database as glycoproteins with known glycosylation sites or probable/potential glycosylation sites. The specificity of this approach was approximately 86% (243/283). When this dataset is compared with what has been published earlier, where lectin affinity purification and hydrazide chemistry were both used to characterize CSF glycoproteins with an ion trap mass spectrometer (LCQ) (Pan et al., 2006), 87 were observed in both datasets; i.e., a 36% overlap of 243 glycoproteins. This overlap is considered reasonable, given that a different database and different technology (LCQ vs. MALDI-TOF-TOF as well as hydrazide chemistry + lectin affinity vs. hydrazide chemistry alone) were used to characterize glycoproteins in two different studies.
Appendix I. Glycopeptides Identified in Human Cerebrospinal Fluid.
| Accessions | Names | Sequence-our | EBI | ISB | ||
|---|---|---|---|---|---|---|
| Identified | Potential | Identified | Potential | |||
| IPI00000877.1 | HYOU1 Hypoxia up-regulated protein 1 precursor | VINETWAWK | Y | Y | ||
| IPI00001662.1 | OPCML Opioid-binding protein/cell adhesion molecule precursor | DYGNYTCVATNK | Y | Y | ||
| IPI00001662.1 | OPCML Opioid-binding protein/cell adhesion molecule precursor | MSTLTFFNVSEK | Y | Y | ||
| IPI00002714.1 | DKK3 Dickkopf-related protein 3 precursor | ASSEVNLANLPPSYHNETNTDTK | Y | Y | ||
| IPI00002714.1 | DKK3 Dickkopf-related protein 3 precursor | ITNNQTGQMVFSETVITSVGDEEGR | Y | Y | ||
| IPI00002714.1 | DKK3 Dickkopf-related protein 3 precursor | VGNNTIHVHR | Y | Y | ||
| IPI00003813.5 | CADM1 Isoform 1 of Cell adhesion molecule 1 precursor | FQLLNFSSSELK | Y | Y | ||
| IPI00003813.5 | CADM1 Isoform 1 of Cell adhesion molecule 1 precursor | VSLTNVSISDEGR | Y | Y | ||
| IPI00003919.1 | QPCT Glutaminyl-peptide cyclotransferase precursor | NYHQPAILNSSALR | Y | Y | ||
| IPI00004413.1 | TNFRSF21 Tumor necrosis factor receptor superfamily member 21 precursor | VLSSIQEGTVPDNTSSAR | Y | Y | ||
| IPI00005517.1 | EFNA5 Ephrin-A5 precursor | YAVYWNSSNPR | Y | Y | ||
| IPI00005794.2 | PGCP 60 kDa protein | IVVYNQPYINYSR | Y | |||
| IPI00006114.4 | SERPINF1 Pigment epithelium-derived factor precursor | VTQNLTLIEESLTSEFIHDIDR | Y | Y | ||
| IPI00006601.5 | CHGB Secretogranin-1 precursor | GHPQEESEESNVSMASLGEK | Y | |||
| IPI00006662.1 | APOD Apolipoprotein D precursor | ADGTVNQIEGEATPVNLT | Y | |||
| IPI00006662.1 | APOD Apolipoprotein D precursor | ADGTVNQIEGEATPVNLTEPAK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | ADGTVNQIEGEATPVNLTEPAKL | Y | |||
| IPI00006662.1 | APOD Apolipoprotein D precursor | ADGTVNQIEGEATPVNLTEPAKLEVK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | CIQANYSLMENGK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | CIQANYSLMENGKI | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | QANYSLMENGK | Y | |||
| IPI00006662.1 | APOD Apolipoprotein D precursor | EATPVNLTEPAK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | EATPVNLTEPAKLEVK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | GTVNQIEGEATPVNLTEPAK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | PVNLTEPAK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | QANYSLMENGK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | QIEGEATPVNLTEPAK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | QIEGEATPVNLTEPAKLEVK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | TVNQIEGEATPVNLTEPAK | Y | Y | ||
| IPI00007199.4 | SERPINA10 Protein Z-dependent protease inhibitor precursor | ETFFNLSK | Y | Y | ||
| IPI00007221.1 | SERPINA5 Plasma serine protease inhibitor precursor | VVGVPYQGNATALFILPSEGK | Y | Y | ||
| IPI00007709.2 | ADAM28 Isoform 1 of ADAM 28 precursor | NLLAPGYTETYYNSTGK | Y | |||
| IPI00009997.1 | B3GNT1 N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase | VAQPGINYALGTNVSYPNNLLR | Y | |||
| IPI00011218.1 | CSF1R Macrophage colony-stimulating factor 1 receptor precursor | HTNYSFSPWHGFTIHR | Y | Y | ||
| IPI00011218.1 | CSF1R Macrophage colony-stimulating factor 1 receptor precursor | VTVQSLLTVETLEHNQTYECR | Y | Y | ||
| IPI00011229.1 | CTSD Cathepsin D precursor | GSLSYLNVTR | Y | Y | ||
| IPI00011732.2 | GFRA2 Isoform 1 of GDNF family receptor alpha-2 precursor | NAIQAFGNGTDVNVSPK | Y | Y | ||
| IPI00012102.1 | GNS N-acetylglucosamine-6-sulfatase precursor | YYNYTLSINGK | Y | Y | ||
| IPI00012440.7 | FUCA2 Plasma alpha-L-fucosidase precursor | SQNDTVTPDVWYTSKPK | Y | Y | ||
| IPI00012887.1 | CTSL1 Cathepsin L1 precursor | YSVANDTGFVDIPK | Y | Y | ||
| IPI00012887.1 | CTSL1 Cathepsin L1 precursor | YSVANDTGFVDIPKQEK | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | FSAGLASNSSWLR | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | GLNLTSTFLR | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | KSVVAPATDGGLNLTSTFLR | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SAGLASNSSWLR | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SVVAPATDGGLN | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SVVAPATDGGLNLT | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SVVAPATDGGLNLTSTF | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SVVAPATDGGLNLTSTFL | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SVVAPATDGGLNLTSTFLR | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | SVVAPATDGGLNLTSTFLRK | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | VVAPATDGGLNLTSTFLR | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | WFSAGLASN | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | WFSAGLASNS | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | WFSAGLASNSS | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | WFSAGLASNSSW | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | WFSAGLASNSSWL | Y | Y | ||
| IPI00013179.1 | PTGDS Prostaglandin-H2 D-isomerase precursor | WFSAGLASNSSWLR | Y | Y | ||
| IPI00013303.2 | LSAMP Limbic system-associated membrane protein precursor | LGVTNASLVLFRPGSVR | Y | Y | ||
| IPI00014048.1 | RNASE1 Ribonuclease pancreatic precursor | SNSSMHITDCR | Y | Y | ||
| IPI00016150.1 | SERPINI1 Neuroserpin precursor | DANLTGLSDNK | Y | Y | ||
| IPI00016150.1 | SERPINI1 Neuroserpin precursor | WVENNTNNLVK | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | EHEGAIYPDNTTDFQR | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | ELHHLQEQNVSNAFLDK | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | ELHHLQEQNVSNAFLDKGEFYIGSK | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | ENLTAPGSDSAVFFEQGTTR | Y | Y | ||
| IPI00019568.1 | F2 Prothrombin precursor (Fragment) | GHVNITR | Y | Y | ||
| IPI00019568.1 | F2 Prothrombin precursor (Fragment) | YPHKPEINSTTHPGADLQENFCR | Y | Y | ||
| IPI00019943.1 | AFM Afamin precursor | DIENFNSTQK | Y | Y | ||
| IPI00019943.1 | AFM Afamin precursor | HNFSHCCSK | Y | Y | ||
| IPI00019943.1 | AFM Afamin precursor | YAEDKFNETTEK | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | CANLVPVPITNATLDR | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | LVPVPITNATLDR | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | PLCANLVPVPITNATLDR | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | QNQCFYNSSYLNVQR | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | FNSTEYQVVTR | Y | Y | ||
| IPI00020986.2 | LUM Lumican precursor | KLHINHNNLTESVGPLPK | Y | Y | ||
| IPI00020986.2 | LUM Lumican precursor | LGSFEGLVNLTFIHLQHNR | Y | Y | ||
| IPI00020986.2 | LUM Lumican precursor | LHINHNNLTESVGPLPK | Y | Y | ||
| IPI00022371.1 | HRG Histidine-rich glycoprotein precursor | IADAHLDRVENTTVY | Y | Y | ||
| IPI00022371.1 | HRG Histidine-rich glycoprotein precursor | VIDFNCTTSSVSSALANTK | Y | Y | ||
| IPI00022395.1 | C9 Complement component C9 precursor | AVNITSENLIDDVVSLIR | Y | Y | ||
| IPI00022395.1 | C9 Complement component C9 precursor | FSYSKNETYQLFLSYSSK | Y | Y | ||
| IPI00022417.4 | LRG1 Leucine-rich alpha-2-glycoprotein precursor | KLPPGLLANFTLLR | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | ANLVPVPITNATLDQITGK | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | CANLVPVPITNATLDQITGK | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | IYNTTYLNVQR | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | LVPVPITNATLDQITGK | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | PITNATLDQITGK | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | PLCANLVPVPITNATLDQITGK | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | QDQCIYNTTYLN | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | QDQCIYNTTYLNVQR | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | QIPLCANLVPVPITNATLDQITGK | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | AALAAFNAQNNGSNFQLEEISR | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | AQNNGSNFQLEEISR | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | KVCQDCPLLAPLNDTR | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | NAQNNGSNFQLEEISR | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | NGSNFQLEEISR | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | VCQDCPLLAPLNDTR | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | ALPQPQNVTSLL | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | ALPQPQNVTSLLG | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | ALPQPQNVTSLLGCT | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | ALPQPQNVTSLLGCTH | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | CSDGWSFDATTLDDNGTMLFFK | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | SWPAVGNCSSALR | Y | Y | ||
| IPI00023019.1 | SHBG Isoform 1 of Sex hormone-binding globulin precursor | LDVDQALNR | Y | Y | ||
| IPI00023648.6 | ISLR Immunoglobulin superfamily containing leucine-rich repeat protein precursor | DLESVPPGFPANVTTLSLSANR | Y | |||
| IPI00023648.6 | ISLR Immunoglobulin superfamily containing leucine-rich repeat protein precursor | FQAFANGSLLIPDFGK | Y | |||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | AAIPSALDTNSSK | Y | Y | ||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | ALGFENATQALGR | Y | Y | ||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | DAGVVCTNETR | Y | Y | ||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | GLNLTEDTYKPR | Y | Y | ||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | TVIRPFYLTNSSGVD | Y | Y | ||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | YKGLNLTEDTYKPR | Y | Y | ||
| IPI00023814.2 | NEO1 Isoform 1 of Neogenin precursor | LPSGMLVISNATEGDGGLYR | Y | Y | ||
| IPI00023814.2 | NEO1 Isoform 1 of Neogenin precursor | TLSDVPSAAPQNLSLEVR | Y | Y | ||
| IPI00023845.1 | KLK6 Kallikrein-6 precursor | DCSANTTSCHILGWGK | Y | Y | ||
| IPI00024035.1 | CDH6 Isoform 1 of Cadherin-6 precursor | EDAQINTTIGSVTAQDPDAAR | Y | Y | ||
| IPI00024572.3 | ASPH aspartate beta-hydroxylase isoform e | YNLSEVLQGK | Y | |||
| IPI00024621.3 | OLFML3 Isoform 1 of Olfactomedin-like protein 3 precursor | IYVLDGTQNDTAFVFPR | Y | Y | ||
| IPI00024966.1 | CNTN2 Contactin-2 precursor | ANSTGILSVR | Y | Y | ||
| IPI00024966.1 | CNTN2 Contactin-2 precursor | GTEILVNSSR | Y | Y | ||
| IPI00024966.1 | CNTN2 Contactin-2 precursor | VPGADAQYFVYSNESVRPYTPFEVK | Y | |||
| IPI00025257.1 | SEMA7A Semaphorin-7A precursor | EDNPDKNPEAPLNVSR | Y | Y | ||
| IPI00025465.1 | OGN Mimecan precursor | CKANDTSYIR | Y | Y | ||
| IPI00026104.1 | IDS Isoform Long of Iduronate 2-sulfatase precursor | EDVQALNISVPYGPIPVDFQR | Y | Y | ||
| IPI00026104.1 | IDS Isoform Long of Iduronate 2-sulfatase precursor | VHAGNFSTIPQYFK | Y | Y | ||
| IPI00026946.2 | NPTX2 Neuronal pentraxin-2 precursor | ANVSNAGLPGDFR | Y | Y | ||
| IPI00027235.1 | ATRN Isoform 1 of Attractin precursor | IDSTGNVTNELR | Y | Y | ||
| IPI00027235.1 | ATRN Isoform 1 of Attractin precursor | NHSCSEGQISIFR | Y | Y | ||
| IPI00027482.1 | SERPINA6 Corticosteroid-binding globulin precursor | AQLLQGLGFNLTER | Y | Y | ||
| IPI00027827.2 | SOD3 Extracellular superoxide dismutase [Cu-Zn] precursor | AKLDAFFALEGFPTEPNSSSR | Y | Y | ||
| IPI00027827.2 | SOD3 Extracellular superoxide dismutase [Cu-Zn] precursor | LDAFFALEGFPTEPNSSSR | Y | Y | ||
| IPI00027851.1 | HEXA Beta-hexosaminidase alpha chain precursor | SAEGTFFINK | Y | Y | ||
| IPI00029260.2 | CD14 Monocyte differentiation antigen CD14 precursor | NVSWATGR | Y | Y | ||
| IPI00029723.1 | FSTL1 Follistatin-related protein 1 precursor | FVEQNETAINITTYPDQENNK | Y | |||
| IPI00029723.1 | FSTL1 Follistatin-related protein 1 precursor | GSNYSEILDK | Y | Y | ||
| IPI00029739.5 | CFH Isoform 1 of Complement factor H precursor | IPCSQPPQIEHGTINSSR | Y | Y | ||
| IPI00029739.5 | CFH Isoform 1 of Complement factor H precursor | ISEENETTCYMGK | Y | Y | ||
| IPI00029739.5 | CFH Isoform 1 of Complement factor H precursor | LNDTLDYECH | Y | Y | ||
| IPI00029751.1 | CNTN1 Isoform 1 of Contactin-1 precursor | ANSTGTLVITDPTR | Y | Y | ||
| IPI00029751.1 | CNTN1 Isoform 1 of Contactin-1 precursor | GKANSTGTLVITDPTR | Y | Y | ||
| IPI00029751.1 | CNTN1 Isoform 1 of Contactin-1 precursor | GNYSCFVSSPSITK | Y | Y | ||
| IPI00029751.1 | CNTN1 Isoform 1 of Contactin-1 precursor | YIITWDHVVALSNESTVTGYK | Y | Y | ||
| IPI00030887.1 | TYRO3 Tyrosine-protein kinase receptor TYRO3 precursor | DLVPATNYSLR | Y | Y | ||
| IPI00031121.2 | CPE Carboxypeptidase E precursor | DLQGNPIANATISVEGIDHDVTSAK | Y | Y | ||
| IPI00031121.2 | CPE Carboxypeptidase E precursor | GNETIVNLIHSTR | Y | Y | ||
| IPI00032179.2 | SERPINC1 Antithrombin III variant | LGACNDTLQQLMEVFK | Y | Y | ||
| IPI00032179.2 | SERPINC1 Antithrombin III variant | SLTFNETYQDISELVYGAK | Y | Y | ||
| IPI00032179.2 | SERPINC1 Antithrombin III variant | WVSNKTEGR | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | HLVIHNEST | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | HLVIHNESTCEQLAK | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | LQAILGVPWKDKNCTSR | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | VIHNESTCEQLAK | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | VYIHPFHLVIHNEST | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | VYIHPFHLVIHNESTCEQLAK | Y | Y | ||
| IPI00032292.1 | TIMP1 Metalloproteinase inhibitor 1 precursor | FVGTPEVNQTTLYQR | Y | Y | ||
| IPI00032292.1 | TIMP1 Metalloproteinase inhibitor 1 precursor | SHNRSEEFLIAGK | Y | Y | ||
| IPI00032328.2 | KNG1 Isoform HMW of Kininogen-1 precursor | HGIQYFNNNTQHSSLFMLNEVK | Y | Y | ||
| IPI00032328.2 | KNG1 Isoform HMW of Kininogen-1 precursor | ITYSIVQTNCSK | Y | Y | ||
| IPI00032328.2 | KNG1 Isoform HMW of Kininogen-1 precursor | LNAENNATFYFK | Y | Y | ||
| IPI00060310.4 | PLD4 Phospholipase D4 | ELGAVIYNCSHLAQDLEK | Y | |||
| IPI00060310.4 | PLD4 Phospholipase D4 | SLQALSNPAANVSVDVK | Y | |||
| IPI00060310.4 | PLD4 Phospholipase D4 | TWPQNFSSHFNR | Y | |||
| IPI00060310.4 | PLD4 Phospholipase D4 | VFIVPVGNHSNIPFSR | Y | |||
| IPI00064667.4 | CNDP1 Beta-Ala-His dipeptidase precursor | AIHLDLEEYRNSSR | Y | Y | ||
| IPI00064667.4 | CNDP1 Beta-Ala-His dipeptidase precursor | AIHLDLEEYRNSSRVEK | Y | Y | ||
| IPI00064667.4 | CNDP1 Beta-Ala-His dipeptidase precursor | LVPHMNVSAVEK | Y | Y | ||
| IPI00073777.1 | PLXDC2 Isoform 2 of Plexin domain-containing protein 2 precursor | VNLSFDFPFYGHFLR | Y | Y | ||
| IPI00152789.4 | SNED1 67 kDa protein | AYNISVFSVK | Y | Y | ||
| IPI00159927.2 | NCAN Neurocan core protein precursor | ANATLLLGPLR | Y | Y | ||
| IPI00160552.3 | TNR Isoform 1 of Tenascin-R precursor | QSVEEEGGIANYNTSSK | Y | Y | ||
| IPI00163207.1 | PGLYRP2 Isoform 1 of N-acetylmuramoyl-L-alanine amidase precursor | GFGVAIVGNYTAALPTEAALR | Y | Y | ||
| IPI00166392.1 | CADM1 Immunoglobulin superfamily member 4 | FQLLNFSSSELK | Y | Y | ||
| IPI00166392.1 | CADM1 Immunoglobulin superfamily member 4 | VSLTNVSISDEGR | Y | Y | ||
| IPI00166729.4 | AZGP1 alpha-2-glycoprotein 1, zinc | DIVEYYNDSNGSHVLQGR | Y1,Y2 | Y1,Y2 | ||
| IPI00166729.4 | AZGP1 alpha-2-glycoprotein 1, zinc | FGCEIENNR | Y | Y | ||
| IPI00167093.4 | CFHR1 complement factor H-related 1 | LQNNENNISCVER | Y | |||
| IPI00168728.1 | IGHM FLJ00385 protein (Fragment) | EEQFNSTFR | Y | |||
| IPI00168728.1 | IGHM FLJ00385 protein (Fragment) | KPREEQFNSTFR | Y | |||
| IPI00168728.1 | IGHM FLJ00385 protein (Fragment) | TKPREEQFNSTFR | Y | |||
| IPI00171411.4 | GOLM1 Golgi membrane protein 1 | AVLVNNITTGER | Y | |||
| IPI00171473.2 | SPON1 Spondin-1 precursor | LTFYGNWSEK | Y | Y | ||
| IPI00176427.1 | CADM4 Cell adhesion molecule 4 precursor | QTLFFNGTR | Y | Y | ||
| IPI00178926.2 | IGJ immunoglobulin J chain | IIVPLNNRENISDPTSPLR | Y | Y | ||
| IPI00215894.1 | KNG1 Isoform LMW of Kininogen-1 precursor | HGIQYFNNNTQHSSLFMLNEVK | Y | Y | ||
| IPI00215894.1 | KNG1 Isoform LMW of Kininogen-1 precursor | HGIQYFNNNTQHSSLFMLNEVKR | Y | Y | ||
| IPI00215894.1 | KNG1 Isoform LMW of Kininogen-1 precursor | ITYSIVQTNCSK | Y | Y | ||
| IPI00215894.1 | KNG1 Isoform LMW of Kininogen-1 precursor | ITYSIVQTNCSKENFLFLTPDCK | Y | Y | ||
| IPI00215894.1 | KNG1 Isoform LMW of Kininogen-1 precursor | KYNSQNQSNNQFVLYR | Y | |||
| IPI00215894.1 | KNG1 Isoform LMW of Kininogen-1 precursor | LNAENNATFYFK | Y | Y | ||
| IPI00216250.5 | CNTNAP4 Cell recognition protein CASPR4 | TNETQTYWGGSSPDLQK | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | ANSTGTLVITDPTR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | GKANSTGTLVITDPTR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | GNYSCFVSSPSITK | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | YIITWDHVVALSNESTVTGYK | Y | Y | ||
| IPI00217376.1 | SCN4B Isoform 1 of Sodium channel subunit beta-4 precursor | WTYNSSDAFK | Y | Y | ||
| IPI00217882.3 | SORT1 Sortilin precursor | DITDLINNTFIR | Y | Y | ||
| IPI00217882.3 | SORT1 Sortilin precursor | HLYTTTGGETDFTNVTSLR | Y | Y | ||
| IPI00218192.2 | ITIH4 Isoform 2 of Inter-alpha-trypsin inhibitor heavy chain H4 precursor | LPTQNITFQTESSVAEQEAEFQSPK | Y | Y | ||
| IPI00218732.3 | PON1 Serum paraoxonase/arylesterase 1 | HANWTLTPLK | Y | Y | ||
| IPI00218732.3 | PON1 Serum paraoxonase/arylesterase 1 | VTQVYAENGTVLQGSTVASVYK | Y | Y | ||
| IPI00242956.4 | FCGBP IgGFc-binding protein precursor | VITVQVANFTLR | Y | |||
| IPI00242956.4 | FCGBP IgGFc-binding protein precursor | YLPVNSSLLTSDCSER | Y | |||
| IPI00290856.4 | LYVE1 Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor | KANQQLNFTEAK | Y | Y | ||
| IPI00291136.4 | COL6A1 Collagen alpha-1(VI) chain precursor | GEDGPAGNGTEGFPGFPGYPGNR | Y | Y | ||
| IPI00291136.4 | COL6A1 Collagen alpha-1(VI) chain precursor | NVTAQICIDK | Y | Y | ||
| IPI00291867.3 | CFI Complement factor I precursor | FLNNGTCTAEGK | Y | Y | ||
| IPI00292071.6 | SCG3 Secretogranin-3 precursor | NKLEKNATDNISK | Y | |||
| IPI00292071.6 | SCG3 Secretogranin-3 precursor | TYPPENKPGQSNYSFVDNLNLLK | Y | |||
| IPI00292732.3 | FMOD fibromodulin precursor | LYLDHNNLTR | Y | Y | ||
| IPI00292946.1 | SERPINA7 Thyroxine-binding globulin precursor | TLYETEVFSTDFSNISAAK | Y | Y | ||
| IPI00294193.4 | ITIH4 Isoform 1 of Inter-alpha-trypsin inhibitor heavy chain H4 precursor | LPTQNITFQTESSVAEQEAEFQSPK | Y | Y | ||
| IPI00294395.1 | C8B Complement component C8 beta chain precursor | EYESYSDFERNVTEK | Y | Y | ||
| IPI00294650.5 | FRZB Secreted frizzled-related protein 3 precursor | SLPWNMTK | Y | Y | ||
| IPI00294776.3 | RELN Isoform 1 of Reelin precursor | APSNVSTIIHILYLPEDAK | Y | Y | ||
| IPI00294776.3 | RELN Isoform 1 of Reelin precursor | HDYILLPEDALTNTTR | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | QNITYLLK | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | SLEVLNLSSNK | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | SLWNMSAANNNIK | Y | Y | ||
| IPI00296165.5 | C1R;ACYP1;C17orf13 Complement C1r subcomponent precursor | EHEAQSNASLDVFLGHTNVEELMK | Y | Y | ||
| IPI00296534.1 | FBLN1 Isoform D of Fibulin-1 precursor | CATPHGDNASLEATFVK | Y | Y | ||
| IPI00296608.6 | C7 Complement component C7 precursor | INNDFNYEFYNSTWSYVK | Y | Y | ||
| IPI00296608.6 | C7 Complement component C7 precursor | NYTLTGR | Y | Y | ||
| IPI00297124.1 | IL6ST Isoform 1 of Interleukin-6 receptor subunit beta precursor | EQYTIINR | Y | Y | ||
| IPI00297124.1 | IL6ST Isoform 1 of Interleukin-6 receptor subunit beta precursor | ETHLETNFTLK | Y | Y | ||
| IPI00297124.1 | IL6ST Isoform 1 of Interleukin-6 receptor subunit beta precursor | LTVNLTNDR | Y | |||
| IPI00297124.1 | IL6ST Isoform 1 of Interleukin-6 receptor subunit beta precursor | NYTIFYR | Y | Y | ||
| IPI00297263.6 | HEG1 Isoform 1 of Protein HEG homolog 1 precursor | SHAASDAPENLTLLAETADAR | Y | Y | ||
| IPI00297646.4 | COL1A1 Collagen alpha-1(I) chain precursor | LMSTEASQNITYHCK | Y | Y | ||
| IPI00298828.3 | APOH Beta-2-glycoprotein 1 precursor | LGNWSAMPSCK | Y | Y | ||
| IPI00298828.3 | APOH Beta-2-glycoprotein 1 precursor | VYKPSAGNNSLYR | Y | Y | ||
| IPI00298971.1 | VTN Vitronectin precursor | NGSLFAFR | Y | Y | ||
| IPI00298971.1 | VTN Vitronectin precursor | NISDGFDGIPDNVDAALALPAHSYSGR | Y | Y | ||
| IPI00298971.1 | VTN Vitronectin precursor | NNATVHEQVGGPSLTSDLQAQSK | Y | Y | ||
| IPI00301395.4 | CPVL Probable serine carboxypeptidase CPVL precursor | QAIHVGNQTFNDGTIVEK | Y | Y | ||
| IPI00301395.4 | CPVL Probable serine carboxypeptidase CPVL precursor | SYAGFLTVNK | Y | Y | ||
| IPI00301512.3 | DPP6 Isoform DPPX-L of Dipeptidyl aminopeptidase-like protein 6 | LAYAAINDSR | Y | Y | ||
| IPI00301579.3 | NPC2 Epididymal secretory protein E1 precursor | GQSYSVNVTFTSNIQSK | Y | Y | ||
| IPI00302641.1 | FAT2 Protocadherin Fat 2 precursor | ASEYTVSIQSNVSK | Y | Y | ||
| IPI00302641.1 | FAT2 Protocadherin Fat 2 precursor | VPENITLYTPILHTQAR | Y | Y | ||
| IPI00303210.3 | ENPP2 Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 precursor | AEGWEEGPPTVLSDSPWTNISGSCK | Y | Y | ||
| IPI00303210.3 | ENPP2 Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 precursor | AIIANLTCK | Y | Y | ||
| IPI00303963.1 | C2 Complement C2 precursor (Fragment) | QSVPAHFVALNGSK | Y | Y | ||
| IPI00307276.1 | ADAMTS4 ADAMTS-4 precursor | EEEIVFPEKLNGSVLPGSGAPAR | Y | |||
| IPI00328609.3 | SERPINA4 Kallistatin precursor | DFYVDENTTVR | Y | Y | ||
| IPI00328609.3 | SERPINA4 Kallistatin precursor | FLNDTMAVYEAK | Y | Y | ||
| IPI00328609.3 | SERPINA4 Kallistatin precursor | SQILEGLGFNLTELSESDVHR | Y | Y | ||
| IPI00328609.3 | SERPINA4 Kallistatin precursor | TTPKDFYVDENTTVR | Y | Y | ||
| IPI00329775.7 | CPB2 Isoform 1 of Carboxypeptidase B2 precursor | KQVHFFVNASDVDNVK | Y | Y | ||
| IPI00329775.7 | CPB2 Isoform 1 of Carboxypeptidase B2 precursor | QVHFFVNASDVDNVK | Y | Y | ||
| IPI00332273.2 | PTPRS Isoform PTPS-MEC of Receptor-type tyrosine-protein phosphatase S precursor | KVEAEALNATAIR | Y | Y | ||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | AENQVNVTCQVR | Y | Y | ||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | GTANLSETIR | Y | Y | ||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | IGNITPADAGTYYCVK | Y | |||
| IPI00333140.8 | DNER Delta and Notch-like epidermal growth factor-related receptor precursor | LVSFEVPQNTSVK | Y | Y | ||
| IPI00333140.8 | DNER Delta and Notch-like epidermal growth factor-related receptor precursor | WDQVEVIPDIACGNASSNSSAGGR | Y1,Y2 | |||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | DGDDEWTSVVVANVSK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | ERPPTFLTPEGNASNK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | ERPPTFLTPEGNASNKEELR | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | FNHTQTIQQK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | GSALHEDIYVLHENGTLEIPVAQK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | LSPYVNYSFR | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | QKDGDDEWTSVVVANVSK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | VISVDELNDTIAANLSDTEFYGAK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | VNVVNSTLAEVHWDPVPLK | Y | Y | ||
| IPI00333776.5 | NRCAM Isoform 1 of Neuronal cell adhesion molecule precursor | YQPINSTHELGPLVDLK | Y | Y | ||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | DKLVLPAKNTTNLK | Y | |||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | LVLPAKNTTNLK | Y | |||
| IPI00377015.5 | EFNA1 Isoform 2 of Ephrin-A1 precursor | HTVFWNSSNPK | Y | Y | ||
| IPI00382750.1 | GNPTG Similar to protein kinase C substrate | YEFCPFHNVTQHEQTFR | Y | Y | ||
| IPI00384938.1 | IGHG1 Putative uncharacterized protein DKFZp686N02209 | TVLHQDWLNGK | Y | |||
| IPI00394992.1 | PGLYRP2 Isoform 2 of N-acetylmuramoyl-L-alanine amidase precursor | GFGVAIVGNYTAALPTEAALR | Y | Y | ||
| IPI00395488.2 | VASN Vasorin precursor | LHEITNETFR | Y | Y | ||
| IPI00399307.2 | PRCP prolylcarboxypeptidase isoform 2 preproprotein | NYSVLYFQQK | Y | Y | ||
| IPI00400826.1 | CLU clusterin isoform 1 | ELPGVCNETMMALWEECKPCLK | Y | Y | ||
| IPI00400826.1 | CLU clusterin isoform 1 | KEDALNETR | Y | Y | ||
| IPI00400826.1 | CLU clusterin isoform 1 | KKEDALNETR | Y | Y | ||
| IPI00400826.1 | CLU clusterin isoform 1 | LANLTQGEDQYYLR | Y | Y | ||
| IPI00400826.1 | CLU clusterin isoform 1 | MLNTSSLLEQLNEQFNWVSR | Y | Y | ||
| IPI00400826.1 | CLU clusterin isoform 1 | QLEEFLNQSSPFYFWMNGDR | Y | Y | ||
| IPI00413016.4 | CADM2 Isoform 1 of Cell adhesion molecule 2 precursor | ELNILFLNK | Y | |||
| IPI00413696.5 | CD47 41 kDa protein | SDAVSHTGNYTCEVTELTR | Y | Y | ||
| IPI00418183.4 | SGCE sarcoglycan, epsilon isoform 2 | LNAINITSALDR | Y | Y | ||
| IPI00418531.4 | GLDN Isoform 1 of Gliomedin | TFSVVQHVNTTYPK | Y | |||
| IPI00419724.2 | SEMA4B semaphorin 4B precursor | FEAEHISNYTALLLSR | Y | |||
| IPI00431645.1 | HP HP protein | MVSHHNLTTGATLINEQWLLTTAK | Y | Y | ||
| IPI00431645.1 | HP HP protein | NLFLNHSENATAK | Y1,Y2 | Y1,Y2 | ||
| IPI00431645.1 | HP HP protein | VSHHNLTTGATLINEQWLLTTAK | Y | Y | ||
| IPI00431645.1 | HP HP protein | VVLHPNYSQVDIGLIK | Y | Y | ||
| IPI00431645.1 | HP HP protein | VVLHPNYSQVDIGLIKL | Y | Y | ||
| IPI00433478.3 | ASPH ASPH protein | YNLSEVLQGK | Y | |||
| IPI00441498.1 | FOLR1 Folate receptor alpha precursor | GWNWTSGFNK | Y | Y | ||
| IPI00456623.2 | BCAN Isoform 1 of Brevican core protein precursor | TLFLFPNQTGFPNK | Y | Y | ||
| IPI00456623.2 | BCAN Isoform 1 of Brevican core protein precursor | VALPAYPASLTDVSLALSELRPNDSGIYR | Y | Y | ||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | ELNILFLNK | Y | |||
| IPI00470696.1 | UNC5D Isoform 1 of Netrin receptor UNC5D precursor | EVFINVTR | Y | Y | ||
| IPI00472011.1 | NEO1 154 kDa protein | TLSDVPSAAPQNLSLEVR | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | GNEANYYSNATTDEHGLVQF | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | IYVLDYLNETQQLTPEVK | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | SLGNVNFTVSAEALESQELCGTEVPSVPEHGR | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | VSNQTLSLFF | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | VSNQTLSLFFTVLQDVPVR | Y | Y | ||
| IPI00478809.3 | F5 Coagulation factor V precursor | TNINSSRDPDNIAAWYLR | Y | Y | ||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | IDVNSWIENFTK | Y | |||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | ISDNNTEFLLNFNEFIDR | Y | Y | ||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | SFSGVLDCGNCSR | Y | |||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | VLKDAVNNITAK | Y | Y | ||
| IPI00513705.1 | NFASC Isoform 1 of Neurofascin precursor | QIVENFSPNQTK | Y | Y | ||
| IPI00513705.1 | NFASC Isoform 1 of Neurofascin precursor | WANITWK | Y | |||
| IPI00513705.1 | NFASC Isoform 1 of Neurofascin precursor | YVAFNGTK | Y | Y | ||
| IPI00513964.1 | SEMA4B Isoform 2 of Semaphorin-4B precursor | FEAEHISNYTALLLSR | Y | |||
| IPI00514397.1 | APOM Apolipoprotein M | TELFSSSCPGGIMLNETGQGYQR | Y | Y | ||
| IPI00549291.4 | IGHM IGHM protein | GLTFQQNASSMCVPDQDTAIR | Y | |||
| IPI00552450.1 | OPCML opioid binding protein/cell adhesion molecule-like isoform b preproprotein | DYGNYTCVATNK | Y | Y | ||
| IPI00552450.1 | OPCML opioid binding protein/cell adhesion molecule-like isoform b preproprotein | MSTLTFFNVSEK | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | ADTHDEILEGLNFNLT | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | FNLTEIPEAQIHEGFQELLR | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | GNATAIFFLPDEGK | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | LGNATAIFFLPDEGK | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIF | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPV | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATA | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATAF | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATAFAM | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATAFAMLSLGTK | Y | |||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | YLGNATAIF | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | YLGNATAIFF | Y | Y | ||
| IPI00553177.1 | SERPINA1 Isoform 1 of Alpha-1-antitrypsin precursor | YLGNATAIFFLPDEGK | Y | Y | ||
| IPI00554518.1 | IL6ST IL6ST nirs variant 4 | ETHLETNFTLK | Y | Y | ||
| IPI00554518.1 | IL6ST IL6ST nirs variant 4 | NYTIFYR | Y | Y | ||
| IPI00554538.3 | TPP1 60 kDa protein | FLSSSPHLPPSSYFNASGR | Y | |||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | QSVEEEGGIANYNTSSK | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | DEGTYTCALHHSGHSPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HENTSSSPIQYEF | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HENTSSSPIQYEFSLTR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | LDCRHENTSSSPIQYEFSLTR | Y | Y | ||
| IPI00555628.1 | NCAM1 Neural cell adhesion molecule 1, 120 kDa isoform variant (Fragment) | DGQLLPSSNYSNIK | Y | |||
| IPI00555628.1 | NCAM1 Neural cell adhesion molecule 1, 120 kDa isoform variant (Fragment) | IYNTPSASYLEVTPDSENDFGNYNCTAVNR | Y | |||
| IPI00556575.1 | FGFR3 Fibroblast growth factor receptor 3 isoform 1 variant (Fragment) | LQVLNASHEDSGAYSCR | Y | Y | ||
| IPI00607580.2 | MEGF8 multiple EGF-like-domains 8 | ALLTNVSSVALGSR | Y | |||
| IPI00607600.1 | APLP1 amyloid precursor-like protein 1 isoform 1 precursor | KVNASVPR | Y | Y | ||
| IPI00607600.1 | APLP1 amyloid precursor-like protein 1 isoform 1 precursor | VNQSLGLLDQN | Y | Y | ||
| IPI00607600.1 | APLP1 amyloid precursor-like protein 1 isoform 1 precursor | VNQSLGLLDQNPHLAQELR | Y | Y | ||
| IPI00607648.1 | NRXN1 Isoform 2 of Neurexin-1-alpha precursor | NTTLFIDQVEAK | Y | Y | ||
| IPI00607648.1 | NRXN1 Isoform 2 of Neurexin-1-alpha precursor | VNSSQVLPVDSGEVK | Y | Y | ||
| IPI00607652.1 | OLFML3 Isoform 2 of Olfactomedin-like protein 3 precursor | IYVLDGTQNDTAFVFPR | Y | |||
| IPI00639937.1 | CFB Complement factor B | SPYYNVSDEISFH | Y | |||
| IPI00639937.1 | CFB Complement factor B | SPYYNVSDEISFHCYDGYTLR | Y | |||
| IPI00641737.1 | HP Haptoglobin precursor | MVSHHNLTTGATLINEQWLLTTAK | Y | Y | ||
| IPI00641737.1 | HP Haptoglobin precursor | NLFLNHSENATAK | Y1,Y2 | Y1,Y2 | ||
| IPI00641737.1 | HP Haptoglobin precursor | VVLHPNYSQVDIGLIK | Y | Y | ||
| IPI00641940.1 | PCDH9 Protocadherin 9 | IVASDSGKPSLNQTALVR | Y | Y | ||
| IPI00642017.1 | IGHA2 Putative uncharacterized protein DKFZp686C02218 (Fragment) | LAGKPTHVNVSVVMAEVDGTC | Y | Y | ||
| IPI00642017.1 | IGHA2 Putative uncharacterized protein DKFZp686C02218 (Fragment) | LSLHRPALEDLLLGSEANLTCTLTGLR | Y | Y | ||
| IPI00642017.1 | IGHA2 Putative uncharacterized protein DKFZp686C02218 (Fragment) | TPLTANITK | Y | Y | ||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | EGHFYYNISEVK | Y | Y | ||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | GAFFPLTERNWSLPNR | Y | Y | ||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | GKEGHFYYNISEVK | Y | |||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | IYSNHSALESLALIPLQAPLK | Y | |||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | NWSLPNR | Y | |||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | RGKEGHFYYNISEVK | Y | |||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | VSNVSCQASVSR | Y | |||
| IPI00643506.3 | C2 Complement component 2 | QSVPAHFVALNGSK | Y | |||
| IPI00643506.3 | C2 Complement component 2 | TMFPNLTDVR | Y | |||
| IPI00643525.1 | C4A Complement component 4A | FSDGLESNSSTQFEVK | Y | Y | ||
| IPI00643525.1 | C4A Complement component 4A | FSDGLESNSSTQFEVKK | Y | Y | ||
| IPI00643525.1 | C4A Complement component 4A | GLNVTLSSTGR | Y | Y | ||
| IPI00643525.1 | C4A Complement component 4A | GLNVTLSSTGRNGFK | Y | Y | ||
| IPI00643525.1 | C4A Complement component 4A | NTTCQDLQIEVTVK | Y | Y | ||
| IPI00643663.1 | PCSK2 Proprotein convertase subtilisin/kexin type 2 | YLEHVQAVITVNATR | Y | Y | ||
| IPI00644276.3 | CNTNAP4 cell recognition protein CASPR4 isoform 2 | TNETQTYWGGSSPDLQK | Y | |||
| IPI00645038.1 | ITIH2 Inter-alpha (Globulin) inhibitor H2 | GAFISNFSMTVDGK | Y | Y | ||
| IPI00654888.4 | KLKB1 Plasma kallikrein precursor | GVNFNVSK | Y | Y | ||
| IPI00654888.4 | KLKB1 Plasma kallikrein precursor | IYPGVDFGGEELNVTFVK | Y | Y | ||
| IPI00655702.3 | NFASC Isoform 5 of Neurofascin precursor | WANITWK | Y | |||
| IPI00655927.1 | PRG4 Isoform B of Proteoglycan-4 precursor | NGTLVAFR | Y | Y | ||
| IPI00656113.2 | SIRPA Signal-regulatory protein alpha | AENQVNVTCQVR | Y | |||
| IPI00656113.2 | SIRPA Signal-regulatory protein alpha | GTANLSETIR | Y | |||
| IPI00656113.2 | SIRPA Signal-regulatory protein alpha | LQLTWLENGNVSR | Y | |||
| IPI00739477.1 | PILRA Isoform 2 of Paired immunoglobulin-like type 2 receptor alpha precursor | LFLNWTEGQK | Y | Y | ||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | IAVQFGPGFSWIANFTK | Y | Y | ||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | VASVININPNTTHSTGSCR | Y | Y | ||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | VQPFNVTQGK | Y | Y | ||
| IPI00743766.2 | FETUB Fetuin-B precursor | GCNDSDVLAVAGFALR | Y | Y | ||
| IPI00743766.2 | FETUB Fetuin-B precursor | VLYLAAYNCTLRPVSK | Y | Y | ||
| IPI00744685.2 | BTD Uncharacterized protein BTD (Fragment) | DVQIIVFPEDGIHGFNFTR | Y | |||
| IPI00744685.2 | BTD Uncharacterized protein BTD (Fragment) | FNDTEVLQR | Y | |||
| IPI00744685.2 | BTD Uncharacterized protein BTD (Fragment) | NPVGLIGAENATGETDPSHSK | Y | |||
| IPI00744685.2 | BTD Uncharacterized protein BTD (Fragment) | WNPCLEPHRFNDTEVLQR | Y | |||
| IPI00744685.2 | BTD Uncharacterized protein BTD (Fragment) | YQFNTNVVFSNNGTLVDR | Y | |||
| IPI00745089.2 | A1BG alpha 1B-glycoprotein precursor | EGDHEFLEVPEAQEDVEATFPVHQPGNYSCSYR | Y | Y | ||
| IPI00745207.1 | B3GNT2 45 kDa protein | DTFFNLSLK | Y | |||
| IPI00748395.2 | SEZ6 seizure related 6 homolog isoform 2 | EGETVTVEGLGGPDPLPLANQSFLLR | Y | |||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | DGEAFEINGTEDGR | Y | Y | ||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | IIPSNNSGTFR | Y | |||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | ISGVNLTQK | Y | Y | ||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | LTWEAGADHNSNISEYIVEFEGNKEEPGR | Y | |||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | VTWKPQGAPVEWEEETVTNHTLR | Y | Y | ||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | YHIYENGTLQINR | Y1,Y2 | Y1,Y2 | ||
| IPI00783987.2 | C3 Complement C3 precursor (Fragment) | TVLTPATNHMGNVTF | Y | Y | ||
| IPI00783987.2 | C3 Complement C3 precursor (Fragment) | TVLTPATNHMGNVTFTIPANR | Y | Y | ||
| IPI00784119.1 | ATP6AP1 Vacuolar ATP synthase subunit S1 precursor | LNASLPALLLIR | Y | Y | ||
| IPI00784169.1 | CD55 Decay-accelerating factor splicing variant 1 | GSQWSDIEEFCNR | Y | |||
| IPI00784432.1 | CBX6 53 kDa protein | VNLSAAPAPVSAVPTGLHSK | Y | |||
| IPI00784807.1 | IGHG2 Putative uncharacterized protein | EEQFNSTFR | Y | Y | ||
| IPI00784807.1 | IGHG2 Putative uncharacterized protein | TKPREEQFNSTFR | Y | Y | ||
| IPI00787050.1 | NPTX1 similar to neuronal pentraxin I precursor | LNSSSQTNSLKDLLQSK | Y | |||
| IPI00788159.1 | DPP7 similar to Dipeptidyl-peptidase 2 precursor | ALAGLVYNASGSEHCYDIYR | Y | |||
| IPI00789795.1 | ADAM22 98 kDa protein | LFEFSLDDLPSEFQQVNITPSK | Y | |||
| IPI00790218.1 | ICOSLG Uncharacterized protein ICOSLG | LFNVTPQDEQK | Y | |||
| IPI00790218.1 | ICOSLG Uncharacterized protein ICOSLG | TVVTYHIPQNSSLENVDSR | Y | |||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | ADTHDEILEGLNFNLT | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | ADTHDEILEGLNFNLTEIPEAQIH | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | FNLTEIPEAQIHEGFQELLR | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | GNATAIFFLPDEGK | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIF | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATA | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATAF | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATAFAMLSLGTK | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | YLGNATAI | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | YLGNATAIF | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | YLGNATAIFF | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | YLGNATAIFFLPDEGK | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | YLGNATAIFFLPDEGKL | Y | Y | ||
| IPI00790784.2 | SERPINA1 Isoform 2 of Alpha-1-antitrypsin precursor | YLGNATAIFFLPDEGKLQHLENELTHDIITK | Y | Y | ||
| IPI00793751.1 | MFAP4 Uncharacterized protein MFAP4 | FNGSVSFFR | Y | |||
| IPI00793751.1 | MFAP4 Uncharacterized protein MFAP4 | VDLEDFENNTAYAK | Y | |||
| IPI00793848.1 | CLU 54 kDa protein | ANLTQGEDQYYLR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | EDALNETR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | EDALNETRESETK | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | EIRHNSTGCLR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | ELPGVCNETMMALWEECKPCLK | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | HNSTGCLR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | KEDALNETR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | KEDALNETRESETK | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | KKEDALNETR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | KKEDALNETRESETK | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | KKKEDALNETR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | KKKEDALNETRESETK | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | LANLTQGEDQYYLR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | LKELPGVCNETMMALWEECKPCLK | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | MLNTSSLLEQLN | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | MLNTSSLLEQLNEQFNWVSR | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | QLEEFLNQS | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | QLEEFLNQSSPF | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | QLEEFLNQSSPFYF | Y | Y | ||
| IPI00793848.1 | CLU 54 kDa protein | QLEEFLNQSSPFYFWMNGDR | Y | Y | ||
| IPI00794403.1 | LUM 23 kDa protein | AFENVTDLQWLILDHNLLENSK | Y | Y | ||
| IPI00794403.1 | LUM 23 kDa protein | KLHINHNNLTESVGPLPK | Y | Y | ||
| IPI00794403.1 | LUM 23 kDa protein | LGSFEGLVNLTFIHLQHNR | Y | Y | ||
| IPI00794403.1 | LUM 23 kDa protein | LHINHNNLTESVGPLPK | Y | Y | ||
| IPI00795624.1 | NELL2 Cerebral protein-12 | QVPGLHNGTK | Y | Y | ||
| IPI00795801.1 | CD109 Isoform 4 of CD109 antigen precursor | INYTVPQSGTFK | Y | Y | ||
| IPI00795801.1 | CD109 Isoform 4 of CD109 antigen precursor | TQDEILFSNSTR | Y | Y | ||
| IPI00795918.1 | NCAM1 neural cell adhesion molecule 1 isoform 2 | DGQLLPSSNYSNIK | Y | Y | ||
| IPI00795918.1 | NCAM1 neural cell adhesion molecule 1 isoform 2 | IYNTPSASYLEVTPDSENDFGNYNCTAVNR | Y | Y | ||
| IPI00796279.1 | SERPINF1 25 kDa protein | VTQNLTLIEESLTSEFIHDIDR | Y | Y | ||
| IPI00796279.1 | SERPINF1 25 kDa protein | VTQNLTLIEESLTSEFIHDIDRELK | Y | Y | ||
| IPI00797025.1 | PRNP Major prion protein | GENFTETDVK | Y | Y | ||
| IPI00797025.1 | PRNP Major prion protein | QHTVTTTTKGENFTETDVK | Y | Y | ||
| IPI00797539.1 | NELL2 80 kDa protein | QVPGLHNGTK | Y | Y | ||
| IPI00798167.1 | PON1 32 kDa protein | HANWTLTPLK | Y | |||
| IPI00798167.1 | PON1 32 kDa protein | VTQVYAENGTVLQGSTVASVYK | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | CGLVPVLAENYNK | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | CGLVPVLAENYNKSDN | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | CGLVPVLAENYNKSDNCEDTPEAGYFAVAVVK | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | GLVPVLAENYNK | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | LVPVLAENYNK | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | PVLAENYNK | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTD | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDC | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGN | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGNF | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGNFC | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGNFCLF | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGNFCLFR | Y | |||
| IPI00798430.1 | TF Transferrin variant (Fragment) | VPVLAENYNK | Y | |||
| IPI00807403.1 | ALCAM Isoform 2 of CD166 antigen precursor | LNLSENYTLSISNAR | Y | Y | ||
| IPI00807403.1 | ALCAM Isoform 2 of CD166 antigen precursor | NATVVWMK | Y | Y | ||
| IPI00815926.1 | IGHG1 IGHG1 protein | TKPREEQYNSTYR | Y | Y | ||
| IPI00829683.1 | FGFR1 fibroblast growth factor receptor 1 isoform 9 precursor | SPHRPILQAGLPANK | Y | Y | ||
| IPI00829767.1 | IGHG2 Uncharacterized protein IGHG2 (Fragment) | EEQFNSTFR | Y | Y | ||
| IPI00829767.1 | IGHG2 Uncharacterized protein IGHG2 (Fragment) | TKPREEQFNSTFR | Y | Y | ||
| IPI00847381.1 | SEPP1 selenoprotein P isoform 2 | EGYSNISYIVVNHQGISSR | Y | Y | ||
| IPI00847589.2 | RELN reelin isoform b | APSNVSTIIHILYLPEDAK | Y | Y | ||
| IPI00847589.2 | RELN reelin isoform b | HDYILLPEDALTNTTR | Y | Y | ||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | FNLTETSEAEIHQSFQH | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | FNLTETSEAEIHQSFQHLLR | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | GAHNTTLTEILK | Y | Y | ||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | GLKFNLTETSEAEIHQSFQHLLR | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | LSLGAHNTTLTEILK | Y | Y | ||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | TLNQSSDELQLSMGN | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | TLNQSSDELQLSMGNAMFVK | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | YTGNASALFILPDQDK | Y | Y | ||
| IPI00848309.1 | SIRPA Isoform 2 of Tyrosine-protein phosphatase non-receptor type substrate 1 precursor | AENQVNVTCQVR | Y | |||
| IPI00848309.1 | SIRPA Isoform 2 of Tyrosine-protein phosphatase non-receptor type substrate 1 precursor | GTANLSETIR | Y | |||
| IPI00848309.1 | SIRPA Isoform 2 of Tyrosine-protein phosphatase non-receptor type substrate 1 precursor | LQLTWLENGNVSR | Y | |||
| IPI00852617.1 | NBL1 neuroblastoma, suppression of tumorigenicity 1 1 | NITQIVGH | Y | |||
| IPI00852617.1 | NBL1 neuroblastoma, suppression of tumorigenicity 1 1 | NITQIVGHSGCEAK | Y | |||
| IPI00852846.1 | NBL1 Neuroblastoma, suppression of tumorigenicity 1 | NITQIVGHSGCEAK | Y | |||
| IPI00853369.1 | PLXNB2 Plexin-B2 precursor | TEAGAFEYVPDPTFENFTGGVK | Y | Y | ||
| IPI00853455.1 | CTSD Protein | GSLSYLNVTR | Y | Y | ||
| IPI00853589.1 | SGCE sarcoglycan, epsilon isoform 3 | LNAINITSALDR | Y | Y | ||
| IPI00855785.1 | FN1 Isoform 15 of Fibronectin precursor | DQCIVDDITYNVNDTFHK | Y | Y | ||
| IPI00855785.1 | FN1 Isoform 15 of Fibronectin precursor | LDAPTNLQFVNETDSTVLVR | Y | Y | ||
| IPI00855785.1 | FN1 Isoform 15 of Fibronectin precursor | WTPLNSSTIIGYR | Y | |||
| IPI00855821.1 | NRXN1-alpha | NTTLFIDQVEAK | Y | Y | ||
| IPI00855821.1 | NRXN1-alpha | SGGNATLQVDSWPVIER | Y | Y | ||
| IPI00855821.1 | NRXN1-alpha | VNSSQVLPVDSGEVK | Y | Y | ||
| IPI00855835.1 | Insulin-like growth factor binding protein 3 isoform b | GLCVNASAVSR | Y | |||
| IPI00855880.2 | SNED1 Isoform 4 of Sushi, nidogen and EGF-like domain-containing protein 1 precursor | AYNISVFSVK | Y | |||
| IPI00855916.1 | Transthyretin | ALGISPFHEHAEVVFTANDSGPR | Y | |||
| IPI00867588.1 | FN1 Isoform 13 of Fibronectin precursor | DQCIVDDITYNVNDTFHK | Y | Y | ||
| IPI00867588.1 | FN1 Isoform 13 of Fibronectin precursor | LDAPTNLQFVNETDSTVLVR | Y | Y | ||
| IPI00871267.1 | L1CAM 140 kDa protein | GYNVTYWR | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | FFPYANGTLGIR | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | GYNVTYWR | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | THNLTDLSPHLR | Y | Y | ||
| IPI00871792.1 | PTPRZ1 265 kDa protein | ESFLQTNYTEIR | Y | Y | ||
| IPI00871792.1 | PTPRZ1 265 kDa protein | TVEINLTNDYR | Y | Y | ||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | LEPNSVDPENITEIFIANQK | Y | |||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | NLTIVDSGLK | Y | |||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | NSNLQHINFTR | Y | |||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | SSPDTQDLYCLNESSK | Y | |||
| IPI00872555.2 | CFI cDNA FLJ76262, highly similar to Homo sapiens I factor (complement) (IF), mRNA | FLNNGTCTAEGK | Y | |||
| IPI00872555.2 | CFI cDNA FLJ76262, highly similar to Homo sapiens I factor (complement) (IF), mRNA | LISNCSK | Y | |||
| IPI00872573.1 | C1RL 48 kDa protein | GFLALYQTVAVNYSQPISEASR | Y | Y | ||
| IPI00873020.1 | PSAP Prosaposin variant | NSTKQEILAALEK | Y | Y | ||
| IPI00873020.1 | PSAP Prosaposin variant | TNSTFVQALVEHVK | Y | Y | ||
| IPI00873201.1 | PSAP Isoform Sap-mu-6 of Proactivator polypeptide precursor | NLEKNSTKQEILAALEK | Y | Y | ||
| IPI00873201.1 | PSAP Isoform Sap-mu-6 of Proactivator polypeptide precursor | NSTKQEILAALEK | Y | Y | ||
| IPI00873201.1 | PSAP Isoform Sap-mu-6 of Proactivator polypeptide precursor | TNSTFVQALVEHVKEECDR | Y | Y | ||
| IPI00873341.1 | PTPRG Uncharacterized protein PTPRG | VEFHWGHSNGSAGSEHSINGR | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | DGDDEWTSVVVANVSK | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | ERPPTFLTPEGNASNK | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | ERPPTFLTPEGNASNKEELR | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | FNHTQTIQQK | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | GSALHEDIYVLHENGTLEIPVAQK | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | QKDGDDEWTSVVVANVSK | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | VISVDELNDTIAANLSDTEFYGAK | Y1,Y2 | Y1,Y2 | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | YQPINSTHELGPLVDLK | Y | Y | ||
| IPI00877792.1 | FGG 50 kDa protein | VDKDLQSLEDILHQVENK | Y | Y | ||
| IPI00877967.1 | F2 36 kDa protein | YPHKPEINSTTHPGADLQENFCR | Y | Y | ||
| IPI00879573.1 | SERPIND1 Heparin cofactor 2 precursor | NLSMPLLPADFHK | Y | Y | ||
| IPI00879665.1 | SEZ6L Seizure related 6 homolog (Mouse)-like | DPYWNDTEPLCR | Y | Y | ||
| IPI00879665.1 | SEZ6L Seizure related 6 homolog (Mouse)-like | SVNLSDGELLSIR | Y | Y | ||
| IPI00879709.2 | C6 complement component 6 precursor | VLNFTTK | Y | |||
| IPI00879931.1 | SERPING1 cDNA FLJ78023, highly similar to Homo sapiens serine (or cysteine) proteinase inhibitor, clade G (C1inhibitor), member 1, (angioedema, hereditary) (SERPING1), mRNA | DTFVNASR | Y | Y | ||
| IPI00879931.1 | SERPING1 cDNA FLJ78023, highly similar to Homo sapiens serine (or cysteine) proteinase inhibitor, clade G (C1inhibitor), member 1, (angioedema, hereditary) (SERPING1), mRNA | GVTSVSQIFHSPDLAIRDTFVNASR | Y | Y | ||
| IPI00879931.1 | SERPING1 cDNA FLJ78023, highly similar to Homo sapiens serine (or cysteine) proteinase inhibitor, clade G (C1inhibitor), member 1, (angioedema, hereditary) (SERPING1), mRNA | VGQLQLSHNLSLVILVPQNLK | Y | Y | ||
| IPI00879931.1 | SERPING1 cDNA FLJ78023, highly similar to Homo sapiens serine (or cysteine) proteinase inhibitor, clade G (C1inhibitor), member 1, (angioedema, hereditary) (SERPING1), mRNA | VLSNNSDANLELINTWVAK | Y | Y | ||
| IPI00884105.1 | LAMP1 Lysosome-associated membrane glycoprotein 1 precursor | GHTLTLNFTR | Y | Y | ||
| IPI00884913.1 | Sex hormone binding globulin (Fragment) | LDVDQALNR | Y | |||
| IPI00884988.1 | APLP2 Isoform 4 of Amyloid-like protein 2 precursor | RNQSLSLLYK | Y | |||
| IPI00887154.2 | LOC100134219 Complement component 4B | FSDGLESNSSTQFEVK | Y | |||
| IPI00887154.2 | LOC100134219 Complement component 4B | GLNVTLSSTGR | Y | |||
| IPI00889714.1 | Fibulin 1 (Fragment) | CATPHGDNASLEATFVK | Y | |||
| IPI00889723.1 | C4A;C4B complement component 4B preproprotein | FSDGLESNSSTQFEVK | Y | Y | ||
| IPI00889740.1 | Fibulin 1 | CATPHGDNASLEATFVK | Y | |||
EBI: European Bioinformatics Institute; ISB: Institute for Systems Biology; N: N-glycosylated site
2. Glycoproteins in brain tissue
An alternative approach to increase the chances to identify proteins of low abundance is to perform targeted proteomics; i.e., identify proteins unique to a disease or disease progression in tissue, followed by confirmation and validation in a body fluid. This concept will be discussed further in a later section (targeted proteomics). To characterize tissue glycoproteins associated with PD and PD progression, particularly those related to development of PD dementia, the advantage of well-characterized PD brains obtained at autopsy was taken. In this study, all PD cases had been given a clinical diagnosis of PD initially, which meant that dementia with Lewy body disease (DLB) cases, a disease overlapping with PD with dementia (PDD) cases pathologically, were excluded from the study. The brain region of interest was the middle frontal gyrus (Figure 1), and the four groups of cases (five per group with matching age, gender, and post-mortem interval) were investigated: normal age-matched control (78.6±4.0; male/female [M/F] ratio=3/2), PD with brainstem Lewy bodies only (77.2±11.3; M/F=3/2), PD with brainstem and limbic Lewy bodies (78.8±8.3; M/F=3/2), and PD with Lewy bodies in neocortex plus brainstem and limbic system (77.0±1.9; M/F=3/2). Glycoproteins were isolated with methods identical to those described for CSF above after iTRAQ labeling. Again, the quantitative data will be published in a separate manuscript that is under preparation.
This investigation revealed 394 non-redundant glycoproteins (Appendix II). In comparison with the existing database, 343 of these proteins were annotated in the UniProtKB/Swiss-Prot and ISB databases as glycoproteins with known glycosylation sites or probable/potential glycosylation sites. The specificity was approximately 87% (343/394). It should be emphasized that this dataset represents the first systematic analysis of glycoproteins in human brain in normal and diseased settings.
Appendix II. Glycopeptides Identified in Human Brain Tissue.
| Accessions | Names | Sequence-our | EBI | ISB | ||
|---|---|---|---|---|---|---|
| Identified | Potential | Identified | Potential | |||
| IPI00000265.2 | C10orf38 UPF0560 protein C10orf38 precursor | LPENTSYSDLTAFLTAASSPSEVDSFPYLR | Y | |||
| IPI00000877.1 | HYOU1 Hypoxia up-regulated protein 1 precursor | LSALDNLLNHSSMFLK | Y | Y | ||
| IPI00000877.1 | HYOU1 Hypoxia up-regulated protein 1 precursor | VFGSQNLTTVK | Y | Y | ||
| IPI00000877.1 | HYOU1 Hypoxia up-regulated protein 1 precursor | VINETWAWK | Y | Y | ||
| IPI00002230.4 | AADACL1 arylacetamide deacetylase-like 1 | LNWTSLLPASFTK | Y | Y | ||
| IPI00002714.1 | DKK3 Dickkopf-related protein 3 precursor | ITNNQTGQMVFSETVITSVGDEEGR | Y | Y | ||
| IPI00002790.3 | SEL1L Isoform 1 of Protein sel-1 homolog 1 precursor | MYSEGSDIVPQSNETALHYFK | Y | Y | ||
| IPI00002897.3 | GABRA3 Gamma-aminobutyric acid receptor subunit alpha-3 precursor | HAPDIPDDSTDNITIFTR | Y | Y | ||
| IPI00003467.3 | GABRB3 Isoform 1 of Gamma-aminobutyric acid receptor subunit beta-3 precursor | LAYSGIPLNLTLDNR | Y | Y | ||
| IPI00003813.5 | CADM1 Isoform 1 of Cell adhesion molecule 1 precursor | FQLLNFSSSELK | Y | Y | ||
| IPI00003813.5 | CADM1 Isoform 1 of Cell adhesion molecule 1 precursor | VSLTNVSISDEGR | Y | Y | ||
| IPI00004440.1 | PTPRN Receptor-type tyrosine-protein phosphatase-like N precursor | HNEQNLSLADVTQQAGLVK | Y | Y | ||
| IPI00005126.1 | EFNB2 Ephrin-B2 precursor | SIVLEPIYWNSSNSK | Y | Y | ||
| IPI00006071.4 | CD38 Isoform 1 of ADP-ribosyl cyclase 1 | HPCNITEEDYQPLMK | Y | Y | ||
| IPI00006071.4 | CD38 Isoform 1 of ADP-ribosyl cyclase 1 | IFDKNSTFGSVEVHNLQPEK | Y | Y | ||
| IPI00006121.1 | IDS Isoform Short of Iduronate 2-sulfatase precursor | EDVQALNISVPYGPIPVDFQR | Y | Y | ||
| IPI00006121.1 | IDS Isoform Short of Iduronate 2-sulfatase precursor | VHAGNFSTIPQYFK | Y | Y | ||
| IPI00006631.6 | SV2B Synaptic vesicle glycoprotein 2B | FINSTFLEQK | Y | Y | ||
| IPI00006631.6 | SV2B Synaptic vesicle glycoprotein 2B | NCTIESTIFYNTDLYEHK | Y | Y | ||
| IPI00006631.6 | SV2B Synaptic vesicle glycoprotein 2B | VFFGEHVYGATINFTMENQIHQHGK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | ADGTVNQIEGEATPVNLTEPAK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | ADGTVNQIEGEATPVNLTEPAKLEVK | Y | Y | ||
| IPI00006662.1 | APOD Apolipoprotein D precursor | CIQANYSLMENGK | Y | Y | ||
| IPI00006967.3 | PCDH9 Protocadherin-9 precursor | NADIVYQLGPNASFFDLDR | Y | Y | ||
| IPI00006967.3 | PCDH9 Protocadherin-9 precursor | YIISPINGTVYLSEKDPVNTK | Y | Y | ||
| IPI00007664.5 | PGCP Plasma glutamate carboxypeptidase precursor | IVVYNQPYINYSR | Y | Y | ||
| IPI00008600.1 | FUT9 Alpha-(1,3)-fucosyltransferase | SGIEHLFNLTLTYR | Y | Y | ||
| IPI00009111.1 | TPBG Trophoblast glycoprotein precursor | NLTEVPTDLPAYVR | Y | Y | ||
| IPI00009111.1 | TPBG Trophoblast glycoprotein precursor | VLHNGTLAELQGLPHIR | Y | Y | ||
| IPI00009890.1 | SERPINE2 Glia-derived nexin precursor | NASEIEVPFVTR | Y | Y | ||
| IPI00009997.1 | B3GNT1 N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase | VAQPGINYALGTNVSYPNNLLR | Y | Y | ||
| IPI00010279.4 | GDE1 Glycerophosphodiester phosphodiesterase 1 | EAVAECLNHNLTIFFDVK | Y | Y | ||
| IPI00010949.3 | SIAE Isoform 1 of Sialate O-acetylesterase precursor | GLLNLTYYQQIQVQK | Y | Y | ||
| IPI00011454.1 | GANAB Isoform 2 of Neutral alpha-glucosidase AB precursor | VNLTLGSIWDK | Y | |||
| IPI00011732.2 | GFRA2 Isoform 1 of GDNF family receptor alpha-2 precursor | NAIQAFGNGTDVNVSPK | Y | Y | ||
| IPI00012102.1 | GNS N-acetylglucosamine-6-sulfatase precursor | YYNYTLSINGK | Y | Y | ||
| IPI00012887.1 | CTSL1 Cathepsin L1 precursor | YSVANDTGFVDIPK | Y | Y | ||
| IPI00013303.2 | LSAMP Limbic system-associated membrane protein precursor | LGVTNASLVLFRPGSVR | Y | Y | ||
| IPI00013744.1 | ITGA2 Integrin alpha-2 precursor | YFFNVSDEAALLEK | Y | Y | ||
| IPI00013897.1 | ADAM10 ADAM 10 precursor | INTTADEKDPTNPFR | Y | Y | ||
| IPI00013897.1 | ADAM10 ADAM 10 precursor | NISQVLEK | Y | Y | ||
| IPI00015688.1 | GPC1 Glypican-1 precursor | SFDDHFQHLLNDSER | Y | Y | ||
| IPI00015872.3 | TSPAN8 Tetraspanin-8 | IVNETLYENTK | Y | Y | ||
| IPI00016848.1 | C20orf103 Uncharacterized protein C20orf103 precursor | ENGTTCLMAEFAAK | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | EHEGAIYPDNTTDFQR | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | ELHHLQEQNVSNAFLDK | Y | Y | ||
| IPI00017601.1 | CP Ceruloplasmin precursor | ENLTAPGSDSAVFFEQGTTR | Y | Y | ||
| IPI00018274.1 | EGFR Isoform 1 of Epidermal growth factor receptor precursor | DSLSINATNIK | Y | Y | ||
| IPI00019988.1 | SGSH N-sulphoglucosamine sulphohydrolase precursor | DAGVLNDTLVIFTSDNGIPFPSGR | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | CANLVPVPITNATLDR | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | LVPVPITNATLDR | Y | Y | ||
| IPI00020091.1 | ORM2 Alpha-1-acid glycoprotein 2 precursor | PLCANLVPVPITNATLDR | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | DNTTCYEFK | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | FNSTEYQVVTR | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | GVTHLNISGLK | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | IETILLNGTDR | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | KLNLDGSNYTLLK | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | LNLDGSNYTLLK | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | LTSCATNASICGDEAR | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | TVPDIDNVTVLDYDAR | Y | Y | ||
| IPI00020557.1 | LRP1 Prolow-density lipoprotein receptor-related protein 1 precursor | WTGHNVTVVQR | Y | Y | ||
| IPI00020747.1 | SCN3B Sodium channel subunit beta-3 precursor | LQWNGSK | Y | Y | ||
| IPI00020987.1 | PRELP Prolargin precursor | IHYLYLQNNFITELPVESFQNATGLR | Y | Y | ||
| IPI00020987.1 | PRELP Prolargin precursor | INGTQICPNDLVAFHDFSSDLENVPHLR | Y | Y | ||
| IPI00020987.1 | PRELP Prolargin precursor | NSFNISNLLVLHLSHNR | Y | Y | ||
| IPI00021091.1 | LGI1 Isoform 1 of Leucine-rich glioma-inactivated protein 1 precursor | ATQLFTNQTDIPNMEDVYAVK | Y | Y | ||
| IPI00021807.2 | GBA Isoform Long of Glucosylceramidase precursor | DLGPTLANSTHHNVR | Y | Y | ||
| IPI00021983.1 | NCSTN Isoform 1 of Nicastrin precursor | ANNSWFQSILR | Y | Y | ||
| IPI00021983.1 | NCSTN Isoform 1 of Nicastrin precursor | DLYEYSWVQGPLHSNETDR | Y | Y | ||
| IPI00021983.1 | NCSTN Isoform 1 of Nicastrin precursor | NISGVVLADHSGAFHNK | Y | Y | ||
| IPI00022229.1 | APOB Apolipoprotein B-100 precursor | FEVDSPVYNATWSASLK | Y | Y | ||
| IPI00022371.1 | HRG Histidine-rich glycoprotein precursor | VIDFNCTTSSVSSALANTK | Y | Y | ||
| IPI00022395.1 | C9 Complement component C9 precursor | AVNITSENLIDDVVSLIR | Y | Y | ||
| IPI00022417.4 | LRG1 Leucine-rich alpha-2-glycoprotein precursor | KLPPGLLANFTLLR | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | LVPVPITNATLDQITGK | Y | Y | ||
| IPI00022429.3 | ORM1 Alpha-1-acid glycoprotein 1 precursor | QDQCIYNTTYLNVQR | Y | Y | ||
| IPI00022431.1 | AHSG Alpha-2-HS-glycoprotein precursor | AALAAFNAQNNGSNFQLEEISR | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | ALPQPQNVTSLLGCTH | Y | Y | ||
| IPI00022488.1 | HPX Hemopexin precursor | SWPAVGNCSSALR | Y | Y | ||
| IPI00022608.1 | SORL1 Sortilin-related receptor precursor | LTIVNSSVLDRPR | Y | Y | ||
| IPI00023542.6 | TMED9 transmembrane emp24 protein transport domain containing 9 | FTFTSHTPGEHQICLHSNSTK | Y | Y | ||
| IPI00023601.1 | HAPLN1 Hyaluronan and proteoglycan link protein 1 precursor | GGNVTLPCK | Y | Y | ||
| IPI00023648.6 | ISLR Immunoglobulin superfamily containing leucine-rich repeat protein precursor | FQAFANGSLLIPDFGK | Y | |||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | ALGFENATQALGR | Y | Y | ||
| IPI00023673.1 | LGALS3BP Galectin-3-binding protein precursor | GLNLTEDTYKPR | Y | Y | ||
| IPI00023807.3 | SEMA4D Semaphorin-4D precursor | AANYTSSLNLPDK | Y | Y | ||
| IPI00023807.3 | SEMA4D Semaphorin-4D precursor | DVNYTQIVVDR | Y | Y | ||
| IPI00023807.3 | SEMA4D Semaphorin-4D precursor | EAVFAVNALNISEK | Y | Y | ||
| IPI00023807.3 | SEMA4D Semaphorin-4D precursor | KDVNYTQIVVDR | Y | Y | ||
| IPI00024035.1 | CDH6 Isoform 1 of Cadherin-6 precursor | EDAQINTTIGSVTAQDPDAAR | Y | Y | ||
| IPI00024036.1 | CDH8 Cadherin-8 precursor | ELSVWHNITIIATEIR | Y | Y | ||
| IPI00024046.1 | CDH13 Cadherin-13 precursor | ANYNLPIMVTDSGKPPMTNITDLR | Y | Y | ||
| IPI00024046.1 | CDH13 Cadherin-13 precursor | DPAGWLNINPINGTVDTTAVLDR | Y | Y | ||
| IPI00024046.1 | CDH13 Cadherin-13 precursor | INNTHALVSLLQNLNK | Y | Y | ||
| IPI00024046.1 | CDH13 Cadherin-13 precursor | NLSVVILGASDK | Y | Y | ||
| IPI00024046.1 | CDH13 Cadherin-13 precursor | NLSVVILGASDKDLHPNTDPFK | Y | Y | ||
| IPI00024046.1 | CDH13 Cadherin-13 precursor | QEDLSVGSVLLTVNATDPDSLQHQTIR | Y | Y | ||
| IPI00024284.4 | HSPG2 Basement membrane-specific heparan sulfate proteoglycan core protein precursor | ALVNFTR | Y | Y | ||
| IPI00024284.4 | HSPG2 Basement membrane-specific heparan sulfate proteoglycan core protein precursor | SLTQGSLIVGDLAPVNGTSQGK | Y | Y | ||
| IPI00024572.3 | ASPH aspartate beta-hydroxylase isoform e | YNLSEVLQGK | Y | |||
| IPI00024766.1 | PLXNC1 Plexin-C1 precursor | SNVIVTGANFTR | Y | |||
| IPI00024966.1 | CNTN2 Contactin-2 precursor | ANSTGILSVR | Y | Y | ||
| IPI00024966.1 | CNTN2 Contactin-2 precursor | VPGADAQYFVYSNESVRPYTPFEVK | Y | Y | ||
| IPI00024966.1 | CNTN2 Contactin-2 precursor | WDPVVPFRNESAVTGYK | Y | Y | ||
| IPI00025297.2 | ENTPD3 Ectonucleoside triphosphate diphosphohydrolase 3 | LQNETAANEVLESIQSYFK | Y | Y | ||
| IPI00026237.1 | MAG Myelin-associated glycoprotein precursor | LGCQASFPNTTLQFEGYASMDVK | Y | Y | ||
| IPI00026237.1 | MAG Myelin-associated glycoprotein precursor | NCTLLLSNVSPELGGK | Y | Y | ||
| IPI00026237.1 | MAG Myelin-associated glycoprotein precursor | SNPEPSVAFELPSRNVTVNESER | Y | Y | ||
| IPI00026270.1 | CPM Carboxypeptidase M precursor | NFPDAFEYNNVSR | Y | Y | ||
| IPI00026946.2 | NPTX2 Neuronal pentraxin-2 precursor | ANVSNAGLPGDFR | Y | Y | ||
| IPI00027078.3 | CPD Carboxypeptidase D precursor | SEGAIQVNFTLVR | Y | Y | ||
| IPI00027230.3 | HSP90B1 Endoplasmin precursor | EEEAIQLDGLNASQIR | Y | Y | ||
| IPI00027230.3 | HSP90B1 Endoplasmin precursor | HNNDTQHIWESDSNEFSVIADPR | Y | Y | ||
| IPI00027230.3 | HSP90B1 Endoplasmin precursor | TDDEVVQREEEAIQLDGLNASQIR | Y | Y | ||
| IPI00027232.3 | IGF1R Insulin-like growth factor 1 receptor precursor | WNPPSLPNGNLSYYIVR | Y | Y | ||
| IPI00027250.1 | GABBR2 Gamma-aminobutyric acid type B receptor subunit 2 precursor | IQDFNYTDHTLGR | Y | Y | ||
| IPI00027482.1 | SERPINA6 Corticosteroid-binding globulin precursor | AQLLQGLGFNLTER | Y | Y | ||
| IPI00027505.2 | ITGAV Isoform 1 of Integrin alpha-V precursor | ANTTQPGIVEGGQVLK | Y | Y | ||
| IPI00027505.2 | ITGAV Isoform 1 of Integrin alpha-V precursor | TAADTTGLQPILNQFTPANISR | Y | Y | ||
| IPI00027851.1 | HEXA Beta-hexosaminidase alpha chain precursor | SAEGTFFINK | Y | Y | ||
| IPI00029343.2 | CNTNAP2 Isoform 1 of Contactin-associated protein-like 2 precursor | GCMESINYNGVNITDLAR | Y | Y | ||
| IPI00029343.2 | CNTNAP2 Isoform 1 of Contactin-associated protein-like 2 precursor | KPGSFANVSIDMCAIIDR | Y | Y | ||
| IPI00029343.2 | CNTNAP2 Isoform 1 of Contactin-associated protein-like 2 precursor | SINLTLDR | Y | Y | ||
| IPI00029343.2 | CNTNAP2 Isoform 1 of Contactin-associated protein-like 2 precursor | TVPVFFNATSYLEVPGR | Y | Y | ||
| IPI00029343.2 | CNTNAP2 Isoform 1 of Contactin-associated protein-like 2 precursor | VGVHINITQTK | Y | Y | ||
| IPI00029533.1 | ITGB8 Integrin beta-8 precursor | NYAIIKPIGFNETAK | Y | Y | ||
| IPI00029739.5 | CFH Isoform 1 of Complement factor H precursor | IPCSQPPQIEHGTINSSR | Y | Y | ||
| IPI00029768.1 | GRIN2A Glutamate [NMDA] receptor subunit epsilon-1 precursor | WENHTLSLR | Y | Y | ||
| IPI00030880.2 | GRIA1 Isoform Flop of Glutamate receptor 1 precursor | ESGANVTGFQLVNYTDTIPAK | Y1,Y2 | Y1,Y2 | ||
| IPI00030887.1 | TYRO3 Tyrosine-protein kinase receptor TYRO3 precursor | DLVPATNYSLR | Y | Y | ||
| IPI00031121.2 | CPE Carboxypeptidase E precursor | DLQGNPIANATISVEGIDHDVTSAK | Y | Y | ||
| IPI00031121.2 | CPE Carboxypeptidase E precursor | GNETIVNLIHSTR | Y | Y | ||
| IPI00032063.6 | LRP1B Similar to Candidate tumor suppressor protein | AFINGTGLETVISR | Y | Y | ||
| IPI00032179.2 | SERPINC1 Antithrombin III variant | SLTFNETYQDISELVYGAK | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | HLVIHNESTCEQLAK | Y | Y | ||
| IPI00032220.3 | AGT Angiotensinogen precursor | VYIHPFHLVIHNESTCEQLAK | Y | Y | ||
| IPI00044823.2 | SLC2A13 Proton myo-inositol cotransporter | ITFKPIAPSGQNATCTR | Y | Y | ||
| IPI00045906.3 | BSCL2 Isoform 3 of Seipin | TDCDSSTTSLCSFPVANVSLTK | Y | Y | ||
| IPI00045928.1 | SLC9A7 Sodium/hydrogen exchanger 7 | AFSTLLVNVSGK | Y | |||
| IPI00047169.5 | SYNPR Synaptoporin | LSVDCVNK | Y | Y | ||
| IPI00047169.5 | SYNPR Synaptoporin | TESNLSIDIAFAYPFR | Y | Y | ||
| IPI00062679.4 | TMEM30A Isoform 2 of Cell cycle control protein 50A | YSLNVTYNYPVHYFDGR | Y | Y | ||
| IPI00064667.4 | CNDP1 Beta-Ala-His dipeptidase precursor | LVPHMNVSAVEK | Y | Y | ||
| IPI00072918.2 | COL6A3 alpha 3 type VI collagen isoform 4 precursor | GPPGVNGTQGFQGCPGQR | Y | Y | ||
| IPI00152850.2 | JAM3 junctional adhesion molecule 3 precursor | IWNVTR | Y | Y | ||
| IPI00152850.2 | JAM3 junctional adhesion molecule 3 precursor | NSSFHLNSETGTLVFTAVHK | Y | Y | ||
| IPI00159927.2 | NCAN Neurocan core protein precursor | ANATLLLGPLR | Y | Y | ||
| IPI00159927.2 | NCAN Neurocan core protein precursor | GTVLCGPPPAVENASLIGAR | Y | Y | ||
| IPI00160552.3 | TNR Isoform 1 of Tenascin-R precursor | CANGTCLCEEGYVGEDCGQR | Y | Y | ||
| IPI00163207.1 | PGLYRP2 Isoform 1 of N-acetylmuramoyl-L-alanine amidase precursor | GFGVAIVGNYTAALPTEAALR | Y | Y | ||
| IPI00165931.7 | PLXNA4 Isoform 1 of Plexin-A4 precursor | SPSYIVCNTTSSDEVLEMK | Y | Y | ||
| IPI00166048.3 | CADM3 Isoform 1 of Cell adhesion molecule 3 precursor | MTQESALIFPFLNK | Y | |||
| IPI00166048.3 | CADM3 Isoform 1 of Cell adhesion molecule 3 precursor | TQESALIFPFLNK | Y | |||
| IPI00167215.6 | HEPACAM Isoform 1 of Hepatocyte cell adhesion molecule precursor | DGKPLLNDSR | Y | Y | ||
| IPI00167215.6 | HEPACAM Isoform 1 of Hepatocyte cell adhesion molecule precursor | TINLTVDVPISR | Y | Y | ||
| IPI00167619.2 | LRTM2 Leucine-rich repeat and transmembrane domain-containing protein 2 precursor | LSALPSWAFANLSSLQR | Y | Y | ||
| IPI00167619.2 | LRTM2 Leucine-rich repeat and transmembrane domain-containing protein 2 precursor | SIFGDLTNLTELQLR | Y | Y | ||
| IPI00168878.1 | TOR1AIP2 Torsin-1A-interacting protein 2 | HLNASNPTEPATIIFTAAR | Y | |||
| IPI00169285.5 | P76 Putative phospholipase B-like 2 precursor | HPDAVAWANLTNAIR | Y | Y | ||
| IPI00169285.5 | P76 Putative phospholipase B-like 2 precursor | SDLNPANGSYPFKALR | Y | Y | ||
| IPI00171385.3 | C3orf39 Uncharacterized glycosyltransferase AGO61 precursor | LNVSHTGVPLGEEYILVFSR | Y | Y | ||
| IPI00171473.2 | SPON1 Spondin-1 precursor | LTFYGNWSEK | Y | Y | ||
| IPI00173947.1 | SV2C Synaptic vesicle glycoprotein 2C | NCTFIDTVFDNTDFEPYK | Y | Y | ||
| IPI00176221.7 | NEGR1 Neuronal growth regulator 1 precursor | GAWLNR | Y | Y | ||
| IPI00176221.7 | NEGR1 Neuronal growth regulator 1 precursor | KLFNGQQGIIIQNFSTR | Y | Y | ||
| IPI00176221.7 | NEGR1 Neuronal growth regulator 1 precursor | LFNGQQGIIIQNFSTR | Y | Y | ||
| IPI00176221.7 | NEGR1 Neuronal growth regulator 1 precursor | SILTVTNVTQEHFGNYT | Y1,Y2 | Y1,Y2 | ||
| IPI00176221.7 | NEGR1 Neuronal growth regulator 1 precursor | SILTVTNVTQEHFGNYTCVAANK | Y1,Y2 | Y1,Y2 | ||
| IPI00176427.1 | CADM4 Cell adhesion molecule 4 precursor | AEAVGETLTLPGLVSADNGTYTCEASNK | Y | Y | ||
| IPI00176427.1 | CADM4 Cell adhesion molecule 4 precursor | QTLFFNGTR | Y | Y | ||
| IPI00182126.3 | FKBP9 FK506-binding protein 9 precursor | YHYNGTLLDGTLFDSSYSR | Y | Y | Y | |
| IPI00182194.7 | ODZ2 Teneurin-2 | NVTSILELR | Y | Y | ||
| IPI00186736.3 | IGSF8 Isoform 3 of Immunoglobulin superfamily member 8 precursor | GETASLLCNISVR | Y | |||
| IPI00186736.3 | IGSF8 Isoform 3 of Immunoglobulin superfamily member 8 precursor | IGPGEPLELLCNVSGALPPAGR | Y | Y | ||
| IPI00215631.1 | VCAN Isoform Vint of Versican core protein precursor | FENQTGFPPPDSR | Y | Y | ||
| IPI00215844.1 | ASAHL Isoform 2 of N-acylethanolamine-hydrolyzing acid amidase precursor | FNVSLDSVPELR | Y | |||
| IPI00216224.1 | ITGA6 Isoform Alpha-6X2B of Integrin alpha-6 precursor | LWNSTFLEEYSK | Y | Y | ||
| IPI00216394.1 | GABRB2 Isoform Long of Gamma-aminobutyric acid receptor subunit beta-2 precursor | LSYNVIPLNLTLDNR | Y | Y | ||
| IPI00216489.3 | ACAN Isoform 2 of Aggrecan core protein precursor | TVYLYPNQTGLPDPLSR | Y | Y | ||
| IPI00216489.3 | ACAN Isoform 2 of Aggrecan core protein precursor | TVYVHANQTGYPDPSSR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | ANSTGTLVITDPTR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | DVYALMGQNVTLECF | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | DVYALMGQNVTLECFALGNPVPDIR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | GKANSTGTLVITDPTR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | GNYSCFVSSPSITK | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | GTEWLVNSSR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | ILIWEDGSLEINNITR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | TIVDNSSASADLVVR | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | YIITWDHVVALSNESTVTGYK | Y | Y | ||
| IPI00216641.1 | CNTN1 Isoform 2 of Contactin-1 precursor | YTCTAQTIVDNSSASADLVVR | Y | Y | ||
| IPI00216762.1 | ECE1 Isoform D of Endothelin-converting enzyme 1 | FFNFSWR | Y | Y | ||
| IPI00216910.1 | FOLH1 Isoform PSMA′ of Glutamate carboxypeptidase 2 | VPYNVGPGFTGNFSTQK | Y | Y | ||
| IPI00217146.1 | SLITRK4 SLIT and NTRK-like protein 4 precursor | GDVFHNLTNLR | Y | |||
| IPI00217766.3 | SCARB2 Lysosome membrane protein 2 | ANIQFGDNGTTISAVSNK | Y | Y | ||
| IPI00217766.3 | SCARB2 Lysosome membrane protein 2 | CNMINGTDGDSFHPLITK | Y | Y | ||
| IPI00217766.3 | SCARB2 Lysosome membrane protein 2 | FFNVTNPEEILR | Y | Y | ||
| IPI00217766.3 | SCARB2 Lysosome membrane protein 2 | NGTNDGDYVFLTGEDSYLNFTK | Y1,Y2 | Y1,Y2 | ||
| IPI00217766.3 | SCARB2 Lysosome membrane protein 2 | TMVFPVMYLNESVHIDK | Y | Y | ||
| IPI00217766.3 | SCARB2 Lysosome membrane protein 2 | YFFNVTNPEEILR | Y | Y | ||
| IPI00217882.3 | SORT1 Sortilin precursor | DITDLINNTFIR | Y | Y | ||
| IPI00217882.3 | SORT1 Sortilin precursor | HLYTTTGGETDFTNVTSLR | Y | Y | ||
| IPI00217882.3 | SORT1 Sortilin precursor | LANNTHQHVFDDLR | Y | Y | ||
| IPI00217987.8 | ITGAM Integrin alpha-M precursor | EFNVTVTVR | Y | Y | ||
| IPI00217987.8 | ITGAM Integrin alpha-M precursor | ELFNITNGAR | Y | Y | ||
| IPI00218192.2 | ITIH4 Isoform 2 of Inter-alpha-trypsin inhibitor heavy chain H4 precursor | LPTQNITFQTESSVAEQEAEFQSPK | Y | Y | ||
| IPI00218646.3 | CYBB Cytochrome b-245 heavy chain | GQTAESLAVHNITVCEQK | Y | Y | ||
| IPI00218725.3 | LAMA2 laminin alpha 2 subunit isoform b precursor | YMQNLTVEQPIEVK | Y | Y | ||
| IPI00218887.1 | PVRL1 Isoform Alpha of Poliovirus receptor-related protein 1 precursor | ADANPPATEYHWTTLNGSLPK | Y | Y | ||
| IPI00218887.1 | PVRL1 Isoform Alpha of Poliovirus receptor-related protein 1 precursor | ESQLNLTVMAK | Y | Y | ||
| IPI00219124.2 | GRIA1 Isoform Flip of Glutamate receptor 1 precursor | TNYTLHVIEMK | Y | Y | ||
| IPI00219249.4 | CNTNAP1 Contactin-associated protein 1 precursor | ANHSLDVSFYFR | Y | Y | ||
| IPI00219249.4 | CNTNAP1 Contactin-associated protein 1 precursor | DVNFTLDGYVQR | Y | Y | ||
| IPI00219249.4 | CNTNAP1 Contactin-associated protein 1 precursor | GHNSTFFGNVNESAVVR | Y1,Y2 | Y1,Y2 | ||
| IPI00219249.4 | CNTNAP1 Contactin-associated protein 1 precursor | TSGNFTIDPDGSGPLKPF | Y | Y | ||
| IPI00219249.4 | CNTNAP1 Contactin-associated protein 1 precursor | VDGQLVNLTLVEGR | Y | Y | ||
| IPI00219249.4 | CNTNAP1 Contactin-associated protein 1 precursor | WDCHSNQTAF | Y | Y | ||
| IPI00220213.1 | TNC Isoform 4 of Tenascin precursor | LLETVEYNISGAER | Y | Y | ||
| IPI00220213.1 | TNC Isoform 4 of Tenascin precursor | LNYSLPTGQWVGVQLPR | Y | Y | ||
| IPI00220213.1 | TNC Isoform 4 of Tenascin precursor | NLTVPGSLR | Y | Y | ||
| IPI00220213.1 | TNC Isoform 4 of Tenascin precursor | QSGVNATLPEENQPVVFNHVYNIK | Y | Y | ||
| IPI00220213.1 | TNC Isoform 4 of Tenascin precursor | VEAAQNLTLPGSLR | Y | Y | ||
| IPI00220277.2 | GRM5 Isoform 2 of Metabotropic glutamate receptor 5 precursor | TNFTGVSGDTILFDENGDSPGR | Y | Y | ||
| IPI00221224.6 | ANPEP Aminopeptidase N | AEFNITLIHPK | Y | Y | ||
| IPI00236554.1 | MPO Isoform H14 of Myeloperoxidase precursor | ALLPFDNLHDDPCLLTNR | Y | Y | ||
| IPI00289329.2 | EPHB3 Ephrin type-B receptor 3 precursor | YAAVNITTNQAAPSEVPTLR | Y | Y | ||
| IPI00289849.6 | ELFN2 Leucine-rich repeat and fibronectin type-III domain-containing protein 6 precursor | FGNLTDLNLTK | Y1,Y2 | Y1,Y2 | ||
| IPI00289870.3 | PCDH7 Isoform C of Protocadherin-7 precursor | IDNLTGELSTSER | Y | Y | ||
| IPI00290456.3 | ICAM5 Intercellular adhesion molecule 5 precursor | AELDLRPHGLGLFENSSAPR | Y | Y | ||
| IPI00290456.3 | ICAM5 Intercellular adhesion molecule 5 precursor | FEEPSCPSNWTWVEGSGR | Y | Y | ||
| IPI00290456.3 | ICAM5 Intercellular adhesion molecule 5 precursor | GGSLWLNCSTNCPRPER | Y | Y | ||
| IPI00290456.3 | ICAM5 Intercellular adhesion molecule 5 precursor | GLGLFENSSAPR | Y | Y | ||
| IPI00290456.3 | ICAM5 Intercellular adhesion molecule 5 precursor | QLVCNVTLGGENR | Y | Y | ||
| IPI00290456.3 | ICAM5 Intercellular adhesion molecule 5 precursor | VLAPGIYVCNATNR | Y | Y | ||
| IPI00291136.4 | COL6A1 Collagen alpha-1(VI) chain precursor | GEDGPAGNGTEGFPGFPGYPGNR | Y | Y | ||
| IPI00291136.4 | COL6A1 Collagen alpha-1(VI) chain precursor | NVTAQICIDK | Y | Y | ||
| IPI00291792.2 | ITGB2 Integrin beta-2 precursor | LNFTGPGDPDSIR | Y | Y | ||
| IPI00292732.3 | FMOD fibromodulin precursor | LYLDHNNLTR | Y | Y | ||
| IPI00293033.5 | NID2 NID2 protein | DYSLTFGAINQTWSYR | Y | Y | ||
| IPI00293033.5 | NID2 NID2 protein | IHQNITYQVCR | Y | Y | ||
| IPI00293074.5 | SLC44A2 Isoform 2 of Choline transporter-like protein 2 | GVLMVGNETTYEDGHGSR | Y | Y | ||
| IPI00293074.5 | SLC44A2 Isoform 2 of Choline transporter-like protein 2 | KNITDLVEGAK | Y | Y | ||
| IPI00293074.5 | SLC44A2 Isoform 2 of Choline transporter-like protein 2 | NITDLVEGAK | Y | Y | ||
| IPI00293088.5 | GAA Lysosomal alpha-glucosidase precursor | GVFITNETGQPLIGK | Y | Y | ||
| IPI00293088.5 | GAA Lysosomal alpha-glucosidase precursor | LENLSSSEMGYTATLTR | Y | Y | ||
| IPI00293088.5 | GAA Lysosomal alpha-glucosidase precursor | NNTIVNELVR | Y | Y | ||
| IPI00293328.3 | P2RX7 P2X purinoceptor 7 | LDDKTTNVSLYPGYNFR | Y | Y | ||
| IPI00293328.3 | P2RX7 P2X purinoceptor 7 | NILPGLNITCTFHK | Y | Y | ||
| IPI00293328.3 | P2RX7 P2X purinoceptor 7 | NIDFPGHNYTTR | Y | Y | ||
| IPI00293328.3 | P2RX7 P2X purinoceptor 7 | PALLNSAENFTVLIK | Y | Y | ||
| IPI00293588.4 | TMEFF1 Isoform 1 of Tomoregulin-1 precursor | SINCSELNVR | Y | |||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | FHVNYTQPL | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | FHVNYTQPLVAVK | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | FLEPYNDSIQAQK | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | FLNVTPNVEVNVECR | Y | |||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | HVNYTQPLVAVK | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | KFHVNYTQPLVAVK | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | TENLDVIVNVSDTESWDQHVQK | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | TQLGNCSGIGDSTHYGY | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | TQLGNCSGIGDSTHYGYSTGQPCVF | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | TQLGNCSGIGDSTHYGYSTGQPCVFIK | Y | Y | ||
| IPI00293971.3 | ATP1B2 Sodium/potassium-transporting ATPase subunit beta-2 | VINFYAGANQSMNVTCAGK | Y1,Y2 | Y1,Y2 | ||
| IPI00294455.1 | UGT8 2-hydroxyacylsphingosine 1-beta-galactosyltransferase precursor | YPGIFNSTTSDAFLQSK | Y | Y | ||
| IPI00294834.6 | ASPH Aspartyl/asparaginyl beta-hydroxylase | LVQLFPNDTSLK | Y | Y | ||
| IPI00295399.4 | CDH10 Cadherin-10 precursor | ELSQWHNLTVIAAEINNPK | Y | Y | ||
| IPI00295494.1 | CCDC39 Coiled-coil domain-containing protein 39 | ATVNRTSSDLEALRK | Y | |||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | QNITYLLK | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | SLEVLNLSSNK | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | SLEVLNLSSNKL | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | SLWNMSAANNNIK | Y | Y | ||
| IPI00295832.1 | OMG Oligodendrocyte-myelin glycoprotein precursor | WSCDHKQNITYLLK | Y | Y | ||
| IPI00296373.3 | GABBR1 Isoform 1C of Gamma-aminobutyric acid type B receptor subunit 1 precursor | AMNSSSFEGVSGHVVFDASGSR | Y | Y | ||
| IPI00296373.3 | GABBR1 Isoform 1C of Gamma-aminobutyric acid type B receptor subunit 1 precursor | LEDFNYNNQTITDQIYR | Y | Y | ||
| IPI00296373.3 | GABBR1 Isoform 1C of Gamma-aminobutyric acid type B receptor subunit 1 precursor | SISNMTSQEFVEK | Y | Y | ||
| IPI00297933.1 | GRIN2B Glutamate [NMDA] receptor subunit epsilon-2 precursor | YLINVTFEGR | Y | Y | ||
| IPI00298237.7 | TPP1 Isoform 1 of Tripeptidyl-peptidase 1 precursor | FLSSSPHLPPSSYFNASGR | Y | Y | ||
| IPI00298281.3 | LAMC1 Laminin subunit gamma-1 precursor | KIPAINQTITEANEK | Y | Y | ||
| IPI00298281.3 | LAMC1 Laminin subunit gamma-1 precursor | LLNNLTSIK | Y | Y | ||
| IPI00298281.3 | LAMC1 Laminin subunit gamma-1 precursor | QVLSYGQNLSFSFR | Y | Y | ||
| IPI00298281.3 | LAMC1 Laminin subunit gamma-1 precursor | TANDTSTEAYNLLLR | Y | Y | ||
| IPI00298281.3 | LAMC1 Laminin subunit gamma-1 precursor | TLAGENQTAFEIEELNR | Y | Y | ||
| IPI00298971.1 | VTN Vitronectin precursor | NISDGFDGIPDNVDAALALPAHSYSGR | Y | Y | ||
| IPI00299063.1 | STIM1 Stromal interaction molecule 1 precursor | LAVTNTTMTGTVLK | Y | Y | ||
| IPI00299299.3 | STCH Stress 70 protein chaperone microsome-associated 60 kDa protein precursor | NSTIEAANLAGLK | Y | |||
| IPI00299652.2 | ADAM11 Isoform Long of ADAM 11 precursor | CLPASAFNFSTCPGSGER | Y | Y | ||
| IPI00301395.4 | CPVL Probable serine carboxypeptidase CPVL precursor | QAIHVGNQTFNDGTIVEK | Y | Y | ||
| IPI00301512.3 | DPP6 Isoform DPPX-L of Dipeptidyl aminopeptidase-like protein 6 | ANYSLQIYPDESHYFTSSSLK | Y | Y | ||
| IPI00301512.3 | DPP6 Isoform DPPX-L of Dipeptidyl aminopeptidase-like protein 6 | LAYAAINDSR | Y | Y | ||
| IPI00301512.3 | DPP6 Isoform DPPX-L of Dipeptidyl aminopeptidase-like protein 6 | LWNVETNTSTVLIEGK | Y | Y | ||
| IPI00303210.3 | ENPP2 Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 precursor | AIIANLTCK | Y | Y | ||
| IPI00304227.4 | CDH11 Isoform 1 of Cadherin-11 precursor | FIFSLPPEIIHNPNFTVR | Y | Y | ||
| IPI00304840.4 | COL6A2 Isoform 2C2 of Collagen alpha-2(VI) chain precursor | GTFTDCALANMTEQIR | Y | Y | ||
| IPI00304840.4 | COL6A2 Isoform 2C2 of Collagen alpha-2(VI) chain precursor | NMTLFSDLVAEK | Y | Y | ||
| IPI00307433.3 | STS Steryl-sulfatase precursor | NYEIIQQPMSYDNLTQR | Y | Y | ||
| IPI00307612.4 | CDH20 Cadherin-20 precursor | NGQHFYYSLAPEAANNPNFTIR | Y | Y | ||
| IPI00328113.2 | FBN1 Fibrillin-1 precursor | NCTDIDECR | Y | Y | ||
| IPI00328113.2 | FBN1 Fibrillin-1 precursor | VLPVNVTDYCQLVR | Y | Y | ||
| IPI00328719.2 | SLC15A2 Oligopeptide transporter, kidney isoform | YHNLSLYTEHSVQEK | Y | Y | ||
| IPI00328829.4 | ITIH5 inter-alpha trypsin inhibitor heavy chain precursor 5 isoform 1 | TLFPNYFNGSEIIIAGK | Y | Y | ||
| IPI00329573.9 | COL12A1 Isoform 1 of Collagen alpha-1(XII) chain precursor | EAGNITTDGYEILGK | Y | Y | ||
| IPI00329573.9 | COL12A1 Isoform 1 of Collagen alpha-1(XII) chain precursor | MLEAYNLTEK | Y | Y | ||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | AENQVNVTCQVR | Y | Y | ||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | GTANLSETIR | Y | Y | ||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | IGNITPADAGTYYCVK | Y | |||
| IPI00332887.5 | SIRPA signal-regulatory protein alpha precursor | LQLTWLENGNVSR | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | DGDDEWTSVVVANVSK | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | ERPPTFLTPEGNASNK | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | ERPPTFLTPEGNASNKEELR | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | GSALHEDIYVLHENGTLEIPVAQK | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | NLNFSTR | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | QKDGDDEWTSVVVANVSK | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | VNVVNSTLAEVHWDPVPLK | Y | Y | ||
| IPI00333777.5 | NRCAM Isoform 2 of Neuronal cell adhesion molecule precursor | YQPINSTHELGPLVDLK | Y | Y | ||
| IPI00335355.3 | SLC6A17 Orphan sodium- and chloride-dependent neurotransmitter transporter NTT4 | DLIPPHVNFSHLTTK | Y | Y | ||
| IPI00337351.3 | MDGA2 MAM domain-containing glycosylphosphatidylinositol anchor protein 2 precursor | FQDSSVFNETLR | Y | |||
| IPI00339364.1 | GGT7 65 kDa protein | AAAVAQDGFNVTHDLAR | Y | Y | ||
| IPI00339364.1 | GGT7 65 kDa protein | RNESHLIDFR | Y | Y | ||
| IPI00375253.2 | MAG myelin associated glycoprotein isoform b precursor | ATAFNLSVEFAPVLLLESH | Y | Y | ||
| IPI00375253.2 | MAG myelin associated glycoprotein isoform b precursor | ATAFNLSVEFAPVLLLESHCAAAR | Y | Y | ||
| IPI00375879.6 | KIAA1467 Uncharacterized protein KIAA1467 | APDSNCSNLLITTR | Y | |||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | DKLVLPAKNTTNLK | Y | Y | ||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | LAMLANNNLQILNINK | Y | Y | ||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | LVLPAKNTTNLK | Y | Y | ||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | PINISCDVK | Y | Y | ||
| IPI00376427.3 | NCAM2 Neural cell adhesion molecule 2 precursor | YNCTATNHIGTR | Y | Y | ||
| IPI00376986.2 | NTRK3 Isoform D of NT-3 growth factor receptor precursor | NPLGTANQTINGHFLK | Y | Y | ||
| IPI00382672.4 | ENTPD1 Isoform Vascular of Ectonucleoside triphosphate diphosphohydrolase 1 | VVNVSDLYK | Y | Y | ||
| IPI00384280.5 | PCYOX1 Prenylcysteine oxidase 1 precursor | GELNTSIFSSR | Y | Y | ||
| IPI00384280.5 | PCYOX1 Prenylcysteine oxidase 1 precursor | GELNTSIFSSRPIDK | Y | Y | ||
| IPI00384454.1 | F3 Tissue factor | NNTFLSLR | Y | Y | ||
| IPI00384454.1 | F3 Tissue factor | SGTTNTVAAYNLTWK | Y | Y | ||
| IPI00384484.1 | GPM6B glycoprotein M6B isoform 2 | SPQTNGTTGVEQICVDIR | Y | Y | ||
| IPI00385291.2 | CD82 CD82 antigen isoform 2 | DYNSSREDSLQDAWDYVQAQVK | Y | |||
| IPI00394770.2 | CSMD2 CSMD2 protein | GFNITFTTFR | Y | Y | ||
| IPI00394820.3 | OLFML1 Olfactomedin-like protein 1 precursor | NNTVWEFANIR | Y | Y | ||
| IPI00395428.1 | SCN1B sodium channel, voltage-gated, type I, beta isoform b | LLFFENYEHNTSVVK | Y | Y | ||
| IPI00395903.1 | TMEM106B Transmembrane protein 106B | LNNITIIGPLDMK | Y | |||
| IPI00396134.3 | P2RX7 P2X purinoceptor | TTNVSLYPGYNFR | Y | Y | ||
| IPI00396411.4 | CLPTM1 Isoform 1 of Cleft lip and palate transmembrane protein 1 | DYYPINESLASLPLR | Y | Y | ||
| IPI00401212.3 | GPM6A glycoprotein M6A isoform 3 | NTTLVEGANLCLDLR | Y | Y | ||
| IPI00409626.2 | PCDH9 protocadherin 9 isoform 1 precursor | ATVTINVTDVNDNPPNIDLR | Y | Y | ||
| IPI00409626.2 | PCDH9 protocadherin 9 isoform 1 precursor | IDPVTGNITLEEKPAPTDVGLHR | Y | Y | ||
| IPI00409626.2 | PCDH9 protocadherin 9 isoform 1 precursor | IVASDSGKPSLNQTALVR | Y | Y | ||
| IPI00409626.2 | PCDH9 protocadherin 9 isoform 1 precursor | LFALNNTTGLITVQR | Y1,Y2 | Y1 | Y2 | |
| IPI00409626.2 | PCDH9 protocadherin 9 isoform 1 precursor | LVVNISDLGYPK | Y | Y | ||
| IPI00409667.1 | PCDHGC3 Isoform 3 of Protocadherin gamma-C3 precursor | ETVPEYNLSITAR | Y | Y | ||
| IPI00409667.1 | PCDHGC3 Isoform 3 of Protocadherin gamma-C3 precursor | VLDANDNAPVFNQSLYR | Y | Y | ||
| IPI00410210.1 | LPHN1 Isoform 2 of Latrophilin-1 precursor | GPDLSNCTSPWVNQVAQK | Y | Y | ||
| IPI00412541.2 | GPR158 Probable G-protein coupled receptor 158 precursor | ILLQDLSSSAPHLANATLETEWFHGLR | Y | Y | ||
| IPI00413690.2 | ARSB arylsulfatase B isoform 2 precursor | CTLIDALNVTR | Y | Y | ||
| IPI00413696.5 | CD47 41 kDa protein | DIYTFDGALNK | Y | Y | ||
| IPI00413696.5 | CD47 41 kDa protein | FVTNMEAQNTTEVYVK | Y | Y | ||
| IPI00413696.5 | CD47 41 kDa protein | GRDIYTFDGALNK | Y | Y | ||
| IPI00413696.5 | CD47 41 kDa protein | SDAVSHTGNYTCEVTELTR | Y | Y | ||
| IPI00418446.4 | ASAH1 N-acylsphingosine amidohydrolase (acid ceramidase) 1 isoform b | TVLENSTSYEEAK | Y | Y | ||
| IPI00418446.4 | ASAH1 N-acylsphingosine amidohydrolase (acid ceramidase) 1 isoform b | ILAPAYFILGGNQSGEGCVITR | Y | Y | ||
| IPI00418446.4 | ASAH1 N-acylsphingosine amidohydrolase (acid ceramidase) 1 isoform b | ILGGNQSGEGCVITR | Y | Y | ||
| IPI00437751.1 | ACE Isoform Somatic-1 of Angiotensin-converting enzyme, somatic isoform precursor | KFDVNQLQNTTIK | Y | Y | ||
| IPI00449669.2 | SSR1 Isoform 2 of Translocon-associated protein subunit alpha precursor | YPQDYQFYIQNFTALPLNTVVPPQR | Y | Y | ||
| IPI00456623.2 | BCAN Isoform 1 of Brevican core protein precursor | LFLFPNQTGFPNK | Y | Y | ||
| IPI00456623.2 | BCAN Isoform 1 of Brevican core protein precursor | TLFLFPNQTGFPNK | Y | Y | ||
| IPI00456623.2 | BCAN Isoform 1 of Brevican core protein precursor | VALPAYPASLTDVSLALSELRPNDSGIYR | Y | Y | ||
| IPI00465308.3 | PIGS Isoform 1 of GPI transamidase component PIG-S | TYNASVLPVR | Y | Y | ||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | ELNILFLNK | Y | |||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | ELNILFLNKTDNGTYR | Y2 | Y1,Y2 | ||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | GSQGQFPLTQNVTVVEGGTAIL | Y | Y | ||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | GSQGQFPLTQNVTVVEGGTAILTCR | Y | Y | ||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | VDQNDNTSLQWSNPAQQTLYFDDK | Y | Y | ||
| IPI00470388.2 | CADM2 Isoform 2 of Cell adhesion molecule 2 precursor | VDQNDNTSLQWSNPAQQTLYFDDKK | Y | Y | ||
| IPI00470529.3 | GPNMB Isoform 1 of Transmembrane glycoprotein NMB precursor | VSVNTANVTLGPQLMEVTVYR | Y | Y | ||
| IPI00470696.1 | UNC5D Isoform 1 of Netrin receptor UNC5D precursor | EVFINVTR | Y | Y | ||
| IPI00472011.1 | NEO1 154 kDa protein | TLSDVPSAAPQNLSLEVR | Y | Y | ||
| IPI00472139.1 | PLXND1 Isoform 2 of Plexin-D1 precursor | ANFTIYDCSR | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | GCVLLSYLNETVTVSASLESVR | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | IYVLDYLNETQQLTPEVK | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | SLGNVNFTVSAEALESQELCGTEVPSVPEHGR | Y | Y | ||
| IPI00478003.1 | A2M Alpha-2-macroglobulin precursor | VSNQTLSLFFTVLQDVPVR | Y | Y | ||
| IPI00478483.3 | LAMC3 Laminin, gamma 3 | LLANLTSLR | Y | Y | ||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | GFSFAFEQLLNYNVSR | Y | Y | ||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | HLVNISVYAFNK | Y1 | Y1,Y2 | ||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | IDVNSWIENFTK | Y | |||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | ISDNNTEFLLNFNEFIDR | Y | Y | ||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | QSCITEQTQYFFDNDSK | Y | |||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | SFSGVLDCGNCSR | Y | |||
| IPI00479514.1 | CACNA2D1 Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | SLDNDNYVFTAPYFNK | Y | |||
| IPI00513767.2 | PTGDS Prostaglandin D2 synthase 21kDa | SVVAPATDGGLNLTSTFLR | Y | Y | ||
| IPI00513767.2 | PTGDS Prostaglandin D2 synthase 21kDa | WFSAGLASNSSWLR | Y | Y | ||
| IPI00513964.1 | SEMA4B Isoform 2 of Semaphorin-4B precursor | FEAEHISNYTALLLSR | Y | Y | ||
| IPI00514424.2 | PPT1 Palmitoyl-protein thioesterase 1 | FLNDSIVDPVDSEWFGFYR | Y | Y | ||
| IPI00514424.2 | PPT1 Palmitoyl-protein thioesterase 1 | NHSIFLADINQER | Y | Y | ||
| IPI00514804.1 | SCN4B Isoform 2 of Sodium channel subunit beta-4 precursor | WTYNSSDAFK | Y | Y | ||
| IPI00550145.3 | OLFM1 NOELIN1_V2 | LDPVSLQTLQTWNTSYPK | Y | Y | ||
| IPI00550145.3 | OLFM1 NOELIN1_V2 | VQNMSQSIEVLDR | Y | Y | ||
| IPI00550918.2 | COL14A1 Isoform 2 of Collagen alpha-1(XIV) chain precursor | SFMVNWTHAPGNVEK | Y | Y | ||
| IPI00552302.3 | NT5E 5′-nucleotidase, ecto | GNVISSHGNPILLNSSIPEDPSIK | Y | Y | ||
| IPI00552450.1 | OPCML opioid binding protein/cell adhesion molecule-like isoform b preproprotein | DYGNYTCVATNK | Y | Y | ||
| IPI00552450.1 | OPCML opioid binding protein/cell adhesion molecule-like isoform b preproprotein | DYGNYTCVATNKL | Y | Y | ||
| IPI00552450.1 | OPCML opioid binding protein/cell adhesion molecule-like isoform b preproprotein | MSTLTFFNVSEK | Y | Y | ||
| IPI00552671.2 | PLXNA1 Plexin-A1 precursor | LSGNLTLLR | Y | Y | ||
| IPI00552671.2 | PLXNA1 Plexin-A1 precursor | YNYTEDPTILR | Y | Y | ||
| IPI00554518.1 | IL6ST IL6ST nirs variant 4 | ETHLETNFTLK | Y | Y | ||
| IPI00554722.1 | LOC442497;SLC3A2 solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 isoform e | DASSFLAEWQNITK | Y | Y | ||
| IPI00554722.1 | LOC442497;SLC3A2 solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 isoform e | LLIAGTNSSDLQQILSLLESNK | Y | Y | ||
| IPI00554722.1 | LOC442497;SLC3A2 solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 isoform e | SLVTQYLNATGNR | Y | Y | ||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | PPKDITISNVTK | Y | Y | ||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | DITISNVTK | Y | Y | ||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | GTNESDSATTQFTTEIDAPK | Y | Y | ||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | IGSYNGTAGDSLSYHQGR | Y | Y | ||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | IGSYNGTAGDSLSYHQGRPF | Y | Y | ||
| IPI00554760.1 | TNR Isoform 2 of Tenascin-R precursor | NCSEPYCPLGCSSR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | ALHHSGHSPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | DEGTYTCALHHSGHSPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HENTSSSPIQY | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HENTSSSPIQYE | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HENTSSSPIQYEF | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HENTSSSPIQYEFSLTR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | HSGHSPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | LDCRHENTSSSPIQYEFSLTR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | SGHSPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | SPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | TCALHHSGHSPPISSQNVTVLR | Y | Y | ||
| IPI00555577.1 | THY1 Thy-1 cell surface antigen variant (Fragment) | TNFTSK | Y | Y | ||
| IPI00555628.1 | NCAM1 Neural cell adhesion molecule 1, 120 kDa isoform variant (Fragment) | DGQLLPSSNYSNIK | Y | Y | ||
| IPI00555628.1 | NCAM1 Neural cell adhesion molecule 1, 120 kDa isoform variant (Fragment) | IYNTPSASYLEVTPDSENDFGNYNCTAVNR | Y | Y | ||
| IPI00555628.1 | NCAM1 Neural cell adhesion molecule 1, 120 kDa isoform variant (Fragment) | PSSNYSNIK | Y | Y | ||
| IPI00555628.1 | NCAM1 Neural cell adhesion molecule 1, 120 kDa isoform variant (Fragment) | RDGQLLPSSNYSNIK | Y | Y | ||
| IPI00604442.2 | SSR2 Putative uncharacterized protein DKFZp686F19123 | IAPASNVSHTVVLRPLK | Y | Y | ||
| IPI00607580.2 | MEGF8 multiple EGF-like-domains 8 | ALLTNVSSVALGSR | Y | Y | ||
| IPI00607652.1 | OLFML3 Isoform 2 of Olfactomedin-like protein 3 precursor | IYVLDGTQNDTAFVFPR | Y | Y | ||
| IPI00607732.1 | NCLN Isoform 2 of Nicalin precursor | VIYNLTEK | Y | Y | ||
| IPI00619903.3 | UGCGL1 UDP-glucose:glycoprotein glucosyltransferase 1 precursor | GTEVNTTVIGENDPIDEVQGFLFGK | Y | |||
| IPI00641150.2 | LAMA1 similar to laminin, alpha 1 precursor | DVAGLSQELLNTSASLSR | Y | Y | ||
| IPI00641524.2 | BTN2A1 Isoform 1 of Butyrophilin subfamily 2 member A1 precursor | GSVALVIHNITAQENGTYR | Y | Y | ||
| IPI00641737.1 | HP Haptoglobin precursor | MVSHHNLTTGATLINEQWLLTTAK | Y | Y | ||
| IPI00641737.1 | HP Haptoglobin precursor | NLFLNHSENATAK | Y | Y | ||
| IPI00641737.1 | HP Haptoglobin precursor | VVLHPNYSQVDIGLIK | Y | Y | ||
| IPI00642378.2 | LASS2 cDNA FLJ75329, highly similar to Homo sapiens LAG1 longevity assurance homolog 2 (S. cerevisiae), transcript variant 2, mRNA | LWLPVNLTWADLEDRDGR | Y | Y | ||
| IPI00642425.1 | ICAM1 Cell surface glycoprotein | ANLTVVLLR | Y | Y | ||
| IPI00643034.2 | PLTP Isoform 1 of Phospholipid transfer protein precursor | EGHFYYNISEVK | Y | Y | ||
| IPI00643384.2 | BGN Uncharacterized protein BGN | LLQVVYLHSNNITK | Y | Y | ||
| IPI00643663.1 | PCSK2 Proprotein convertase subtilisin/kexin type 2 | NPEAGVATTDLYGNCTLR | Y | Y | ||
| IPI00643663.1 | PCSK2 Proprotein convertase subtilisin/kexin type 2 | YLEHVQAVITVNATR | Y | Y | ||
| IPI00644458.1 | TM9SF3 SM-11044 binding protein | IVDVNLTSEGK | Y | Y | ||
| IPI00644480.1 | LPHN2 Latrophilin 2 | SLGQFLSTENATIK | Y | Y | ||
| IPI00645060.1 | PBXIP1 Isoform 2 of Pre-B-cell leukemia transcription factor-interacting protein 1 | LQGLENWGQDPGVSANASK | Y | |||
| IPI00645194.1 | ITGB1 integrin beta 1 isoform 1A precursor | DTCTQECSYFNITK | Y | Y | ||
| IPI00645484.1 | SV2A Isoform 2 of Synaptic vesicle glycoprotein 2A | LINSTFLHNK | Y | Y | ||
| IPI00645484.1 | SV2A Isoform 2 of Synaptic vesicle glycoprotein 2A | NCTFINTVFYNTDLFEYK | Y | Y | ||
| IPI00645484.1 | SV2A Isoform 2 of Synaptic vesicle glycoprotein 2A | VEHVTFNFTLENQIHR | Y | Y | ||
| IPI00646891.2 | GRIN1 Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | LVQVGIYNGTHVIPNDR | Y | Y | ||
| IPI00646891.2 | GRIN1 Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | NVTALLMEAK | Y | Y | ||
| IPI00646891.2 | GRIN1 Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | QNVSLSILK | Y | Y | ||
| IPI00647704.1 | IGHA1;IGHV3OR16-13 cDNA FLJ41552 fis, clone COLON2004478, highly similar to Protein Tro alpha1 H,myeloma | LAGKPTHVNVSVVMAEVDGTCY | Y | Y | ||
| IPI00647704.1 | IGHA1;IGHV3OR16-13 cDNA FLJ41552 fis, clone COLON2004478, highly similar to Protein Tro alpha1 H,myeloma | LSLHRPALEDLLLGSEANLTCTLTGLR | Y | Y | ||
| IPI00647704.1 | IGHA1;IGHV3OR16-13 cDNA FLJ41552 fis, clone COLON2004478, highly similar to Protein Tro alpha1 H,myeloma | PALEDLLLGSEANLTCTLTGLR | Y | Y | ||
| IPI00654584.5 | NPTN Isoform 4 of Neuroplastin precursor | ANATIEVK | Y | Y | ||
| IPI00654584.5 | NPTN Isoform 4 of Neuroplastin precursor | DSPVLPVTLQCNLTSSSH | Y | Y | ||
| IPI00655702.3 | NFASC Isoform 5 of Neurofascin precursor | HNFGPGTDFVVEYIDSNHTK | Y | Y | ||
| IPI00655702.3 | NFASC Isoform 5 of Neurofascin precursor | IHESAPDEQSIWNVTVLPNSK | Y | Y | ||
| IPI00655702.3 | NFASC Isoform 5 of Neurofascin precursor | WANITWK | Y | Y | ||
| IPI00656113.2 | SIRPA Signal-regulatory protein alpha | LLVNVSAHR | Y | Y | ||
| IPI00658202.1 | CDH2 Uncharacterized protein CDH2 | SNISILR | Y | Y | ||
| IPI00735310.1 | LAMA4 Isoform 2 of Laminin subunit alpha-4 precursor | NLTEVVPQLLDQLR | Y | Y | ||
| IPI00737429.3 | ODZ4 Teneurin-4 | IFPSGNVTNILELR | Y | |||
| IPI00737429.3 | ODZ4 Teneurin-4 | LTNVTFPTGQVSSFR | Y | |||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | IAVQFGPGFSWIANFTK | Y | Y | ||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | VASVININPNTTHSTGSCR | Y | Y | ||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | VQPFNVTQGK | Y | Y | ||
| IPI00739827.1 | LAMP2 Isoform LAMP-2B of Lysosome-associated membrane glycoprotein 2 precursor | WQMNFTVR | Y | Y | ||
| IPI00743064.1 | LCN2 Uncharacterized protein LCN2 | SYNVTSVLFR | Y | Y | ||
| IPI00743104.2 | ITGA1 Integrin alpha-1 precursor | VYVYALNQTR | Y | Y | ||
| IPI00743203.2 | LAMB2 Similar to S-laminin | LALNLTLR | Y | Y | ||
| IPI00743203.2 | LAMB2 Similar to S-laminin | NTSAASTAQLVEATEELR | Y | Y | ||
| IPI00743302.2 | ICAM5 intercellular adhesion molecule 5 precursor | VELMPLPPWQPVGENFTLSCR | Y | Y | ||
| IPI00743517.1 | PTPRS protein tyrosine phosphatase, receptor type, sigma isoform 2 precursor | KVEAEALNATAIR | Y | Y | ||
| IPI00744685.2 | BTD Uncharacterized protein BTD (Fragment) | DVQIIVFPEDGIHGFNFTR | Y | Y | ||
| IPI00744835.1 | PSAP Isoform Sap-mu-9 of Proactivator polypeptide precursor | DNATEEEILVYLEK | Y | Y | ||
| IPI00744835.1 | PSAP Isoform Sap-mu-9 of Proactivator polypeptide precursor | NLEKNSTKQEILAALEK | Y | Y | ||
| IPI00744835.1 | PSAP Isoform Sap-mu-9 of Proactivator polypeptide precursor | NSTKQEILAALEK | Y | Y | ||
| IPI00745954.2 | GRM7 Isoform 3 of Metabotropic glutamate receptor 7 precursor | YDIFQYQTTNTSNPGYR | Y | Y | ||
| IPI00746595.3 | MOG Uncharacterized protein MOG | NATGMEVGWYRPPFSR | Y | Y | ||
| IPI00747849.2 | ATP1B1 Isoform 1 of Sodium/potassium-transporting ATPase subunit beta-1 | LGNCSGLNDETYGYK | Y | Y | ||
| IPI00759642.1 | CD163 Isoform 2 of Scavenger receptor cysteine-rich type 1 protein M130 precursor | EDAAVNCTDISVQK | Y | |||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | IIPSNNSGTFR | Y | |||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | VTWKPQGAPVEWEEETVTNHTLR | Y | Y | ||
| IPI00783390.1 | CHL1 Isoform 1 of Neural cell adhesion molecule L1-like protein precursor | YHIYENGTLQINR | Y | Y | ||
| IPI00783665.2 | LAMA5 Laminin subunit alpha-5 precursor | DNATLQATLHAAR | Y | Y | ||
| IPI00783665.2 | LAMA5 Laminin subunit alpha-5 precursor | GVHNASLALSASIGR | Y | Y | ||
| IPI00783665.2 | LAMA5 Laminin subunit alpha-5 precursor | LNASIADLQSQLR | Y | Y | ||
| IPI00783698.4 | TMEM87A Isoform 1 of Transmembrane protein 87A precursor | LFQNCSELFK | Y | Y | ||
| IPI00783987.2 | C3 Complement C3 precursor (Fragment) | TVLTPATNHMGNVTFTIPANR | Y | Y | ||
| IPI00784119.1 | ATP6AP1 Vacuolar ATP synthase subunit S1 precursor | ILFWAQNFSVAYK | Y | Y | ||
| IPI00784119.1 | ATP6AP1 Vacuolar ATP synthase subunit S1 precursor | LNASLPALLLIR | Y | Y | ||
| IPI00784119.1 | ATP6AP1 Vacuolar ATP synthase subunit S1 precursor | QKQPVSPVIHPPVSYNDTAPR | Y | Y | ||
| IPI00784119.1 | ATP6AP1 Vacuolar ATP synthase subunit S1 precursor | QPVSPVIHPPVSYNDTAPR | Y | Y | ||
| IPI00784119.1 | ATP6AP1 Vacuolar ATP synthase subunit S1 precursor | SPVIHPPVSYNDTAPR | Y | Y | ||
| IPI00784147.1 | NPTXR;CBX6 Uncharacterized protein NPTXR | VNLSAAPAPVSAVPTGLHSK | Y | Y | ||
| IPI00784169.1 | CD55 Decay-accelerating factor splicing variant 1 | GSQWSDIEEFCNR | Y | Y | ||
| IPI00784543.1 | KIAA0090 Isoform 2 of Uncharacterized protein KIAA0090 precursor | FINYNQTVSR | Y | Y | ||
| IPI00787965.2 | ATP1B3 similar to Sodium/potassium-transporting ATPase subunit beta-3 | NLTVCPDGALFEQK | Y | Y | ||
| IPI00788159.1 | DPP7 similar to Dipeptidyl-peptidase 2 precursor | ALAGLVYNASGSEHCYDIYR | Y | Y | ||
| IPI00788189.1 | FCGBP similar to Fc fragment of IgG binding protein | VITVQVANFTLR | Y | Y | ||
| IPI00789795.1 | ADAM22 98 kDa protein; ADAM22 Isoform 5 of ADAM 22 precursor | TLNCSGGHVK | Y | Y | ||
| IPI00789973.1 | GRIN1 Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | KLVQVGIYNGTHVIPNDR | Y | Y | ||
| IPI00789973.1 | GRIN1 Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | TMNFTYEVHLVADGK | Y | Y | ||
| IPI00791304.1 | C20orf3 Chromosome 20 open reading frame 3 | AGPNGTLFVADAYK | Y | Y | ||
| IPI00791304.1 | C20orf3 Chromosome 20 open reading frame 3 | NMSFVNDLTVTQDGR | Y | Y | ||
| IPI00791516.1 | CD59 13 kDa protein | TAVNCSSDFDACLITK | Y | Y | ||
| IPI00793495.1 | C6orf27 G7c protein | TFVNPSFSLTSNLSR | Y | |||
| IPI00793688.1 | CD276 60 kDa protein | TALFPDLLAQGNASLR | Y | Y | ||
| IPI00793751.1 | MFAP4 Uncharacterized protein MFAP4 | VDLEDFENNTAYAK | Y | Y | ||
| IPI00793751.1 | MFAP4 Uncharacterized protein MFAP4 | FNGSVSFFR | Y | Y | ||
| IPI00793829.1 | GRM3 99 kDa protein | INFTAPFNPNK | Y | Y | ||
| IPI00794214.1 | BCAM Lutheran glycoprotein | TQNFTLLVQGSPELK | Y | |||
| IPI00794423.1 | SLC1A2 Solute carrier family 1 | VLVAPPPDEEANATSAVVSLLNETVTEVPEETK | Y1,Y2 | Y1,Y2 | ||
| IPI00795030.1 | LASS6 LASS6 protein | FWLPHNVTWADLK | Y | |||
| IPI00795150.1 | BSG 46 kDa protein(IPI00019906) | ILLTCSLNDSATEVTGHR | Y | Y | ||
| IPI00795326.1 | LINGO1 Isoform 2 of Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 precursor | LIPLGVFTGLSNLTK | Y | Y | ||
| IPI00795504.1 | ALCAM 62 kDa protein | KLGDCISEDSYPDGNITWYR | Y | Y | ||
| IPI00795504.1 | ALCAM 62 kDa protein | TVNSLNVSAISIPEHDEADEISDENR | Y | Y | ||
| IPI00795633.1 | CLU CLU | LANLTQGEDQYYLR | Y | Y | ||
| IPI00795633.1 | CLU CLU | QLEEFLNQSSPF | Y | Y | ||
| IPI00795720.1 | CD63 13 kDa protein | NNHTASILDR | Y | Y | ||
| IPI00795801.1 | CD109 Isoform 4 of CD109 antigen precursor | TQDEILFSNSTR | Y | Y | ||
| IPI00795830.1 | AHSG 29 kDa protein | VCQDCPLLAPLNDTR | Y | Y | ||
| IPI00796279.1 | SERPINF1 25 kDa protein | VTQNLTLIEESLTSEFIHDIDR | Y | Y | ||
| IPI00797025.1 | PRNP Major prion protein | GENFTETDVK | Y | Y | ||
| IPI00797503.1 | ITGA7 106 kDa protein | LWNSTFLEEYSAVK | Y | Y | ||
| IPI00798430.1 | TF Transferrin variant (Fragment) | CGLVPVLAENYNK | Y | Y | ||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGNF | Y | Y | ||
| IPI00798430.1 | TF Transferrin variant (Fragment) | QQQHLFGSNVTDCSGNFCLFR | Y | Y | ||
| IPI00807403.1 | ALCAM Isoform 2 of CD166 antigen precursor | IIISPEENVTLTCTAENQLER | Y | Y | ||
| IPI00807403.1 | ALCAM Isoform 2 of CD166 antigen precursor | LGDCISEDSYPDGNITWYR | Y | Y | ||
| IPI00807403.1 | ALCAM Isoform 2 of CD166 antigen precursor | LNLSENYTLSISNAR | Y | Y | ||
| IPI00807403.1 | ALCAM Isoform 2 of CD166 antigen precursor | NATVVWMK | Y | Y | ||
| IPI00828205.1 | IGHM IGHM protein | GLTFQQNASSMCVPDQDTAIR | Y | Y | ||
| IPI00829767.1 | IGHG2 Uncharacterized protein IGHG2 (Fragment) | EEQFNSTFR | Y | Y | ||
| IPI00829867.1 | GBA GBA protein | TYTYADTPDDFQLHNFSLPEEDTK | Y | Y | ||
| IPI00844079.1 | PTPRC Isoform 1 of Leukocyte common antigen precursor | YANITVDYLYNK | Y | Y | ||
| IPI00844348.1 | PON2 39 kDa protein | HTNMNLTQLK | Y | Y | ||
| IPI00845399.1 | KIAA1946 Isoform 2 of UPF0560 protein KIAA1946 precursor | QYLSQAVVEVFVNYTK | Y | Y | ||
| IPI00847414.1 | DPP10 dipeptidyl peptidase 10 isoform short | WINDTDVVYK | Y | Y | ||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | FNLTETSEAEIHQSFQHLLR | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | LSLGAHNTTLTEILK | Y | Y | ||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | TLNQSSDELQLSMGNAMFVK | Y | |||
| IPI00847635.1 | SERPINA3 Isoform 1 of Alpha-1-antichymotrypsin precursor | YTGNASALFILPDQDK | Y | Y | ||
| IPI00853369.1 | PLXNB2 Plexin-B2 precursor | ALSNISLR | Y | Y | ||
| IPI00853369.1 | PLXNB2 Plexin-B2 precursor | SINVTGQGFSLIQR | Y | Y | ||
| IPI00853369.1 | PLXNB2 Plexin-B2 precursor | TEAGAFEYVPDPTFENFTGGVK | Y | Y | ||
| IPI00853589.1 | SGCE sarcoglycan, epsilon isoform 3 | LNAINITSALDR | Y | Y | ||
| IPI00854766.1 | TXNDC15 Isoform 2 of Thioredoxin domain-containing protein 15 precursor | IFIFNQTGIEAK | Y | |||
| IPI00855821.1 | NRXN1-alpha | SGGNATLQVDSWPVIER | Y | Y | ||
| IPI00869004.1 | SERPINA1 Isoform 3 of Alpha-1-antitrypsin precursor | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | Y | Y | ||
| IPI00869004.1 | SERPINA1 Isoform 3 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIA | Y | Y | ||
| IPI00869004.1 | SERPINA1 Isoform 3 of Alpha-1-antitrypsin precursor | QLAHQSNSTNIFFSPVSIATAF | Y | Y | ||
| IPI00869004.1 | SERPINA1 Isoform 3 of Alpha-1-antitrypsin precursor | YLGNATAIF | Y | Y | ||
| IPI00869004.1 | SERPINA1 Isoform 3 of Alpha-1-antitrypsin precursor | YLGNATAIFFLPDEGK | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | FKLEWLGNCSGLNDETYGYK | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | LAVQFTNLTMDTEIR | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | LEWLGNCSGL | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | LEWLGNCSGLN | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | LEWLGNCSGLNDETYGYK | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | VLGFKPKPPKNESLETYPVMK | Y | Y | ||
| IPI00871221.1 | ATP1B1 Isoform 2 of Sodium/potassium-transporting ATPase subunit beta-1 | YLQPLLAVQFTNLTMDTEIR | Y | Y | ||
| IPI00871253.1 | PTPRK Mutant receptor type protein tyrosine phosphatase K | GPLANPIWNVTGFTGR | Y | Y | ||
| IPI00871253.1 | PTPRK Mutant receptor type protein tyrosine phosphatase K | IAVDWESLGYNITR | Y | Y | ||
| IPI00871326.1 | PLXNA1 plexin A1 | VNVSEDCPQILPSTQIYVPVGVVKPITLAAR | Y | Y | ||
| IPI00871339.1 | CACNA2D2 129 kDa protein | AAEDWTENPEPFNASFYR | Y | |||
| IPI00871339.1 | CACNA2D2 129 kDa protein | AGFEYAFDQLQNSNITR | Y | |||
| IPI00871339.1 | CACNA2D2 129 kDa protein | NYTWVPIR | Y | |||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | GPWQEQIVSDPFLVVSNTSTFVPYEIK | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | DHVVVPANTTSVILSGLR | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | DHVVVPANTTSVILSGLRPY | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | FFPYANGTLGIR | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | GYNVTYWR | Y | Y | ||
| IPI00871467.1 | L1CAM cDNA FLJ76744, highly similar to Homo sapiens L1 cell adhesion molecule (L1CAM), transcript variant 1, mRNA | THNLTDLSPHLR | Y | Y | ||
| IPI00871501.1 | SLC44A1 Uncharacterized protein SLC44A1 | CAPVNISCYAK | Y | |||
| IPI00871501.1 | SLC44A1 Uncharacterized protein SLC44A1 | FAEINGSALCSYNLKPSEYTTSPK | Y | |||
| IPI00871510.1 | GRIA2 Isoform 3 of Glutamate receptor 2 precursor | INYTINIMELK | Y | Y | ||
| IPI00871510.1 | GRIA2 Isoform 3 of Glutamate receptor 2 precursor | IQFGGANVSGFQIVDYDDSLVSK | Y | Y | ||
| IPI00871570.1 | SIDT1 SID1 transmembrane family member 1 precursor | VYVNSSSENLNYPVLVVVR | Y | Y | ||
| IPI00871792.1 | PTPRZ1 265 kDa protein | ESFLQTNYTEIR | Y | Y | ||
| IPI00871792.1 | PTPRZ1 265 kDa protein | TVEINLTNDYR | Y | Y | ||
| IPI00871938.1 | PTGFRN 103 kDa protein | ELDLTCNITTDR | Y | Y | ||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | LEPNSVDPENITEIFIANQK | Y | Y | ||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | NLTIVDSGLK | Y | Y | ||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | NSNLQHINFTR | Y | Y | ||
| IPI00872117.1 | NTRK2 Tyrosine-protein kinase receptor (Fragment) | SSPDTQDLYCLNESSK | Y | Y | ||
| IPI00872343.1 | SLC2A3 54 kDa protein | IIKEFINK | Y | Y | ||
| IPI00872375.2 | SLC2A1 Uncharacterized protein SLC2A1 (Fragment) | VIEEFYNQTWVHR | Y | Y | ||
| IPI00872579.1 | PCDH1 Isoform 2 of Protocadherin-1 precursor | ANDSDQGANAEIEYTFHQAPEVVR | Y | Y | ||
| IPI00872579.1 | PCDH1 Isoform 2 of Protocadherin-1 precursor | YGTALVHLYVNETLANR | Y | Y | ||
| IPI00872773.1 | ERO1L Uncharacterized protein ERO1L | WGHNITEFQQR | Y | Y | ||
| IPI00872795.1 | PPAP2A 42 kDa protein | INCSDGYIEYYICR | Y | Y | ||
| IPI00873151.1 | ABCA2 270 kDa protein | LHPEALNLSLDELPPALR | Y | Y | ||
| IPI00873201.1 | PSAP Isoform Sap-mu-6 of Proactivator polypeptide precursor | TNSTFVQALVEHVK | Y | Y | ||
| IPI00873210.1 | FN1 263 kDa protein | LDAPTNLQFVNETDSTVLVR | Y | Y | ||
| IPI00873446.1 | NRCAM Isoform 5 of Neuronal cell adhesion molecule precursor | VISVDELNDTIAANLSDTEFYGAK | Y1,Y2 | Y1,Y2 | ||
| IPI00873846.1 | DPP10 Isoform 1 of Inactive dipeptidyl peptidase 10 | AGVNYTMQVYPDEGHNVSEK | Y | Y | ||
| IPI00873846.1 | DPP10 Isoform 1 of Inactive dipeptidyl peptidase 10 | LNIETNATTLLLENTTFVTFK | Y1,Y2 | Y1,Y2 | ||
| IPI00873889.1 | LAMA2 Uncharacterized protein LAMA2 | VSQAESHAAQLNDSSAVLDGILDEAK | Y | Y | ||
| IPI00874147.1 | CXADR Uncharacterized protein CXADR (Fragment) | SGDASINVTNLQLSDIGTYQCK | Y | Y | ||
| IPI00874212.1 | CREG1 27 kDa protein | IVTPEEYYNVT | Y | |||
| IPI00874212.1 | CREG1 27 kDa protein | LNITNIWVLDYFGGPK | Y | |||
| IPI00876857.1 | TTYH3 Isoform 2 of Protein tweety homolog 3 | VWDTAVGLNHTAEPSLQTLER | Y | Y | ||
| IPI00877100.1 | ACE Isoform Somatic-2 of Angiotensin-converting enzyme, somatic isoform precursor | ELYEPIWQNFTDPQLR | Y | Y | ||
| IPI00877110.1 | SLC12A5 Isoform 1 of Solute carrier family 12 member 5 | FLNATCDEYFTR | Y | |||
| IPI00877110.1 | SLC12A5 Isoform 1 of Solute carrier family 12 member 5 | NNVTEIQGIPGAASGLIK | Y | |||
| IPI00877115.1 | SLC39A12 Isoform 4 of Zinc transporter ZIP12 | QDEDSSFLSQNETEDILAFTR | Y | |||
| IPI00877792.1 | FGG 50 kDa protein | VDKDLQSLEDILHQVENK | Y | Y | ||
| IPI00878568.1 | RTN4R Protein | DLGNLTHLFLHGNR | Y | Y | ||
| IPI00879665.1 | SEZ6L Seizure related 6 homolog (Mouse)-like | DPYWNDTEPLCR | Y | Y | ||
| IPI00879883.1 | RNF13 15 kDa protein | DILAYNFENASQTFDDLPAR | Y | |||
| IPI00879883.1 | RNF13 15 kDa protein | DNSSGTFIVLIR | Y | |||
| IPI00879931.1 | SERPING1 cDNA FLJ78023, highly similar to Homo sapiens serine (or cysteine) proteinase inhibitor, clade G (C1inhibitor), member 1, (angioedema, hereditary) (SERPING1), mRNA | VLSNNSDANLELINTWVAK | Y | Y | ||
| IPI00880178.1 | C19orf63 Isoform 3 of UPF0510 protein C19orf63 precursor | GHEVEDVDLELFNTSVQLQPPTTAPGPETAAFIER | Y | |||
| IPI00884105.1 | LAMP1 Lysosome-associated membrane glycoprotein 1 precursor | ENTSDPSLVIAFGR | Y | Y | ||
| IPI00884105.1 | LAMP1 Lysosome-associated membrane glycoprotein 1 precursor | GHTLTLNFTR | Y | Y | ||
| IPI00884105.1 | LAMP1 Lysosome-associated membrane glycoprotein 1 precursor | SSCGKENTSDPSLVIAFGR | Y | Y | ||
| IPI00889518.1 | MOG Myelin oligodendrocyte glycoprotein isoform alpha1 variant (Fragment) | ISPGKNATGMEVGWYRPPFSR | Y | Y | ||
| IPI00889723.1 | C4A;C4B complement component 4B preproprotein | FSDGLESNSSTQFEVK | Y | Y | ||
| IPI00889723.1 | C4A;C4B complement component 4B preproprotein | GLNVTLSSTGR | Y | Y | ||
EBI: European Bioinformatics Institute; ISB: Institute for Systems Biology; N: N-glycosylated site
3. Gene Ontology analysis
Over the last few years, a Gene Ontology (GO) method has been used to study datasets generated by proteomic analysis (Kitsou et al., 2008; Pan et al., 2007a; Pan et al., 2007b; Shi et al., 2008) to provide insight into the underlying biology (Alexa et al., 2006). GO analysis, either based on cellular components (CC) or biological processes (BP), detects over-represented GO categories (Alexa et al., 2006). When the glycoproteins identified in human CSF and tissues were classified by GO analysis, it was apparent that a majority of the proteins belong to either the extracellular compartment or are associated with the plasma membrane (Figure 2). This is entirely consistent with the claim that most membrane proteins are glycosylated, and that a significant portion of glycoproteins are designated for secretion into the extracellular fluid and thereby enter blood or CSF (Yang et al., 2005).
Figure 2.
GO analysis of glycoproteins identified in human brain (A) and CSF (B), to clearly emphasize the fact that a majority of the proteins are distributed to extracellular and membrane compartments.
4. A Brief discussion of overlapped proteins
As indicated earlier, one of the major goals to isolate glycoproteins is to reveal CNS-specific proteins that are low in abundance in body fluids with the potential to serve as biomarkers for disease diagnosis or disease progression. To this end, there are a few features of the data presented above that must be stressed: 1) isolation of glycoproteins significantly increased the portion of proteins related to CNS function and/or structure (a partial list of those proteins is shown in Table 1), 2) the overlap between the CSF and tissue proteomes is also improved significantly over the general profiling, where only 140 proteins were found in tissue and CSF general profiles that account for 9% of ∼1,500 identified CSF proteins. When glycoproteins were analyzed, 98 proteins were seen in brain tissue (a total of 343 proteins) and CSF (a total of 243 proteins) to account for 43% of CSF glycoproteins. Furthermore, several of the overlapping proteins identified with glycoprotein isolation are likely related to PD pathogenesis. For example, ceruloplasmin and transferrin, both regulate iron metabolism, were reported to be deregulated in PD patients (Dexter et al., 1989; Riederer et al., 1989).
Table 1.
A Partial list of overlapped glycoproteins between human CSF and brain tissue
| Protein IPI | Description | Comments (source: UniPortKB/Swiss-Prot database) |
|---|---|---|
| IPI00002714.1 | Dickkopf-related protein 3 precursor | Inhibitor of Wnt signaling pathway (Potential). Highest expression in heart, brain, and spinal cord |
| IPI00003813.5 | Isoform 1 of cell adhesion molecule 1 precursor | May act as a synaptic cell adhesion molecule that drives synapse assembly. May be involved in neuronal migration, axon growth, pathfinding, and fasciculation on the axons of differentiating neurons. |
| IPI00009997.1 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase | Can initiate the synthesis or the elongation of the linear poly-N-acetyllactosaminoglycans. In the adult, highly expressed in heart, brain, skeletal muscle and kidney |
| IPI00011732.2 | Isoform 1 of GDNF family receptor alpha-2 precursor | Receptor for neurturin. Mediates the NRTN-induced autophosphorylation and activation of the RET receptor. Also able to mediate GDNF signaling through the RET tyrosine kinase receptor. Isoform 1 is found in brain and placenta |
| IPI00013303.2 | Limbic system-associated membrane protein precursor | Mediates selective neuronal growth and axon targeting. Contributes to the guidance of developing axons and remodeling of mature circuits in the limbic system. Essential for normal growth of the hippocampal mossy fiber projection (By similarity) |
| IPI00017601.1 | Ceruloplasmin precursor | Defects in CP are the cause of aceruloplasminemia. It is an autosomal recessive disorder of iron metabolism characterized by iron accumulation in the brain/visceral organs. |
| IPI00020557.1 | Prolow-density lipoprotein receptor-related protein 1 precursor | May modulate cellular events, such as APP metabolism, kinase-dependent intracellular signaling, neuronal calcium signaling as well as neurotransmission |
| IPI00024035.1 | Isoform 1 of cadherin-6 precursor | Cadherins are calcium dependent cell adhesion proteins. They preferentially interact with themselves in a homophilic manner in connecting cells; cadherins may thus contribute to the sorting of heterogeneous cell types. |
| IPI00024966.1 | Contactin-2 precursor | Attached to the neuronal membrane by a GPI-anchor and is also released from neurons. May play a role in the initial growth and guidance of axons. May be involved in cell adhesion |
| IPI00026946.2 | Neuronal pentraxin-2 precursor | Likely to play role in the modification of cellular properties that underlie long-term plasticity. Binds to agar matrix in a calcium-dependent manner |
| IPI00030887.1 | Tyrosine-protein kinase Receptor TYRO3 precursor | May be involved in cell adhesion processes, particularly in the central nervous system |
| IPI00031121.2 | Carboxypeptidase E precursor | Removes residual C-terminal Arg or Lys remaining after initial endoprotease cleavage during prohormone processing. Processes proinsulin. Neuropeptide signaling pathway |
| IPI00064667.4 | Beta-Ala-His dipeptidase p recursor | Preferential hydrolysis of the beta-Ala-|-His dipeptide (carnosine), and also anserine, Xaa-|-His dipeptides and other dipeptides including homocarnosine. |
| IPI00159927.2 | Neurocan core protein precursor | May modulate neuronal adhesion and neurite growth during development by binding to neural cell adhesion molecules (NG-CAM and N-CAM). Chondroitin sulfate proteoglycan; binds to hyaluronic acid |
| IPI00160552.3 | Isoform 1 of tenascin-R precursor | Neural extracellular matrix (ECM) protein involved in interactions with different cells and matrix components |
| IPI00171473.2 | Spondin-1 precursor | Cell adhesion protein that promotes the attachment of spinal cord and sensory neuron cells and the outgrowth of neurites in vitro. May contribute to the growth and guidance of axons in both the spinal cord and the PNS (By similarity). Major factor for vascular smooth muscle cell |
| IPI00176427.1 | Cell adhesion molecule 4 precursor | Involved in the cell-cell adhesion. Has calcium- and magnesium-independent cell-cell adhesion activity. May have tumor- suppressor activity. |
| IPI00216641.1 | Isoform 2 of contactin-1 precursor | Contactins mediate cell surface interactions during nervous system development. Interaction with TNR induces a repulsion of neurons and an inhibition of neurite outgrowth |
| IPI00217882.3 | Sortilin precursor | Promotes neuronal apoptosis by mediating endocytosis of the proapoptotic precursor forms of BDNF (proBDNF) and NGFB (proNGFB). Also acts as a receptor for neurotensin. |
| IPI00295832.1 | Oligodendrocyte-myelin glycoprotein precursor | Cell adhesion molecule contributing to the interactive process required for myelination in the central nervous system. |
| IPI00301512.3 | Isoform DPPX-L of Dipeptidyl aminopeptidase-like protein 6 | May be involved in the physiological processes of brain function. Has no dipeptidyl aminopeptidase activity. May modulate the cell surface expression and the activity of the potassium channel KCND2. |
| IPI00303210.3 | Isoform 2 of ectonucleotide pyrophosphatase/phosphodiest erase family member 2 precursor | Involved in several motility- related processes such as angiogenesis and neurite outgrowth |
| IPI00332887.5 | signal-regulatory protein alpha precursor | Supports adhesion of cerebellar neurons, neurite outgrowth and glial cell attachment. |
| IPI00376427.3 | Neural cell adhesion molecule 2 precursor | May play important roles in selective fasciculation and zone-to-zone projection of the primary olfactory axons |
| IPI00413696.5 | 41 kDa protein | Plays an important role in memory formation and synaptic plasticity in the hippocampus |
| IPI00456623.2 | Isoform 1 of brevican core protein precursor | May play a role in the terminally differentiating and the adult nervous system during postnatal development. Could stabilize interactions between hyaluronan (HA) and brain proteoglycans. |
| IPI00470696.1 | Isoform 1 of netrin receptor UNC5D precursor | Receptor for netrin. May be involved in axon guidance by mediating axon repulsion of neuronal growth cones in the developing nervous system upon ligand binding. |
| IPI00479514.1 | Voltage-dependent calcium channel subunit alpha-2/delta-1 precursor | The alpha-2/delta subunit of voltage-dependent calcium channels regulates calcium current density and activation/inactivation kinetics of the calcium channel. Plays an important role in excitation-contraction coupling |
| IPI00513964.1 | Isoform 2 of semaphorin-4B precursor | Inhibits axonal extension by providing local signals to specify territories inaccessible for growing axons |
| IPI00552450.1 | Opioid binding protein/cell adhesion molecule-like isoform b preproprotein | Binds opioids in the presence of acidic lipids; probably involved in cell contact |
| IPI00554760.1 | Isoform 2 of tenascin-R precursor | Neural extracellular matrix (ECM) protein involved in interactions with different cells and matrix components. These interactions can influence cellular behavior by either evoking a stable adhesion and differentiation, or repulsion and inhibition of neurite growth |
| IPI00655702.3 | Isoform 5 of neurofascin precursor | Cell adhesion, ankyrin-binding protein which may be involved in neurite extension, axonal guidance, synaptogenesis, myelination and neuron-glial cell interactions |
| IPI00783390.1 | Isoform 1 of neural cell adhesion molecule L1-like protein precursor | Extracellular matrix and cell adhesion protein that plays a role in nervous system development and in synaptic plasticity. |
| IPI00797025.1 | Major prion protein | PrP is found in high quantity in the brain of humans and animals infected with neurodegenerative diseases known as transmissible spongiform encephalopathies or prion diseases, like: Creutzfeldt-Jakob disease |
| IPI00807403.1 | Isoform 2 of CD166 antigen precursor | Cell adhesion molecule that binds to CD6. Involved in neurite extension by neurons via heterophilic and homophilic interactions. May play a role in the binding of T- and B-cells to activated leukocytes, as well as in interactions between cells of the nervous system. |
| IPI00855821.1 | Isoform 2 of neurexin-1-alpha precursor | Neuronal cell surface protein that may be involved in cell recognition and cell adhesion. May mediate intracellular signaling. |
| IPI00873446.1 | Isoform 5 of neuronal cell adhesion molecule precursor | Cell adhesion, ankyrin-binding protein involved in neuron-neuron adhesion. May play a role in the molecular assembly of the nodes of Ranvier |
Besides the proteins known to be important to PD pathogenesis, others such as neuroserpin, neural cell adhesion molecule, and neuronal pentraxin II are critical to CNS function, and some have been linked to other neurodegenerative diseases. For example, one of the overlapping proteins, neuroserpin, is a member of the serpin family of serine protease inhibitors. Tissue-distribution analysis reveals a predominantly neuronal expression during the late stages of neurogenesis and, in the adult brain, in areas where synaptic changes are associated with learning and memory (synaptic plasticity). To this end, it should be mentioned that synaptic dysfunction appears to be one of the major early signs of PD progression in human cortex (Pisani et al., 2005). Another example, neural cell adhesion molecule, is involved in signal transduction (Niethammer et al., 2002), and promotes neurite outgrowth and fasciculation (Rutishauser & Edelman, 1980). In the SNpc of PD patients, polysialated-neural cell-adhesion molecule-positive immature neurons were detected. The polysialated neural cell adhesion molecule is a marker of immature, migrating neuroblasts (Yoshimi et al., 2005). The third example, neuronal pentraxin II, was recently reported to be highly up-regulated in PD and is a novel component of Lewy bodies (Moran et al., 2008). Neuronal pentraxin II is also known as the neuronal activity-regulated protein, which is secreted and involved in long-term neuronal plasticity (Hsu & Perin, 1995).
V. Future perspectives
From the analysis of glycoproteins of human CSF and brain tissue, it is obvious that even a focused analysis of glycoproteins remains inadequate for an extensive characterization of CNS-specific proteins. When comparing glycoproteins identified in brain tissue (Appendix II) with those identified in CSF (Appendix I), we found, as expected, that proteins related to the CNS structurally and/or functionally are more frequently identified/quantified in tissue. Because of dynamic issues and a common technical caveat associated with MS-based proteomics, absence of a protein only means that it is not detected, but not necessarily absent, in a particular analysis. We believe that it is critical to examine the tissue proteins unique to a disease process (PD diagnosis and progression in this case) in body fluid by targeted analysis. In our opinion, this approach is critical to the CNS-based disease, given that the CNS is highly organized and specialized with each neurodegenerative disorder that involves selective brain regions. For example, AD predominantly affects the cerebral cortex and hippocampus, whereas PD usually damages brainstem structures, particularly during the early stages of the diseases before other brain regions are involved (Braak et al., 2000; Sudo et al., 2005; Wenk, 2003). Therefore, the pathology-specific proteins could be so diluted in CSF that they are difficult to detect even when glycoproteins are isolated first.
Targeted analysis of proteomics - first identify unique proteins in the CNS, followed by confirmation/validation of known proteins in CSF or plasma in this case - indicates a progression away from unbiased profiling toward a multi-phase technology that allows key elements that uniquely represent a specific biological condition to be analyzed (Aebersold, 2003; Pan et al., 2005). The technology uses isotope dilution followed by MS analysis (Anderson, 2005; Anderson & Hunter, 2006; Anderson et al., 2004; Gerber et al., 2003; Pan et al., 2005), in which test-samples are supplemented (spiked) with synthetic peptides that serve as the signature markers to identify and quantify native peptides (target) within each sample. To date, few investigations have been reported that use the concept of candidate-based targeted quantitative proteomics to study selected peptides/proteins for biomarker verification/validation via ESI or MALDI based platforms. For the ESI approach, a hybrid triple-quadrupole/ion trap mass spectrometer was used to identify and quantify a selected group of targeted proteins within human plasma (Anderson & Hunter, 2006). Alternatively, an off-line LC MALDI-TOF/TOF platform was established to monitor a panel of targeted glycopeptides/glycoproteins in human serum, in conjunction with a sample preparation strategy that extracted deglycosylated N-linked glycopeptides from human serum (Pan et al., 2005). These early investigations have demonstrated the feasibility and advantages of the MS-based targeted quantitative proteomics to simultaneously identify and quantify a panel of selected peptides/proteins in a complex milieu, and consequently could be applied for biomarker verification/validation of AD and PD. Figure 3 demonstrates the basic concepts and work flow to validate a protein of interest in an LC-MALDI format. In fact, we have recently applied this technology to confirm/validate a subset of proteins identified in a previous nonbiased proteomics profiling (Abdi et al., 2006)] unique to AD and PD, respectively, in CSF (Pan et al., 2008). A project is also underway to use this platform to cross-examine the proteins identified in brain tissue with CSF (and vice versa), and eventually in human plasma.
Figure 3.
The illustration of mass spectrometry-based targeted quantitative analysis to detect N-linked glycopeptides in body fluids. Synthetic peptides with stable isotope labeling are used as internal standards for the quantification of endogenous glycopeptides. As an example, N-linked glycopeptide AQLLQGLGFN*L#TER (Corticosteroid-binding globulin) was extracted from human serum with hydrazide chemistry-based solid-phase extraction and detected with an LC MALDI TOF/TOF platform with targeted approach. (Note: # indicates the amino acid that was stable isotope labeled (13C and 15N) in reference peptides; * indicates enzyme-catalyzed conversion of asparagines to aspartic acid at the site of carbohydrate attachment.)
VI. Concluding remarks
The development of technologies from gel electrophoresis-based approaches to high-resolution MS-based approaches for protein identification and quantification has revolutionized protein biomarker discovery critical to disease diagnosis and disease progression monitoring, as well as greatly facilitated studies to reveal the molecular events that underlie neurodegenerative diseases. Among these studies, protein glycosylation and glycoproteomics are growing fields of interest due to the relationship between glycosylation degree/type and the health status of cells. The discovery and identification of glycosylated peptides and proteins and the analyses of their glyco-structures are increasingly important in diagnosis and treatment of neurodegenerative diseases. However, it is obvious that the complete characterization of glycoproteins remains a major challenge in the years to come, largely because of the enormous dynamic range of typical human samples as well as the heterogeneity of human beings. Thus, effective and in-depth protein identification of glycoproteins involved in neurodegenerative disorders requires a concerted approach, including improved glycoprotein enrichment, extensive separation of proteins/peptides, high-resolution tandem mass spectrometric analysis, at profiling and targeted modes, and state-of-the-art bioinformatics.
Acknowledgments
The proteomics analysis of human CSF and brain tissue is supported by NIH grants (ES012703; NS057567; AG025327; AG033398; NS060252; AG05136; AG08671), an award by Michael J. Fox Foundation as well as the Chen-Mei Shaw and the Nancy and Buster Alvord Endowments.
References
- Aarsland D, Andersen K, Larsen JP, Lolk A, Kragh-Sorensen P. Prevalence and characteristics of dementia in Parkinson disease: an 8-year prospective study. Arch Neurol. 2003;60:387–392. doi: 10.1001/archneur.60.3.387. [DOI] [PubMed] [Google Scholar]
- Aarsland D, Andersen K, Larsen JP, Lolk A, Nielsen H, Kragh-Sorensen P. Risk of dementia in Parkinson's disease: a community-based, prospective study. Neurology. 2001;56:730–736. doi: 10.1212/wnl.56.6.730. [DOI] [PubMed] [Google Scholar]
- Aarsland D, Larsen JP, Karlsen K, Lim NG, Tandberg E. Mental symptoms in Parkinson's disease are important contributors to caregiver distress. Int J Geriatr Psychiatry. 1999;14:866–874. [PubMed] [Google Scholar]
- Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, Nixon R, Nutt J, Chung K, Zabetian C, Samii A, Lin M, Hattan S, Pan C, Wang Y, Jin J, Zhu D, Li GJ, Liu Y, Waichunas D, Montine TJ, Zhang J. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- Aebersold R. Constellations in a cellular universe. Nature. 2003;422:115–116. doi: 10.1038/422115a. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. J Proteome Res. 2005;4:1104–1109. doi: 10.1021/pr050027n. [DOI] [PubMed] [Google Scholar]
- Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Arakawa T, Kita Y, Niikura T. A rescue factor for Alzheimer's diseases: discovery, activity, structure, and mechanism. Curr Med Chem. 2008;15:2086–2098. doi: 10.2174/092986708785747616. [DOI] [PubMed] [Google Scholar]
- Bahl JM, Jensen SS, Larsen MR, Heegaard NH. Characterization of the human cerebrospinal fluid phosphoproteome by titanium dioxide affinity chromatography and mass spectrometry. Anal Chem. 2008;80:6308–6316. doi: 10.1021/ac800835y. [DOI] [PubMed] [Google Scholar]
- Borghi R, Marchese R, Negro A, Marinelli L, Forloni G, Zaccheo D, Abbruzzese G, Tabaton M. Full length alpha-synuclein is present in cerebrospinal fluid from Parkinson's disease and normal subjects. Neurosci Lett. 2000;287:65–67. doi: 10.1016/s0304-3940(00)01153-8. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Bratzke H, Hamm-Clement J, Sandmann-Keil D, Rub U. Staging of the intracerebral inclusion body pathology associated with idiopathic Parkinson's disease (preclinical and clinical stages) J Neurol. 2002;249:III/, 1–5. doi: 10.1007/s00415-002-1301-4. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Braak H, Rub U, Sandmann-Keil D, Gai WP, de Vos RA, Jansen Steur EN, Arai K, Braak E. Parkinson's disease: affection of brain stem nuclei controlling premotor and motor neurons of the somatomotor system. Acta Neuropathol. 2000;99:489–495. doi: 10.1007/s004010051150. [DOI] [PubMed] [Google Scholar]
- Bundy JL, Fenselau C. Lectin and carbohydrate affinity capture surfaces for mass spectrometric analysis of microorganisms. Anal Chem. 2001;73:751–757. doi: 10.1021/ac0011639. [DOI] [PubMed] [Google Scholar]
- Bunkenborg J, Pilch BJ, Podtelejnikov AV, Wisniewski JR. Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics. 2004;4:454–465. doi: 10.1002/pmic.200300556. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Tsuji T, Shiozaki A, Kohno R, Yoshizato K, Shimohama S, Aksenov M, Pierce WM, Booze R. Proteomic analysis of oxidatively modified proteins in Alzheimer's disease brain: insights into neurodegeneration. Cell Mol Biol (Noisy-le-grand) 2003;49:747–751. [PubMed] [Google Scholar]
- Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- Casu B, Guerrini M, Torri G. Structural and conformational aspects of the anticoagulant and anti-thrombotic activity of heparin and dermatan sulfate. Curr Pharm Des. 2004;10:939–949. doi: 10.2174/1381612043452794. [DOI] [PubMed] [Google Scholar]
- Catalina MI, Koeleman CA, Deelder AM, Wuhrer M. Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun Mass Spectrom. 2007;21:1053–1061. doi: 10.1002/rcm.2929. [DOI] [PubMed] [Google Scholar]
- Chiang PK, Lam MA, Luo Y. The many faces of amyloid beta in Alzheimer's disease. Curr Mol Med. 2008;8:580–584. doi: 10.2174/156652408785747951. [DOI] [PubMed] [Google Scholar]
- Colangelo CM, Williams KR. Isotope-coded affinity tags for protein quantification. Methods Mol Biol. 2006;328:151–158. doi: 10.1385/1-59745-026-X:151. [DOI] [PubMed] [Google Scholar]
- Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Cookson MR. The biochemistry of Parkinson's disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- Cummings RD, Kornfeld S. Fractionation of asparagine-linked oligosaccharides by serial lectin-Agarose affinity chromatography. A rapid, sensitive, and specific technique. J Biol Chem. 1982;257:11235–11240. [PubMed] [Google Scholar]
- D'Ascenzo M, Choe L, Lee KH. iTRAQPak: an R based analysis and visualization package for 8-plex isobaric protein expression data. Brief Funct Genomic Proteomic. 2008;7:127–135. doi: 10.1093/bfgp/eln007. [DOI] [PubMed] [Google Scholar]
- Dexter DT, Wells FR, Lees AJ, Agid F, Agid Y, Jenner P, Marsden CD. Increased nigral iron content and alterations in other metal ions occurring in brain in Parkinson's disease. J Neurochem. 1989;52:1830–1836. doi: 10.1111/j.1471-4159.1989.tb07264.x. [DOI] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Durham M, Regnier FE. Targeted glycoproteomics: serial lectin affinity chromatography in the selection of O-glycosylation sites on proteins from the human blood proteome. J Chromatogr A. 2006;1132:165–173. doi: 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- El-Agnaf OM, Salem SA, Paleologou KE, Curran MD, Gibson MJ, Court JA, Schlossmacher MG, Allsop D. Detection of oligomeric forms of alpha-synuclein protein in human plasma as a potential biomarker for Parkinson's disease. Faseb J. 2006;20:419–425. doi: 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- Farrer M, Chan P, Chen R, Tan L, Lincoln S, Hernandez D, Forno L, Gwinn-Hardy K, Petrucelli L, Hussey J, Singleton A, Tanner C, Hardy J, Langston JW. Lewy bodies and parkinsonism in families with parkin mutations. Ann Neurol. 2001;50:293–300. doi: 10.1002/ana.1132. [DOI] [PubMed] [Google Scholar]
- Finehout EJ, Franck Z, Choe LH, Relkin N, Lee KH. Cerebrospinal fluid proteomic biomarkers for Alzheimer's disease. Ann Neurol. 2007;61:120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- Foltynie T, Brayne CE, Robbins TW, Barker RA. The cognitive ability of an incident cohort of Parkinson's patients in the UK. The CamPaIGN study. Brain. 2004;127:550–560. doi: 10.1093/brain/awh067. [DOI] [PubMed] [Google Scholar]
- Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Krokhin O, Antonovici M, Ens W, Standing KG, Beavis RC, Wilkins JA. Lectin affinity as an approach to the proteomic analysis of membrane glycoproteins. J Proteome Res. 2004;3:841–850. doi: 10.1021/pr049937f. [DOI] [PubMed] [Google Scholar]
- Goldstein IJ, Hollerman CE, Smith EE. Protein-Carbohydrate Interaction. Ii. Inhibition Studies on the Interaction of Concanavalin a with Polysaccharides. Biochemistry. 1965;4:876–883. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]
- Gulcicek EE, Colangelo CM, McMurray W, Stone K, Williams K, Wu T, Zhao H, Spratt H, Kurosky A, Wu B. Proteomics and the analysis of proteomic data: an overview of current protein-profiling technologies. Curr Protoc Bioinformatics. 2005;Chapter 13(Unit 13 11) doi: 10.1002/0471250953.bi1301s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI, Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- Hanisch FG. O-glycosylation of the mucin type. Biol Chem. 2001;382:143–149. doi: 10.1515/BC.2001.022. [DOI] [PubMed] [Google Scholar]
- Hanisch FG, Jovanovic M, Peter-Katalinic J. Glycoprotein identification and localization of O-glycosylation sites by mass spectrometric analysis of deglycosylated/alkylaminylated peptide fragments. Anal Biochem. 2001;290:47–59. doi: 10.1006/abio.2000.4955. [DOI] [PubMed] [Google Scholar]
- Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16:288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Haqqani AS, Kelly JF, Stanimirovic DB. Quantitative protein profiling by mass spectrometry using isotope-coded affinity tags. Methods Mol Biol. 2008;439:225–240. doi: 10.1007/978-1-59745-188-8_16. [DOI] [PubMed] [Google Scholar]
- Haynes PA, Aebersold R. Simultaneous detection and identification of O-GlcNAc-modified glycoproteins using liquid chromatography-tandem mass spectrometry. Anal Chem. 2000;72:5402–5410. doi: 10.1021/ac000512w. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconj J. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Hirotani M, Maita C, Niino M, Iguchi-Ariga S, Hamada S, Ariga H, Sasaki H. Correlation between DJ-1 levels in the cerebrospinal fluid and the progression of disabilities in multiple sclerosis patients. Mult Scler. 2008;14:1056–1060. doi: 10.1177/1352458508093616. [DOI] [PubMed] [Google Scholar]
- Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4:628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Perin MS. Human neuronal pentraxin II (NPTX2): conservation, genomic structure, and chromosomal localization. Genomics. 1995;28:220–227. doi: 10.1006/geno.1995.1134. [DOI] [PubMed] [Google Scholar]
- Hwang HY, Olson SK, Esko JD, Horvitz HR. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 2003;423:439–443. doi: 10.1038/nature01634. [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- Irungu J, Go EP, Zhang Y, Dalpathado DS, Liao HX, Haynes BF, Desaire H. Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J Am Soc Mass Spectrom. 2008;19:1209–1220. doi: 10.1016/j.jasms.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakowec MW, Petzinger GM, Sastry S, Donaldson DM, McCormack A, Langston JW. The native form of alpha-synuclein is not found in the cerebrospinal fluid of patients with Parkinson's disease or normal controls. Neurosci Lett. 1998;253:13–16. doi: 10.1016/s0304-3940(98)00599-0. [DOI] [PubMed] [Google Scholar]
- Jankovic J. Parkinson's disease. A half century of progress. Neurology. 2001;57:S1–3. [PubMed] [Google Scholar]
- Jin J, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol Cell Proteomics. 2006;5:1193–1204. doi: 10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- Jobst KA, Barnetson LP, Shepstone BJ. Int Psychogeriatr. Vol. 9. 1997. Accurate prediction of histologically confirmed Alzheimer's disease and the differential diagnosis of dementia: the use of NINCDS-ADRDA and DSM-III-R criteria, SPECT, X-ray CT, and APO E4 medial temporal lobe dementias. The Oxford Project to Investigate Memory and Aging; pp. 191–222. discussion 247-152. [PubMed] [Google Scholar]
- Johansen PG, Marshall RD, Neuberger A. Carbohydrates in protein. 3 The preparation and some of the properties of a glycopeptide from hen's-egg albumin. Biochem J. 1961;78:518–527. doi: 10.1042/bj0780518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Kaji H, Yamauchi Y, Takahashi N, Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat Protoc. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- Kameyama A, Nakaya S, Ito H, Kikuchi N, Angata T, Nakamura M, Ishida HK, Narimatsu H. Strategy for simulation of CID spectra of N-linked oligosaccharides toward glycomics. J Proteome Res. 2006;5:808–814. doi: 10.1021/pr0503937. [DOI] [PubMed] [Google Scholar]
- Kamra A, Gupta MN. Crosslinked concanavalin A-O-(diethylaminoethyl)-cellulose--an affinity medium for concanavalin A-interacting glycoproteins. Anal Biochem. 1987;164:405–410. doi: 10.1016/0003-2697(87)90511-2. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Achord DT, Sly WS. Phosphohexosyl components of a lysosomal enzyme are recognized by pinocytosis receptors on human fibroblasts. Proc Natl Acad Sci U S A. 1977;74:2026–2030. doi: 10.1073/pnas.74.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Kitsou E, Pan S, Zhang J, Shi M, Zabeti A, Dickson D, Albin R, Gearing M, Kashima D, Wang Y, Beyer R, Zhou Y, Pan C, Caudle W, Zhang J. Identification of proteins in human substantia nigra. PROTEOMICS - CLINICAL APPLICATIONS. 2008;2:776–782. doi: 10.1002/prca.200800028. [DOI] [PubMed] [Google Scholar]
- Korolainen MA, Goldsteins G, Alafuzoff I, Koistinaho J, Pirttila T. Proteomic analysis of protein oxidation in Alzheimer's disease brain. Electrophoresis. 2002;23:3428–3433. doi: 10.1002/1522-2683(200210)23:19<3428::AID-ELPS3428>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Krokhin O, Ens W, Standing KG, Wilkins J, Perreault H. Site-specific N-glycosylation analysis: matrix-assisted laser desorption/ionization quadrupole-quadrupole time-of-flight tandem mass spectral signatures for recognition and identification of glycopeptides. Rapid Commun Mass Spectrom. 2004;18:2020–2030. doi: 10.1002/rcm.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Sato Y, Suzuki Y, Goto-Inoue N, Toda T, Suzuki M, Hisanaga S, Suzuki A, Endo T. Analysis of glycopeptides using lectin affinity chromatography with MALDI-TOF mass spectrometry. Anal Chem. 2008;80:3693–3698. doi: 10.1021/ac800070d. [DOI] [PubMed] [Google Scholar]
- Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- Kurogochi M, Nishimura S. Structural characterization of N-glycopeptides by matrix-dependent selective fragmentation of MALDI-TOF/TOF tandem mass spectrometry. Anal Chem. 2004;76:6097–6101. doi: 10.1021/ac049294n. [DOI] [PubMed] [Google Scholar]
- Kussmann M, Lassing U, Sturmer CA, Przybylski M, Roepstorff P. Matrix-assisted laser desorption/ionization mass spectrometric peptide mapping of the neural cell adhesion protein neurolin purified by sodium dodecyl sulfate polyacrylamide gel electrophoresis or acidic precipitation. J Mass Spectrom. 1997;32:483–493. doi: 10.1002/(SICI)1096-9888(199705)32:5<483::AID-JMS502>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Larsen MR, Cordwell SJ, Roepstorff P. Graphite powder as an alternative or supplement to reversed-phase material for desalting and concentration of peptide mixtures prior to matrix-assisted laser desorption/ionization-mass spectrometry. Proteomics. 2002;2:1277–1287. doi: 10.1002/1615-9861(200209)2:9<1277::AID-PROT1277>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Lattard V, Fondeur-Gelinotte M, Gulberti S, Jacquinet JC, Boudrant J, Netter P, Magdalou J, Ouzzine M, Fournel-Gigleux S. Purification and characterization of a soluble form of the recombinant human galactose-beta1,3-glucuronosyltransferase I expressed in the yeast Pichia pastoris. Protein Expr Purif. 2006;47:137–143. doi: 10.1016/j.pep.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Leverenz JB, Umar I, Wang Q, Montine TJ, McMillan PJ, Tsuang DW, Jin J, Pan C, Shin J, Zhu D, Zhang J. Proteomic identification of novel proteins in cortical lewy bodies. Brain Pathol. 2007;17:139–145. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BE, Katzen HL. Early cognitive changes and nondementing behavioral abnormalities in Parkinson's disease. Adv Neurol. 2005;96:84–94. [PubMed] [Google Scholar]
- Levin Y, Schwarz E, Wang L, Leweke FM, Bahn S. Label-free LC-MS/MS quantitative proteomics for large-scale biomarker discovery in complex samples. J Sep Sci. 2007;30:2198–2203. doi: 10.1002/jssc.200700189. [DOI] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Liu F, Zaidi T, Iqbal K, Grundke-Iqbal I, Merkle RK, Gong CX. Role of glycosylation in hyperphosphorylation of tau in Alzheimer's disease. FEBS Lett. 2002;512:101–106. doi: 10.1016/s0014-5793(02)02228-7. [DOI] [PubMed] [Google Scholar]
- Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochnit G, Geyer R. An optimized protocol for nano-LC-MALDI-TOF-MS coupling for the analysis of proteolytic digests of glycoproteins. Biomed Chromatogr. 2004;18:841–848. doi: 10.1002/bmc.399. [DOI] [PubMed] [Google Scholar]
- Lowe J, Graham L, Leigh PN. Disorders of movement and system degeneration. In: Graham DI, Lantos PL, editors. Neuropathology. 1997. pp. 281–366. [Google Scholar]
- Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- Madera M, Mechref Y, Klouckova I, Novotny MV. High-sensitivity profiling of glycoproteins from human blood serum through multiple-lectin affinity chromatography and liquid chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;845:121–137. doi: 10.1016/j.jchromb.2006.07.067. [DOI] [PubMed] [Google Scholar]
- Madera M, Mechref Y, Klouckova I, Novotny MV. Semiautomated high-sensitivity profiling of human blood serum glycoproteins through lectin preconcentration and multidimensional chromatography/tandem mass spectrometry. J Proteome Res. 2006;5:2348–2363. doi: 10.1021/pr060169x. [DOI] [PubMed] [Google Scholar]
- Madera M, Mechref Y, Novotny MV. Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides. Anal Chem. 2005;77:4081–4090. doi: 10.1021/ac050222l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder K, Tang MX, Cote L, Stern Y, Mayeux R. The frequency and associated risk factors for dementia in patients with Parkinson's disease. Arch Neurol. 1995;52:695–701. doi: 10.1001/archneur.1995.00540310069018. [DOI] [PubMed] [Google Scholar]
- Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer's mice: understanding the interface between physiology and disease. PLoS ONE. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- Monzo A, Bonn GK, Guttman A. Boronic acid-lectin affinity chromatography. 1. Simultaneous glycoprotein binding with selective or combined elution. Anal Bioanal Chem. 2007;389:2097–2102. doi: 10.1007/s00216-007-1627-y. [DOI] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Moran LB, Hickey L, Michael GJ, Derkacs M, Christian LM, Kalaitzakis ME, Pearce RK, Graeber MB. Neuronal pentraxin II is highly upregulated in Parkinson's disease and a novel component of Lewy bodies. Acta Neuropathol. 2008;115:471–478. doi: 10.1007/s00401-007-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle W, Michalski JC. Glycomics and mass spectrometry. Curr Pharm Des. 2005;11:2615–2645. doi: 10.2174/1381612054546897. [DOI] [PubMed] [Google Scholar]
- Mormann M, Paulsen H, Peter-Katalinic J. Electron capture dissociation of O-glycosylated peptides: radical site-induced fragmentation of glycosidic bonds. Eur J Mass Spectrom(Chichester, Eng) 2005;11:497–511. doi: 10.1255/ejms.738. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Burger MM. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J Biol Chem. 1974;249:3116–3122. [PubMed] [Google Scholar]
- Nalivaeva NN, Turner AJ. Post-translational modifications of proteins: acetylcholinesterase as a model system. Proteomics. 2001;1:735–747. doi: 10.1002/1615-9861(200106)1:6<735::AID-PROT735>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Delling M, Sytnyk V, Dityatev A, Fukami K, Schachner M. Cosignaling of NCAM via lipid rafts and the FGF receptor is required for neuritogenesis. J Cell Biol. 2002;157:521–532. doi: 10.1083/jcb.200109059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novogrodsky A, Lotan R, Ravid A, Sharon N. Peanut agglutinin, a new mitogen that binds to galactosyl sites exposed after neuraminidase treatment. J Immunol. 1975;115:1243–1248. [PubMed] [Google Scholar]
- Nussbaum M, Treves TA, Inzelberg R, Rabey JM, Korczyn AD. Survival in Parkinson's disease: the effect of dementia. Parkinsonism Relat Disord. 1998;4:179–181. doi: 10.1016/s1353-8020(98)00039-x. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Osorio C, Sullivan PM, He DN, Mace BE, Ervin JF, Strittmatter WJ, Alzate O. Mortalin is regulated by APOE in hippocampus of AD patients and by human APOE in TR mice. Neurobiol Aging. 2007;28:1853–1862. doi: 10.1016/j.neurobiolaging.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Pan S, Rush J, Peskind ER, Galasko D, Chung K, Quinn J, Jankovic J, Leverenz JB, Zabetian C, Pan C, Wang Y, Oh JH, Gao J, Zhang J, Montine T, Zhang J. Application of Targeted Quantitative Proteomics Analysis in Human Cerebrospinal Fluid Using a Liquid Chromatography Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Tandem Mass Spectrometer (LC MALDI TOF/TOF) Platform. J Proteome Res. 2008;7:720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- Pan S, Shi M, Jin J, Albin RL, Lieberman A, Gearing M, Lin B, Pan C, Yan X, Kashima DT, Zhang J. Proteomics identification of proteins in human cortex using multidimensional separations and MALDI tandem mass spectrometer. Mol Cell Proteomics. 2007a;6:1818–1823. doi: 10.1074/mcp.M700158-MCP200. [DOI] [PubMed] [Google Scholar]
- Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin J, Li JG, Zhu D, Pan C, Zhang J. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- Pan S, Zhang H, Rush J, Eng J, Zhang N, Patterson D, Comb MJ, Aebersold R. High throughput proteome screening for biomarker detection. Mol Cell Proteomics. 2005;4:182–190. doi: 10.1074/mcp.M400161-MCP200. [DOI] [PubMed] [Google Scholar]
- Pan S, Zhu D, Quinn JF, Peskind ER, Montine TJ, Lin B, Goodlett DR, Taylor G, Eng J, Zhang J. A combined dataset of human cerebrospinal fluid proteins identified by multi-dimensional chromatography and tandem mass spectrometry. Proteomics. 2007b;7:469–473. doi: 10.1002/pmic.200600756. [DOI] [PubMed] [Google Scholar]
- Pisani A, Centonze D, Bernardi G, Calabresi P. Striatal synaptic plasticity: implications for motor learning and Parkinson's disease. Mov Disord. 2005;20:395–402. doi: 10.1002/mds.20394. [DOI] [PubMed] [Google Scholar]
- Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5:1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker GJ, Pergantis SA, Blok-Tip L, Langridge JI, Kleen A, Thomas-Oates JE. Mass spectrometric determination of the sites of O-glycan attachment with low picomolar sensitivity. Anal Biochem. 1998;257:149–160. doi: 10.1006/abio.1997.2548. [DOI] [PubMed] [Google Scholar]
- Riederer P, Sofic E, Rausch WD, Schmidt B, Reynolds GP, Jellinger K, Youdim MB. Transition metals, ferritin, glutathione, and ascorbic acid in parkinsonian brains. J Neurochem. 1989;52:515–520. doi: 10.1111/j.1471-4159.1989.tb09150.x. [DOI] [PubMed] [Google Scholar]
- Rite I, Arguelles S, Venero JL, Garcia-Rodriguez S, Ayala A, Cano J, Machado A. Proteomic identification of biomarkers in the cerebrospinal fluid in a rat model of nigrostriatal dopaminergic degeneration. J Neurosci Res. 2007;85:3607–3618. doi: 10.1002/jnr.21452. [DOI] [PubMed] [Google Scholar]
- Robertson LA, Moya KL, Breen KC. The potential role of tau protein O-glycosylation in Alzheimer's disease. J Alzheimers Dis. 2004;6:489–495. doi: 10.3233/jad-2004-6505. [DOI] [PubMed] [Google Scholar]
- Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102:285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- Rutishauser U, Edelman GM. Effects of fasciculation on the outgrowth of neurites from spinal ganglia in culture. J Cell Biol. 1980;87:370–378. doi: 10.1083/jcb.87.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Valero J, Barquero MS, Marcos A, McLean CA, Small DH. Altered glycosylation of acetylcholinesterase in lumbar cerebrospinal fluid of patients with Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2000;69:664–667. doi: 10.1136/jnnp.69.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Valero J, Fodero LR, Sjogren M, Andreasen N, Amici S, Gallai V, Vanderstichele H, Vanmechelen E, Parnetti L, Blennow K, Small DH. Glycosylation of acetylcholinesterase and butyrylcholinesterase changes as a function of the duration of Alzheimer's disease. J Neurosci Res. 2003;72:520–526. doi: 10.1002/jnr.10599. [DOI] [PubMed] [Google Scholar]
- Saez-Valero J, Sberna G, McLean CA, Small DH. Molecular isoform distribution and glycosylation of acetylcholinesterase are altered in brain and cerebrospinal fluid of patients with Alzheimer's disease. J Neurochem. 1999;72:1600–1608. doi: 10.1046/j.1471-4159.1999.721600.x. [DOI] [PubMed] [Google Scholar]
- Saez-Valero J, Small DH. Acetylcholinesterase and butyrylcholinesterase glycoforms are biomarkers of Alzheimer's disease. J Alzheimers Dis. 2001;3:323–328. doi: 10.3233/jad-2001-3307. [DOI] [PubMed] [Google Scholar]
- Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- Schrag A, Jahanshahi M, Quinn N. How does Parkinson's disease affect quality of life? A comparison with quality of life in the general population. Mov Disord. 2000;15:1112–1118. doi: 10.1002/1531-8257(200011)15:6<1112::aid-mds1008>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Sharon N, Lis H. Lectins as cell recognition molecules. Science. 1989;246:227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- Shi M, Jin J, Wang Y, Beyer RP, Kitsou E, Albin RL, Gearing M, Pan C, Zhang J. Mortalin: a protein associated with progression of Parkinson disease? J Neuropathol Exp Neurol. 2008;67:117–124. doi: 10.1097/nen.0b013e318163354a. [DOI] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Siddique N, Siddique T. Genetics of amyotrophic lateral sclerosis. Phys Med Rehabil Clin N Am. 2008;19:429–439. doi: 10.1016/j.pmr.2008.05.001. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihlbom C, Davidsson P, Emmett MR, Marshall AG, L NC. Glycoproteomics of cerebrospinal fluid in neurodegenerative disease. Int J Mass Spectrom. 2004;234:145–152. [Google Scholar]
- Silveyra MX, Cuadrado-Corrales N, Marcos A, Barquero MS, Rabano A, Calero M, Saez-Valero J. Altered glycosylation of acetylcholinesterase in Creutzfeldt-Jakob disease. J Neurochem. 2006;96:97–104. doi: 10.1111/j.1471-4159.2005.03514.x. [DOI] [PubMed] [Google Scholar]
- Simonsen AH, McGuire J, Podust VN, Hagnelius NO, Nilsson TK, Kapaki E, Vassilopoulos D, Waldemar G. A novel panel of cerebrospinal fluid biomarkers for the differential diagnosis of Alzheimer's disease versus normal aging and frontotemporal dementia. Dement Geriatr Cogn Disord. 2007;24:434–440. doi: 10.1159/000110576. [DOI] [PubMed] [Google Scholar]
- Song X, Bandow J, Sherman J, Baker JD, Brown PW, McDowell MT, Molloy MP. iTRAQ experimental design for plasma biomarker discovery. J Proteome Res. 2008;7:2952–2958. doi: 10.1021/pr800072x. [DOI] [PubMed] [Google Scholar]
- Spiro RG. Glycoproteins. Adv Protein Chem. 1973;27:349–467. doi: 10.1016/s0065-3233(08)60451-9. [DOI] [PubMed] [Google Scholar]
- Spiro RG. Periodate Oxidation of the Glycoprotein Fetuin. J Biol Chem. 1964;239:567–573. [PubMed] [Google Scholar]
- Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Murphy MJ, Jeffery J. Cerebrospinal fluid: the role of biochemical analysis. Br J Hosp Med (Lond) 2008;69:218–221. doi: 10.12968/hmed.2008.69.4.28977. [DOI] [PubMed] [Google Scholar]
- Stephens E, Sugars J, Maslen SL, Williams DH, Packman LC, Ellar DJ. The N-linked oligosaccharides of aminopeptidase N from Manduca sexta: site localization and identification of novel N-glycan structures. Eur J Biochem. 2004;271:4241–4258. doi: 10.1111/j.1432-1033.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- Sudo S, Shiozawa M, Cairns NJ, Wada Y. Aberrant accentuation of neurofibrillary degeneration in the hippocampus of Alzheimer's disease with amyloid precursor protein 717 and presenilin-1 gene mutations. J Neurol Sci. 2005;234:55–65. doi: 10.1016/j.jns.2005.03.043. [DOI] [PubMed] [Google Scholar]
- Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6:141–149. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kitajima K, Inoue S, Inoue Y. N-glycosylation/deglycosylation as a mechanism for the post-translational modification/remodification of proteins. Glycoconj J. 1995;12:183–193. doi: 10.1007/BF00731318. [DOI] [PubMed] [Google Scholar]
- Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28:641–653. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Bertolini M, Pigman W. Serine and threonine glycosidic linkages in bovine submaxillary mucin. Biochem Biophys Res Commun. 1964;16:404–409. doi: 10.1016/0006-291x(64)90366-3. [DOI] [PubMed] [Google Scholar]
- Thomas B, Beal MF. Parkinson's disease. Hum Mol Genet 16 Spec No. 2007;2:R183–194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- Tokuda T, Salem SA, Allsop D, Mizuno T, Nakagawa M, Qureshi MM, Locascio JJ, Schlossmacher MG, El-Agnaf OM. Decreased alpha-synuclein in cerebrospinal fluid of aged individuals and subjects with Parkinson's disease. Biochem Biophys Res Commun. 2006;349:162–166. doi: 10.1016/j.bbrc.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Tumani H, Teunissen C, Sussmuth S, Otto M, Ludolph AC, Brettschneider J. Cerebrospinal fluid biomarkers of neurodegeneration in chronic neurological diseases. Expert Rev Mol Diagn. 2008;8:479–494. doi: 10.1586/14737159.8.4.479. [DOI] [PubMed] [Google Scholar]
- Varki A, Kornfeld S. Structural studies of phosphorylated high mannose-type oligosaccharides. J Biol Chem. 1980;255:10847–10858. [PubMed] [Google Scholar]
- Verbeek MM, De Jong D, Kremer HP. Brain-specific proteins in cerebrospinal fluid for the diagnosis of neurodegenerative diseases. Ann Clin Biochem. 2003;40:25–40. doi: 10.1258/000456303321016141. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Tanji K, Mori F, Takahashi H. The Lewy body in Parkinson's disease: molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology. 2007;27:494–506. doi: 10.1111/j.1440-1789.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu SL, Hancock WS. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap--Fourier transform mass spectrometry. Glycobiology. 2006;16:514–523. doi: 10.1093/glycob/cwj091. [DOI] [PubMed] [Google Scholar]
- Waragai M, Wei J, Fujita M, Nakai M, Ho GJ, Masliah E, Akatsu H, Yamada T, Hashimoto M. Increased level of DJ-1 in the cerebrospinal fluids of sporadic Parkinson's disease. Biochem Biophys Res Commun. 2006;345:967–972. doi: 10.1016/j.bbrc.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Wenk GL. Neuropathologic changes in Alzheimer's disease. J Clin Psychiatry. 2003;64 9:7–10. [PubMed] [Google Scholar]
- Wiener MC, van Hoek AN. A lectin screening method for membrane glycoproteins: application to the human CHIP28 water channel (AQP-1) Anal Biochem. 1996;241:267–268. doi: 10.1006/abio.1996.0411. [DOI] [PubMed] [Google Scholar]
- Williams-Gray CH, Foltynie T, Brayne CE, Robbins TW, Barker RA. Evolution of cognitive dysfunction in an incident Parkinson's disease cohort. Brain. 2007;130:1787–1798. doi: 10.1093/brain/awm111. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Hokke CH, Deelder AM. Glycopeptide analysis by matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry reveals novel features of horseradish peroxidase glycosylation. Rapid Commun Mass Spectrom. 2004;18:1741–1748. doi: 10.1002/rcm.1546. [DOI] [PubMed] [Google Scholar]
- Xiong L, Andrews D, Regnier F. Comparative proteomics of glycoproteins based on lectin selection and isotope coding. J Proteome Res. 2003;2:618–625. doi: 10.1021/pr0340274. [DOI] [PubMed] [Google Scholar]
- Yahara I, Edelman GM. Restriction of the mobility of lymphocyte immunoglobulin receptors by concanavalin A. Proc Natl Acad Sci U S A. 1972;69:608–612. doi: 10.1073/pnas.69.3.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Totani K, Ohkura T, Takasaki S, Goldstein IJ, Kobata A. Carbohydrate binding properties of complex-type oligosaccharides on immobilized Datura stramonium lectin. J Biol Chem. 1987;262:1602–1607. [PubMed] [Google Scholar]
- Yang YR, Liu SL, Qin ZY, Liu FJ, Qin YJ, Bai SM, Chen ZY. Comparative proteomics analysis of cerebrospinal fluid of patients with Guillain-Barre syndrome. Cell Mol Neurobiol. 2008;28:737–744. doi: 10.1007/s10571-007-9257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053:79–88. [PubMed] [Google Scholar]
- Yang Z, Hancock WS, Chew TR, Bonilla L. A study of glycoproteins in human serum and plasma reference standards (HUPO) using multilectin affinity chromatography coupled with RPLC-MS/MS. Proteomics. 2005;5:3353–3366. doi: 10.1002/pmic.200401190. [DOI] [PubMed] [Google Scholar]
- Yoshimi K, Ren YR, Seki T, Yamada M, Ooizumi H, Onodera M, Saito Y, Murayama S, Okano H, Mizuno Y, Mochizuki H. Possibility for neurogenesis in substantia nigra of parkinsonian brain. Ann Neurol. 2005;58:31–40. doi: 10.1002/ana.20506. [DOI] [PubMed] [Google Scholar]
- Yuan X, Desiderio DM. Human cerebrospinal fluid peptidomics. J Mass Spectrom. 2005;40:176–181. doi: 10.1002/jms.737. [DOI] [PubMed] [Google Scholar]
- Yuan X, Desiderio DM. Proteomics analysis of phosphotyrosyl-proteins in human lumbar cerebrospinal fluid. J Proteome Res. 2003;2:476–487. doi: 10.1021/pr025589a. [DOI] [PubMed] [Google Scholar]
- Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15:881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- Zhang J. Proteomics of human cerebrospinal fluid - the good, the bad, and the ugly. Proteomics Clin Appl. 2007;1:805–819. doi: 10.1002/prca.200700081. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sokal I, Peskind ER, Quinn JF, Jankovic J, Kenney C, Chung KA, Millard SP, Nutt JG, Montine TJ. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129:526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, Mann M. Integrated analysis of the cerebrospinal fluid peptidome and proteome. J Proteome Res. 2008;7:386–399. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]