Abstract
With the advent of induced pluripotent stem cell (iPSC) technology, it is now feasible to generate iPSCs with a defined genotype or disease state. When coupled with direct differentiation of defined lineage, such as hepatic endoderm (HE). iPSC would revolutionise the way we study human liver biology and generate efficient “off the shelf” models of human liver disease.
Here we show the `proof of concept' that iPSC lines representing both male and female sexes and two ethnic origins can be differentiated to HE at efficiencies of between 70–90%, using a method mimicking a physiological condition. iPSC-derived HE exhibited hepatic morphology, and expressed the hepatic markers, Albumin and E-Cadherin as assessed by immuno-histochemistry. They also expressed alpha fetal protein (AFP), HNF4a, and a metabolic marker, Cyp7A1, demonstrating a definitive endodermal lineage differentiation. Furthermore, iPSC-derived hepatocytes produced and secreted the plasma proteins, fibrinogen, fibronectin, transthyretin (TTR) and AFP, an essential feature for functional HE. Additionally iPSC-derived HE supported both CYP1A2 and 3A4 metabolism, which is essential for drug and toxicology testing.
Conclusion
This work is first to demonstrate the efficient generation of hepatic endodermal lineage from human iPSC that exhibits key attributes of hepatocytes, and the potential application of iPSC-derived HE in studying human liver biology. In particular, iPSC from individuals representing highly polymorphic variants in metabolic genes and different ethnic groups will provide pharmaceutical development and toxicology studies a unique opportunity to revolutionise predictive drug toxicology assays and allow the creation of in vitro hepatic disease models.
INTRODUCTORY STATEMENT
Human iPSCs are reprogrammed mature somatic fibroblasts which represent a pluripotent cell population able to generate all primary cell types in vitro [1–3]. The ability to derive iPSCs from an indefinite range of genotypes makes them an attractive resource on which to model liver function reflecting the complexity of polygenic influences on metabolism in vitro. Another facet of iPSC technology is the ability to study the impact of gene polymorphisms in a native chromatin setting and model with precision, gene interactions. Therefore iPSC-derived models hold great potential to develop a detailed understanding of human liver disease and metabolism including drug toxicity (for a review see Dalgetty et al [4]). Any methods which might streamline and standardize the process of drug and toxicology testing, which currently relies on primary human hepatocytes, would represent a significant development. Therefore an iPSC resource representative of polymorphic variants and ethnic groups; unhindered by quality and supply, would revolutionise predictive drug toxicology assays and impact on drug attrition.
Presently primary human hepatocytes (PHHs) are the golden standard cell type used in predictive drug toxicology. Unfortunately PHHs are a scare, heterogeneous and expensive resource which function only short term in vitro. The generation of HE from iPSCs has the potential to fulfil the major challenge to acquire the reliable and clonal source of functional human hepatocyte cells for biotechnology purposes. To date efficient models of deriving hepatic endoderm from iPSCs have not been described or developed. Capitalising on our recent investigations that hESCs can be stimulated to form hepatic endoderm [5], we have developed a parallel methodology for iPSCs, here we describe the generation of functional hepatic endoderm from multiple human iPSCs lines that can potentially model human drug metabolism.
EXPERIMENTAL PROCEDURES
Generation of human iPS cells
iPS cells from diabetic North American Indian (JDM-iPS1) and female Caucasian (PGP9-iPS1) were reported previously [6–7]. 1×105 fibroblasts of normal male Caucasian (ATCC: CRL-2465) were plated in one well of a six-well plate and infected with four individual retrovirus each containing a single reprogramming factor, Oct4, Sox2, Klf4 and c-MYC, was used at a MOI of 10 [1]. After 3 days of infection, cells were split into 10cm plates pre-seeded with irradiated mouse embryonic fibroblasts (MEFs) and cultured under human ES culture medium conditions until colonies appeared. Colonies were picked, replated onto irradiated MEFs and expanded for characterization.
Cell Culture and Differentiation
iPS cell colonies were maintained in hES medium (80% KO/DMEM, 20% KO Serum Replacement, 10ng/ml bFGF, 1 mM L-glutamine, 100mM non-essential amino acids, 100uM 2-mercaptoethanol, 50U/ml penicillin, and 50mg/ml streptomycin (Invitrogen)) on an irradiated mouse embryonic feeder layer (CF-1, VHbio). Before HE differentiation, iPSCs were cultured on Matrigel™ (BD Biosciences). iPSCs were differentiated to hepatocyte like cells using Activin A and Wnt3a (R&D systems) on Matrigel™ (BD biosciences). Although the differentiation protocol was similar to that of Hay et al [5], one major modification was required in order to generate human hepatic endoderm from human iPSCs. In brief, after iPSCs were passaged onto matrigel and cultured in MEF-CM until a confluence of 50–70% was attained, MEF-CM was then replaced with RPMI/B27 and iPSCs treated with Activin A and Wnt3a for 3 days and required a further 2 day incubation in Activin A (100ng/ml) alone before hepatic endoderm was specified using established conditions as follows: Cells were cultured in SR/DMSO (KO-DMEM containing 20% serum replacement (SR), 1mM glutamine, 1% non-essential amino acids, 0.1mM β-mercaptoethanol and 1% DMSO). The final maturation Step involved culturing the cells in L-15 medium which was supplemented with 8.3% fetal calf serum, 8.3% tryptose phosphate broth, 10μM Hydrocortisone 21-hemisuccinate, 1μM insulin, 2mM glutamine and supplemented with 10ng/ml Hepatocyte growth factor and 20ng/ml Oncostatin M [5]. For further information see supplementary Figure 2.
Flow Cytometry
Cells were resuspended at 1× 107 cells/ml in FACS-PBS (PBS supplemented with 0.1% BSA and 0.1% sodium azide). Aliquots of 1 × 106 cells were incubated for 40 minutes at 4°C with optimum concentration (determined by titration), of primary antibody to SSEA-1 (mouse IgM), SSEA-4 (mouse IgG3) (DHSB, Iowa, USA), TRA-1-60 (anti-mouse IgM, Chemicon) and EpCAM-APC (CD326, Biolegend). Cells were washed twice to remove unbound antibody and resuspended in 100μl FACS-PBS. Binding of primary antibody was detected using optimum concentration (determined by titration) of appropriate isotype specific fluorochrome labeled secondary antibody or Avidin: anti-mouse IgM – PE and anti-rat IgM-PE and anti-mouse IgG3-FITC (Jackson Labs) and Streptavidin (Caltag Medsystems). After incubation for 40m at 4°C cells were washed twice and finally re-suspended in 250μl. Unstained cells and cells labeled with secondary antibody alone were included as controls. Dead and apoptotic cells and debris were excluded from analysis using an electronic `live gate on forward scatter and side scatter parameters. Data for up to 25,000 `live' events were acquired for each sample using a FACSCaliber cytometer equipped with a 488nm and 633nm lasers and analysed using CellQuest software (Becton Dickinson).
RT-PCR
RNA was isolated using RNeasy kit (Qiagen, West Sussex, U.K., http://www1.qiagen.com) following manufacturer's instruction and DNA was removed by the treatment with DNase (Qiagen). cDNA was synthesised using 2 μg total RNA with reverse transcriptase (Roche Diagnostics) in a 20–25 μl volume. Polymerase chain reaction (PCR) was carried out as previously described [1, 5]. Primer sequences and PCR conditions are provided in supplemental online data Table 1. qPCR carried out as Hay et al. [5].
Assay for Teratoma Formation
For teratoma formation, cells were harvested, washed once with DMEM/F12 and mixed with Matrigel (BD Biosciences) and collagen [8–9]. 1×106 cells were injected intramuscularly into immune-compromised NSG mice. Teratomas formed within 6–8 weeks and paraffin sections were stained with Masson Trichromatic for all histological determinations.
Immunocytochemistry
Cells were fixed with chilled Methanol (−20 °C) for 10 minutes, washed with PBS, and blocked with 10% goat serum and 0.02%–0.1% Triton X-100 for 1 hour. The cells were then incubated with primary antibody at the appropriate dilution at 4°C overnight. Secondary antibody was applied for 30 minutes after washing with PBS. The cells were finally mounted with Mowiol (Calbiochem) and then visualized and captured using a Leica DMIRB.
ELISA
ELISAs were carried out as previously described [5].
Cytochrome p450 Assay
CYP1A2 and CYP3A4 activity was assessed using the pGlo kit (Cat nos. V8771, V8901 Promega) according to manufacturer's instruction for non-lytic CYP450 activity estimation. CYP activities are expressed as relative light units (RLU/ml) per of media, normalised against percentage of hepatocyte like cells.
RESULTS
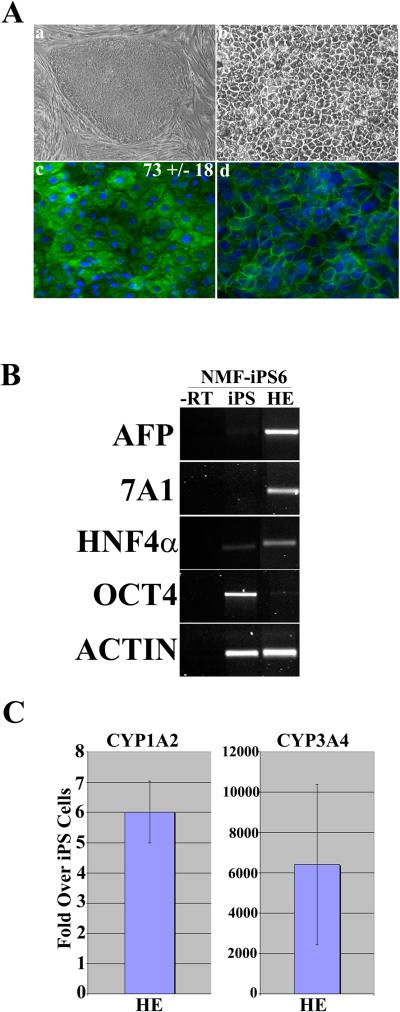
We have previously demonstrated a successfully generation of iPSCs that are capable of self renewal and differentiation by retroviral transduction of four transcription factors [1]. As iPSCs colonies appeared, they where manually disaggregated and plated onto a feeder layer and sequentially passaged (Supplementary Figure 1 and [6–7]). The derived iPSC lines were characterised using a number of stem cell criteria: cell morphology, stem cell gene expression, stem cell surface expression of SSEA 3, 4 and Tra 1–60 and absence of SSEA1 and teratoma formation in vivo (Supplementary Figure 1 and [6–7]). By applying the method we had used for differentiating, we attempted to generate hepatic endodermal lineage from human iPSCs. We initially focused our efforts on iPS cell line derived from normal adult Caucasian male, NMF-iPS6 (Figure 1Aa). NMF-iPS6 were differentiated towards hepatic endoderm via physiologically relevant conditions; treatment with Wnt3a/Activin A, Activin A followed by DMSO and a final maturation step with hepatic growth factor and Oncostatin M [5] (Figure 1Ab). Differentiation of iPS cells into hepatic endoderm was associated with a dramatic change in cellular morphology similar to hepatocyte differentiation. Hepatic phenotype was assessed by the albumin production (Figure 1Ac) and E-Cadherin (Figure 1Ad) confirmed by immunofluorescence. We observed an efficiency of hepatic endoderm generation of between 70–90%, as assessed by albumin positive cells (Figure 1Ac). HE derived from the male Caucasian iPSCs (NMF-iPS6) expressed a number of key hepatic transcripts as assessed by RT-PCR, namely AFP, HNF4. In addition, we observed the expression of the endodermal markers, Sox17 and CXCR4 [10] at day 5 in the procedure (data not shown) and CYP7A1 (Figures 1 and 2) which demonstrates both a definitive endoderm origin and importantly is not derived from yolk sac [11]. Additionally, upon differentiation, pluripotent marker OCT4 which expressed in iPS cells became down-regulated (Figure 1B). One of the immediate potential applications of iPSC-derived HE is the human drug toxicity assessment, and therefore we investigated the expression of two key adult cytochrome p450's, CYP1A2 and CYP3A4. Both enzymes were induced in HE cells compared with undifferentiated iPS cells, ~6 fold increase in CYP1A2 and ~6000 fold increase in CYP3A4 levels (Figure 1C).
Figure 1. Derivation of hepatic endoderm from iPSCs.
(Aa) Phase contrast microscropy demonstrating the typical iPSC colony morphology and (Ab) iPSC-derived hepatic endoderm (HE) following 14 days in the differentiation procedure (Magnification x20). (Ac) iPSC-derived HE stains positive for albumin at day 14 in the differentiation procedure. The numbers represent the efficiency of the procedure +/− SE. (Ad) iPSC-derived HE stains positive for E-Cadherin at day 14 in the differentiation procedure (B) RT-PCR gene expression of iPSCs and iPSC-derived hepatic endoderm. iPSC-derived hepatic endoderm on (lane 1) express the markers AFP, CYP7A1 and HNF4 alpha, but not OCT-4. iPSCs strongly express OCT4. Potentially due to spontaneous differentiation found in iPSC culture. PCR reactions were controlled using a –RT control (lane 4) and details of primers and cycles can be found in the supplementary methods section (C) CYP1A2 and CYP3A4.
Figure 2. Derivation of HE from 2 other iPSC lines.
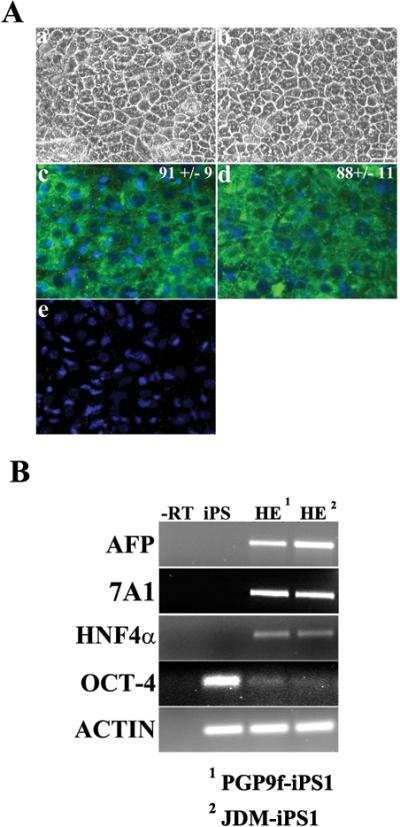
(Aa,b) Phase contrast microscropy demonstrating the typical iPSC HE morphology following 14 days in the differentiation procedure (Magnification x20). (Ac,d) IPSC-derived HE stains positive for albumin at day 14 in the differentiation procedure. The numbers represent the efficiency of the procedure +/− SE. (B) RT-PCR gene expression of iPSCs and iPSC-derived hepatic endoderm. iPSC-derived hepatic endoderm on (lane 1) express the markers AFP, and HNF4 alpha, but not OCT-4. iPSCs strongly express OCT4. Potentially due to spontaneous differentiation found in iPSC culture. PCR reactions were controlled using a –RT control (-RT) and details of primers and cycles can be found in the supplementary methods section
In addition to the male Caucasian NMF-iPS cell line, we also applied HE differentiation protocol to iPS cells derived from diabetic North American Indian (JDM-iPS1) and female Caucasian (PGP9f-iPS1) (Figure 2Aab). Both iPS cell lines differentiated into HE with similar efficiencies as male Caucasian NMF-iPS6 cell line. HE differentiation was assessed by cell morphology and albumin staining (Figure 2A cde). When we analysed hepatic gene expression in the iPSC-derived HE, both lines exhibited similar gene expression patterns as observed from NMF-iPS6 cells, indicating hepatic identity (Figure 2B).
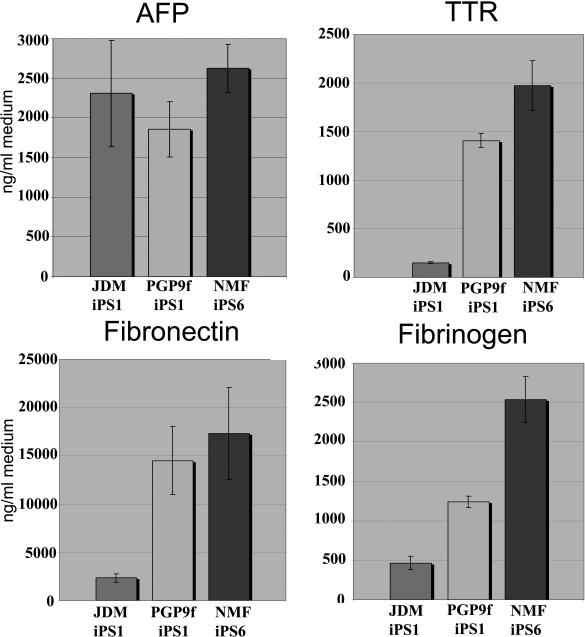
Although iPSC-derived HE expressed a number of liver genes, we were also keen to assess their liver specific function in culture. An important functional marker for HE is the production and export of serum proteins. We assessed iPSC-HE production of these key serum proteins and measured their levels by ELISA. In all lines tested, we detected substantial amounts of AFP, TTR, Fibronectin and Fibrinogen at levels equivalent to those reported for HE derived from hESCs [5].
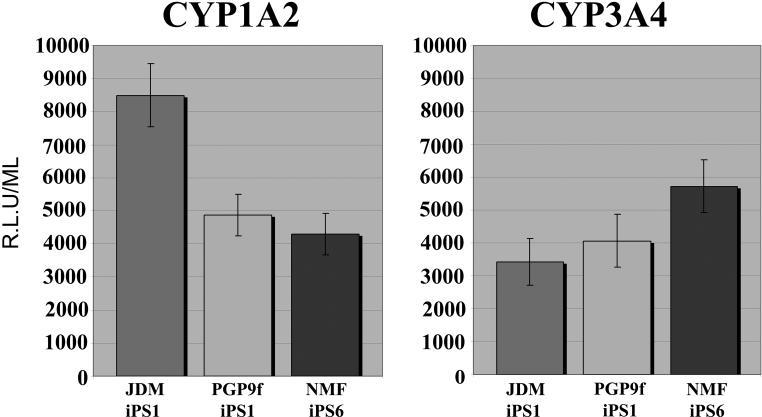
In order to further functionally validate, iPSC-derived HE were assessed for their metabolic ability. The cytochrome P450 enzymes are critical in drug metabolism, and CYP1A2 and CYP3A4 are the key enzymes. The function of these P450 components were examined and importantly all lines exhibited CYP1A2 and CYP3A4 activity as assessed by the generation of a luminescent metabolite (Figure 4). CYP1A2 metabolism was similar between lines PGP9f-iPS1 and NMF-iPS6 but was higher in line JDM-iPS1, while we observed only slight variation with CYP3A4 metabolism in all 3 lines tested.
Figure 4. iPSC-derived hepatic endoderm displays cytochrome P450 metabolism.
iPSC-derived hepatic endoderm were incubated with hepatocyte culture medium supplemented with 50 μM of CYP3A4 or CYP1A2 pGlo TM substrates (Promega) as per manufacturers instructions. 4 hours post-treatment 50 μl of culture medium was removed and read on a luminometer (POLARstar optima). CYP1A2 and CYP3A4 activity is expressed as relative light units (R.L.U.)/ml of tissue culture medium. (n=6). CYP1A2 activity was similar on cells maintained on both ECMs. Luciferase activity is expressed as relative light units (R.L.U.)/ml of tissue culture media. JDM-iPS1 is presented by the first bar in all graphs, PGP9f-iPS1 is represented by the middle bar in each graph and NMF-iPS6 is represented by the last bar in each graph.
DISCUSSION
Here we demonstrate for the first time the derivation of HE from human iPSC of both sexes and two ethnicities. The iPSC-derived HE was functionally equivalent to hESC-derived HE and interestingly all iPSC lines tested so far showed higher efficientcy to form functional HE. The generic ability of iPSCs to form HE in response to our model [5] has not been observed with hESC in deriving efficient levels of HE. Therefore one could speculate that this is due to the consistent manner in which the iPSCs were reprogrammed may play an important role in their developmental potential. It also suggests that iPSC may prove a more valuable and uniform starting material for derivation of HE, than from human embryonic stem cells which show dramatic line to line variability in susceptibility to individual lineage differentiation.
Such a resource has the ability to revolutionise the manner in which we define drug metabolism, model liver disease and human liver development. Because iPSC-derived HE can be differentiated ex-vivo, the unlimited supply of ethically and genetically diverse HE models can be obtained. This will become a powerful resource allowing the study of ethnic/polymorphic variation on xenobiotic metabolism involving poor metabolisers (e.g. CYP2C9/Warfarin) and disease genotypes (e.g. alpha 1-antitrypsin). In addition, the ability to model liver development in vitro will allow the development of novel biomarkers for both disease and the identification of stage specific makers during the differentiation process [12].
Identification and reprogramming of human fibroblasts displaying metabolically different features for key polymorphisms, an iPSC library could be developed. Presently the ability to model the human liver and disease using hESCs or PHHs is limited by the number of stem cell lines available and the ability to produce functional HE from individual ESC lines. Therefore, by applying iPSC HE technology we predict these will by-pass the issues associated with hESCs and PHHs.
In conclusion: Our studies provide a proof of concept that multiple iPSCs lines can be efficiently differentiated to functioning HE. In addition provides a novel approach that overcomes the current limitations associated with PHHs and hESCs. We predict that this technology will be applicable to iPSC lines derived from healthy and diseased patients from different ethnic backgrounds allowing the creation of a library. The development of such a resource is essential in the identification and testing of new medicines and the modelling of disease.
Supplementary Material
iPSCs were generated using established conditions [1] and characterised for A. stem cell gene expression by PCR (Nanog., Sox2 and hTert expression), lane 1 NMF-iPS6, lane 2 RT negative control and lane 3 parental line. B. stem cell surface marker expression (SSEA4, Tra-160 and EpCAM) and absence of the murine stem cell marker SSEA1. C. stem cell pluripotency was assessed by teratoma formation in nude mice. Note lines JDM-iPS1 and PGP9f-iPS1 have been fully characterised see [6–7].
iPSCs were differentiated to HE using a 4 step differentiation protocol. A. Schematic of the differentiation procedure. Abbreviations: iPS induced pluripotent stem cell; bFGF, fibroblast growth factor; FCS, fetal calf serum; MEF-CM, irradiated mouse embryonic fibroblast conditioned media; HGF, hepatocyte growth factor; KOSR, KnockOut Serum Replacement; OSM, oncostatin M. B. Morphological changes in iPSCs are they are differentiated to hepatic endoderm.
Figure 3. iPSC-derived hepatic endoderm produces hepatic specific serum proteins.
iPSC-derived hepatic endoderm was maintained in 1ml of hepatocyte culture medium for 24 hours. The following morning culture supernatants were harvested and serum protein production measured by ELISA and quoted as ng/ml of tissue culture medium per 24 hours. Note background has been removed from the presented data. Each line is represented as a different coloured bar: JDM-iPS1 is presented by the first bar in all graphs, PGP9ff-iPS1 is repsresented by the middle bar in each graph and NMF-iPS6 is represented by the last bar in each graph. In all lines the serum protein tested was detected. fibrinogen and transthyretin (TTR), fibronectin and alpha-fetoprotein (AFP) (n=6).
ACKNOWLEDGEMENTS
We would like to thank Dr.Val Wilson for the analysis of the teratoma data. Antibodies used for Flow Cytometry were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. Ian Wilmut was supported by Scottish Funding Council. David Hay was supported by a RCUK Fellowship and John Iredale is supported by a MRC programme grant.
REFERENCES
- 1.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2007;318:1917–1920. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalgetty DM, Medine CN, Iredale JP, Hay DC. Progress and fuure challenges in stem cell-derived liver technologies. Am. J. Physiol. Gastrointest Liver Physiol. 2009;297 doi: 10.1152/ajpgi.00138.2009. doi:10.1152/ajpgi.00138.2009. [DOI] [PubMed] [Google Scholar]
- 5.Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc. Nat. Acad. Sci. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, et al. Targeted and genomescale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prokhorova TA, Harkness LM, Frandsen U, Ditzel N, Burns JS, Schroeder HD, et al. Teratoma Formation by Human Embryonic Stem Cells is site-dependent and enhanced by the presence of Matrigel. Stem Cells & Dev. 2007 doi: 10.1089/scd.2007.0266. 10.1089/scd.2007.0266. [DOI] [PubMed] [Google Scholar]
- 9.Kleinman HK, McGarvey ML, Liotta LA, Robey PG, Tryggvason K, Martin GR. Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochem. 1982;21:6188. doi: 10.1021/bi00267a025. [DOI] [PubMed] [Google Scholar]
- 10.Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, et al. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 11.Asahina K, Fujimori H, Shimizu-Saito K, Kumashiro Y, Okamura K, Tanaka Y, et al. Expression of the liver-specific gene Cyp7a1 reveals hepatic differentiation in embryoid bodies derived from mouse embryonic stem cells. Genes Cells. 2004;9:1297–1308. doi: 10.1111/j.1365-2443.2004.00809.x. [DOI] [PubMed] [Google Scholar]
- 12.Gadue P, Gouon-Evans V, Cheng X, Wandzioch E, Zaret KS, Grompe M, et al. The Generation of Monoclonal Antibodies Specific for Cell Surface Molecules Expressed on Early Mouse Endoderm. Stem Cells. 2009 doi: 10.1002/stem.147. EPub. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
iPSCs were generated using established conditions [1] and characterised for A. stem cell gene expression by PCR (Nanog., Sox2 and hTert expression), lane 1 NMF-iPS6, lane 2 RT negative control and lane 3 parental line. B. stem cell surface marker expression (SSEA4, Tra-160 and EpCAM) and absence of the murine stem cell marker SSEA1. C. stem cell pluripotency was assessed by teratoma formation in nude mice. Note lines JDM-iPS1 and PGP9f-iPS1 have been fully characterised see [6–7].
iPSCs were differentiated to HE using a 4 step differentiation protocol. A. Schematic of the differentiation procedure. Abbreviations: iPS induced pluripotent stem cell; bFGF, fibroblast growth factor; FCS, fetal calf serum; MEF-CM, irradiated mouse embryonic fibroblast conditioned media; HGF, hepatocyte growth factor; KOSR, KnockOut Serum Replacement; OSM, oncostatin M. B. Morphological changes in iPSCs are they are differentiated to hepatic endoderm.