Abstract
Background
The predominant sterol in the membranes of the alga Chlamydomonas reinhardtii is ergosterol, which is commonly found in the membranes of fungi, but is rarely found in higher plants. Higher plants and fungi synthesize sterols by different pathways, with plants producing cycloartenol as a precursor to end-product sterols, while non-photosynthesizing organisms like yeast and humans produce lanosterol as a precursor. Analysis of the C. reinhardtii genome sequence reveals that this algae is also likely to synthesize sterols using a pathway resembling the higher plant pathway, indicating that its sterols are synthesized somewhat differently than in fungi. The work presented here seeks to establish experimental evidence to support the annotated molecular function of one of the sterol biosynthetic genes in the Chlamydomonas genome.
Methodology/Principal Findings
A gene with homology to the yeast sterol C-5 desaturase, ERG3, is present in the Chlamydomonas genome. To test whether the ERG3 ortholog of C. reinhardtii encodes a sterol C-5 desaturase, Saccharomyces cerevisiae ERG3 knockout strains were created and complemented with a plasmid expressing the Chlamydomonas ERG3. Expression of C. reinhardtii ERG3 cDNA in erg3 null yeast was able to restore ergosterol biosynthesis and reverse phenotypes associated with lack of ERG3 function.
Conclusions/Significance
Complementation of the yeast erg3 null phenotypes strongly suggests that the gene annotated as ERG3 in C. reinhardtii functions as a sterol C-5 desaturase.
Introduction
Sterols are isoprenoid-derived molecules found in the membranes of eukaryotic organisms and have been shown to play an important role in membrane fluidity and permeability [1]–[4]. While sterols are essential components of eukaryotic membranes, the specific sterols found in different organisms vary [5]. Cholesterol is the predominant sterol in vertebrates and ergosterol is the most common sterol found in fungi. Plants have a variety of sterols including sitosterol, 24-methyl cholesterol and stigmasterol [6].
Two major pathways for sterol biosynthesis have been found in eukaryotes. Fungi and vertebrates synthesize sterols with lanosterol as an intermediate (Fig. 1), while plants synthesize sterols using cycloartenol as an intermediate [5]. In these pathways the biosynthetic steps from isopentanyl PP to squalene epoxide are the same. However, the cyclization of squalene epoxide is where the two pathways diverge producing either cycloartenol or lanosterol [7]. Nes et al., have identified cycloartenol in fungal-like organisms Prototheca wickerhamii and Dictyostelium discoidium that have been presumed to be descendents of algae after the evolutionary loss of the chloroplast [8]. It is now scientifically accepted that red algae, green algae and diatoms make cycloartenol while dinoflagellates have been reported to make lanosterol [9], [10].
Figure 1. Schematic diagram of the putative pathway of ergosterol biosynthesis in S. cerevisiae and C. reinhardtii.
In the green alga C. reinhardtii, the predominant sterols are ergosterol and 7-dehydroporiferasterol [11]. These two sterols are commonly found in fungi, but not so often with higher plants. However, bioinformatics evidence supports the idea that C. reinhardtii uses the cycloartenol pathway, as genes coding for orthologs of cycloartenol cyclase and cyclopropyl isomerase, two key enzymes in the cycloartenol pathway, are found in the C. reinhardtii genome [12] (Brumfield and Moroney, unpublished results). So while C. reinhardtii synthesizes ergosterol, a sterol normally associated with the fungal biosynthetic pathway, it appears to use a pathway that more closely resembles that of higher plants. Earlier studies of ergosterol deficient mutants in C. reinhardtii provide evidence for the final few steps of ergosterol biosynthesis in this alga [13], [14]. However, the steps from squalene epoxide to ergosta- 5,7,24 (28)-trienol have not been determined. A goal of this work is to further elucidate the sterol biosynthetic pathway in C. reinhardtii.
With the publication of the Chlamydomonas genome, genes likely to be associated with sterol biosynthesis in C. reinhardtii can be identified [12]. While C. reinhardtii offers many advantages as an experimental organism, it is still difficult to obtain targeted knock-out mutants of a desired gene. In addition, the fact that mutants earlier in the pathway have not been identified implies that a complete loss of sterol might be lethal to the alga under normal growth conditions. An alternative approach is to use Sacchromyces cerevisiae to study the function of genes involved in sterol biosynthesis in C. reinhardtii. The post-squalene biosynthesis pathway of ergosterol in yeast has been well defined [15], [16]. Enzymes of the ergosterol biosynthetic pathway have been found to be major targets for drug interactions, and a number of antifungal drugs on the market were derived specifically to target ergosterol biosynthesis [17]–[19]. Much of what is known about the sterol pathway in Arabidopsis has been worked out by complementing yeast strains defective in specific steps of sterol biosynthesis with the Arabidopsis ortholog [20]. We have previously used this approach to study phospholipid biosynthesis in C. reinhardtii [21], [22].
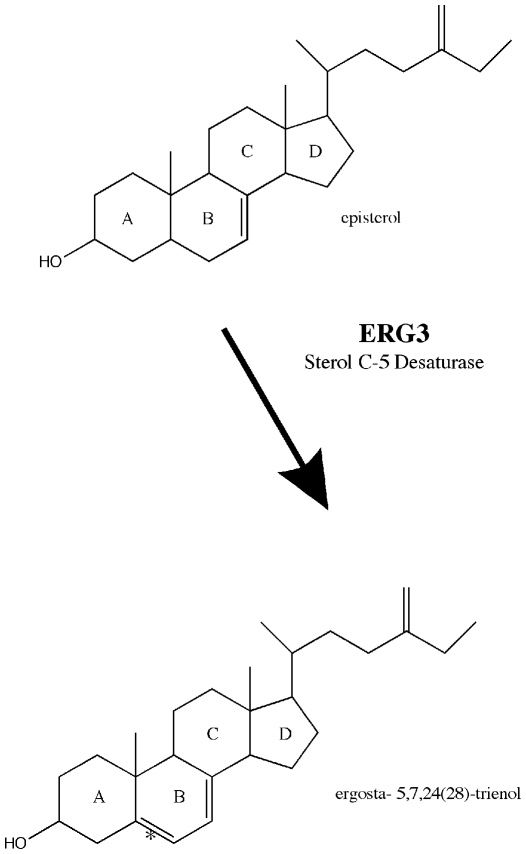
One gene essential to ergosterol biosynthesis is ERG3, which encodes the C-5 sterol desaturase responsible for introducing a double bond at C-5 in the B ring of episterol [23] (Fig. 2). This enzyme is sensitive to cyanide and requires iron as well as molecular oxygen for its activity [15]. ERG3 in yeast is also associated with NAD(P)H-cytochrome b/cytochrome b 5 [23]. It has been previously demonstrated that ERG3 is required for the breakdown of respiratory substrates when cells are heme deficient [24]. Deletion of ERG3 in S. cerevisiae produces haploid cells that overaccumulate the ergosterol precursor, episterol [25]. Episterol and ergosta-7, 22-dien- 3-beta-ol are reported as possible substrates for the C-5 sterol desaturase [26]. Cloning and sequencing of ERG3 has been reported in several model systems including Arabidopsis thaliana [27] and Homo sapiens [28]. In yeast, ERG3 is a non-essential gene except under environments of heme-deficiency [24]. It has been suggested that Erg3 protein is a critical target in ergosterol biosynthesis [29], and when other ergosterol biosynthetic enzymes are mutated, ERG3 expression is directly affected and regulated by these mutations [15].
Figure 2. Schematic diagram of the reaction catalyzed by Erg3p in yeast.
Erg3p is responsible for introducing a double bond at the C-5 carbon (denoted by the star) of the B-ring of episterol to produce ergosta- 5,7,24(28)- trienol. This step is the second to last step in the biosynthetic pathway to ergosterol.
In this study, we begin to characterize the function of ERG3 in C. reinhardtii by complementation in S. cerevisiae ergosterol mutants. Questions remain as to whether algae produce sterols for other purposes, such as hormonal regulators [30], or if they function strictly for membrane integrity. Understanding the dynamics of this pathway may provide further evidence for hormonal regulation in C. reinhardtii, and the important cellular functions that may arise from the production of these sterols and their biosynthetic precursors.
Results
Sequence Analysis of the ERG3 Gene
A comparison of the Saccharomyces cerevisiae ERG3 to the Chlamydomonas genome revealed several genes with significant protein sequence homology. The gene with the highest homology had a 21% sequence identity to the ERG3 sterol desaturase of yeast. This Chlamydomonas gene also had a high similarity to ERG3 genes annotated in higher plants. The gene with the next highest homology had a sequence identity of only 16%, and appears to be more closely related to the sequence of the yeast ERG25 methyl sterol oxidase. Based on both the higher similarity of the 21% homologous gene to yeast and plant ERG3, and the higher similarity of the 16% homologous gene to ERG25, we chose the former gene for further analysis, and tentatively annotated it as Chlamydomonas ERG3 (Accession No. XP_001701457).
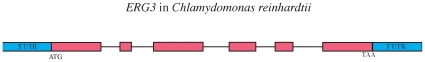
In yeast, ERG3 is a C-5 sterol desaturase that adds a double bond in the ring structure of episterol to produce in the ergosterol biosynthetic pathway [31] (Fig. 2). In the proposed pathway for ergosterol biosynthesis in C. reinhardtii (Fig. 1), ERG3 would catalyze the conversion of episterol to ergosta- 5,7,24 (28)-trienol. ERG3 in C. reinhardtii is located on chromosome 16 of the genome [12], and the predicted amino acid sequence of ERG3 aligns well with other C-5 sterol desaturases (Table 1 and Fig. 3). The ERG3 gene in C. reinhardtii encodes a protein of 351 amino acids [12]. The cDNA is approximately 1.1kb long and is composed of six exons according to Version 4.0 of the Chlamydomonas Genome [12] (Fig 4). Like other known Erg3 proteins, the homologous gene in C. reinhardtii codes for four putative histidine metal binding domains [28] (Fig. 3). A genome database (http://genomeportal.jgi-psf.org/Chlre4/Chlre4.home.html) predicts that Erg3 protein has 3 transmembrane helices. The first helix is thought to be made up of amino acids 10–32, while the second and third helices correspond to amino acids 110–132 and 191–213 [12], [32], respectively (Fig. 3).
Table 1. Alignment scores among sterol C-5 desaturases of various organisms generated from ClustalW.
| AtERG3 | ZmERG3 | CrERG3 | HsERG3 | RnERG3 | ScERG3 | |
| AtERG3 | - | 65 | 46 | 27 | 27 | 24 |
| ZmERG3 | 65 | - | 48 | 24 | 25 | 24 |
| CrERG3 | 46 | 48 | - | 19 | 19 | 21 |
| HsERG3 | 27 | 24 | 19 | - | 82 | 43 |
| RnERG3 | 27 | 25 | 19 | 82 | - | 44 |
| ScERG3 | 24 | 24 | 21 | 43 | 44 | - |
Arabidopsis thaliana (AtERG3); Zea mays (ZmERG3); Chlamydomonas reinhardtii (CrERG3); Homo sapiens (HsERG3); Rattus norvegicus (RnERG3); Saccharomyces cerevisiae (ScERG3).
Figure 3. Amino acid sequence alignment of Sterol C-5 desaturase in different organisms.
Arabidopsis thaliana (AtERG3) NCBI Accession Number CAA62079; Zea mays (ZmERG3) ACG38774; Chlamydomonas reinhardtii (CrERG3) XP_001701457; Homo sapiens (HsERG3) BAA33729; Rattus norvegicus (RnERG3) NP_446094; Sacchromyces cerevisiae (ScERG3) NP_013157. Conserved amino acid sequences shared by all organisms are denoted by a star. The highlighted area corresponds to the putative histidine-containing metal binding domain. Dashed lines indicate gaps in the alignment. The bold, italicized font corresponds to the three putative transmembrane spanning regions of the C. reinhardtii ERG3 protein.
Figure 4. Schematic diagram of sterol C-5 desaturase in C. reinhardtii as annotated in the JGI Chlamydomonas Genome Version 4.0.
ERG3 has six exons and five introns.
Deletion of ERG3 by Homologous Recombination
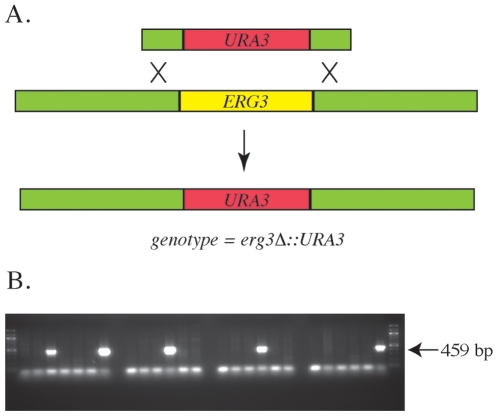
While ERG3 from Chlamydomonas aligned well with putative plant ERG3 genes (Fig. 3), its alignment with the characterized ERG3 protein of yeast showed a lesser degree of identity and similarity. To provide evidence that Chlamydomonas ERG3 functions as a C-5 desaturase, we designed experiments to complement a yeast erg3Δ mutant. To generate the null mutation in yeast, the ERG3 gene was knocked out by homologous recombination and replaced with the URA3 marker in a diploid yeast strain (Fig. 5A). PCR analysis was used to identify positive yeast recombinants that possessed the ERG3 mutation, and these isolates were sporulated to obtain haploid erg3Δ strains (Fig. 5B). Yeast lacking a functional ERG3 are viable, as the gene is non-essential under aerobic conditions [33]. In addition, yeast lacking ERG3 have an increased sensitivity to cycloheximide as previously described [34]. Haploid isolates obtained from yeast ERG3 knockouts demonstrated increased sensitivity to cycloheximide when grown under optimal conditions in rich media.
Figure 5. Deletion of the ERG3 gene in yeast.
(A) Schematic diagram of homologous recombination strategy used to delete ERG3 in Sacchromyces cerevisiae. PCR primers were designed to amplify the URA3 marker gene containing the flanking regions of ERG3. (B) Ura+ isolates were screened by PCR to verify proper integration of URA3 and deletion of ERG3. Only correctly integrated isolates would show a 459 base pair product using primers internal to URA3 and upstream of the flanking homology. Stars denote the positive isolates selected for further analysis. PCR was also used to verify the downstream end of flanking region homology with URA3 (data not shown).
Complementation of ERG3 Mutants
To test the function of the C. reinhardtii ERG3 gene, complementation experiments using yeast haploid strains deficient in ERG3 were conducted. Yeast erg3Δ strains display a marked hypersensitivity to low non-lethal levels of cycloheximide [34]. In these experiments, plasmids expressing the yeast ERG3 ORF, C. reinhardtii ERG3 ORF and the empty vector were transformed into the erg3Δ knockout strains (Fig. 6A and Table 2). The selectable marker LEU2 was used to select for transformants on minimal media lacking leucine (Fig. 6A), and Leu+ isolates were then screened for resistance to low levels of cycloheximide (Fig. 6B). Mutant erg3Δ strains transformed with the pDD1193, the empty vector, remained hypersensitive to cycloheximide, as they displayed very little growth on plates containing the drug (Fig. 6B). Transformants containing plasmid pDD1192 expressing the yeast ERG3 ORF were completely resistant to low levels of cycloheximide (Fig. 6B). Transforming the mutants with pDD1191, expressing the C. reinhardtii ERG3 ORF, showed enhanced resistance to the drug, demonstrating that the algal gene could complement the loss of the yeast ERG3 (Fig. 6B).
Figure 6. Phenotypic complementation of yeast erg3Δ strains.
Known phenotypes of yeast erg3 null mutants (hypersensitivity to cycloheximide and inability to grow on acetate) were complemented by plasmids expressing either S. cerevisiae ERG3 or C. reinhardtii ERG3 cDNA. A) S. cerevisiae ADH1 promoter plasmid used to express ERG3 genes. Haploid yeast carrying the erg3Δ::URA3 allele were transformed with the designated plasmids and plated on dextrose minimal media lacking leucine (YMD minus Leu) to select for transformants. Single colony isolates were then re-streaked on minimal media lacking leucine and containing cycloheximide (0.13 µg/mL) (B) minimal media lacking leucine and containing acetate as sole carbon and energy source (C), and on YMD lacking as overall growth control (D). Yeast mutant for ERG3 and containing the vector plasmid lacking an insert cannot grow on cycloheximide or acetate, while cells containing the plasmid expressing either yeast or C. reinhardtii ERG3 are able to grow. Each pair of plates represents erg3Δ yeast sporulated from two independently isolated diploid knockout isolates, 1+2 are DDY4259 and DDY4260 transformed with the vector only, 3+4 are DDY4261 and DDY4262, the same strains transformed with ADH1-yeast ERG3, and 5+6 are DDY4263 and DDY4264 with ADH1-C.reinhardtii ERG3. For the second panel, 7+8 are DDY4253 and DDY4254 transformed with the vector only, 9+10 are DDY4255 and DDY4256 transformed with ADH1-yeast ERG3, and 11+12 are DDY4257 and DDY4258 with ADH1-C.reinhardtii ERG3.
Table 2. Yeast strains used in this work.
| Strains | Genotype | Source |
| DDY 4259 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD1193 | This study |
| DDY 4260 | MAT a ADE2 his3 leu2 lys2Δ trp1 ura3 erg3Δ::URA3 pDD1193 | This study |
| DDY 4253 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD1193 | This study |
| DDY 4254 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD1193 | This study |
| DDY 4261 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD 1192 | This study |
| DDY 4262 | MAT a ADE2 his3 leu2 lys2Δ trp1 ura3 erg3Δ::URA3 pDD1192 | This study |
| DDY 4255 | MAT a ade2 his3 leu2 lys2Δ trp1 ura3 erg3Δ::URA3 pDD1192 | This study |
| DDY 4256 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD1192 | This study |
| DDY 4263 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD1191 | This study |
| DDY 4264 | MAT a ADE2 his3 leu2 lys2Δ trp1 ura3 erg3Δ::URA3 pDD1191 | This study |
| DDY 4257 | MAT a ade2 his3 leu2 lys2Δ trp1 ura3 erg3Δ::URA3 pDD1191 | This study |
| DDY 4258 | MAT ADE2 his3 leu2 LYS2 trp1 ura3 erg3Δ::URA3 pDD1191 | This study |
Previous work had also reported that yeast ERG3 mutants are viable but cannot grow on non-fermentable carbon sources [24], [33], [35]. We used this phenotype as a second test of the ability of the Chlamydomonas ERG3 to complement yeast erg3Δ strains. Consistent with the earlier observations, transformants expressing C. reinhardtii or yeast ERG3 ORFs are able to grow on acetate as a sole carbon source, while the mutants transformed with the empty vector control cannot grow (Fig. 6C). All strains (Table 3) showed normal growth on minimal dextrose media minus leucine (Fig. 6D).
Table 3. Plasmids used in this study.
| Plasmid | Description | Source |
| pDD 1191 | 1.1 kb Hind III fragment containing ERG3 open reading frame from C. reinhardtii inserted into pDD 1193 | This study |
| pDD 1192 | 1.1 kb Hind III fragment containing ERG3 open reading frame from S. cerevisiae inserted into pDD 1193 | This study |
| pDD 1193 | LEU2 marked S. cerevisiae ADH1 promoter vector. | This study |
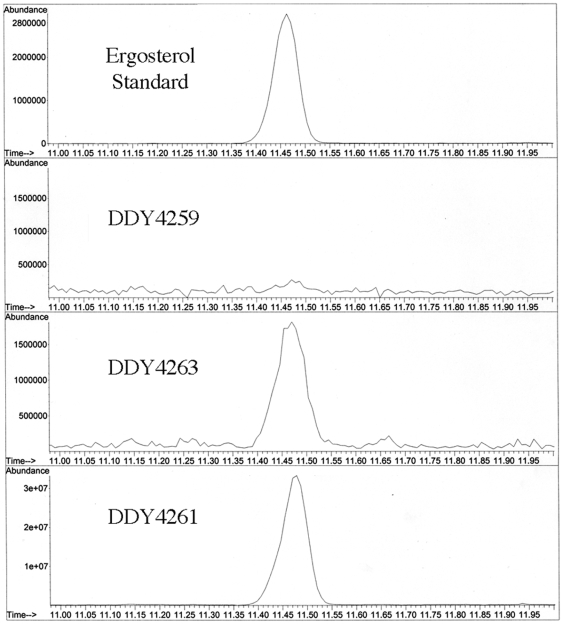
To further characterize the degree of complementation, qualitative GC/MS analysis was performed to verify the presence of ergosterol in the complemented yeast transformants. Cultures of erg3Δ yeast expressing S. cerevisiae ERG3 or C. reinhardtii ERG3 were grown to similar cell densities, collected, and lipids were extracted and derivatized for analyses. Selected ion monitoring for ion 363 of ergosterol was used to search the total ion chromatograph for ergosterol in the samples (Fig. 7). When analyzing the total ion chromatograph data for specific ions of ergosterol, the knockout strain with the vector control was found to have a marked reduction in the level of ergosterol in the membranes compared to the strains complemented with the ERG3 ORF from C. reinhardtii and S. cerevisiae (Fig. 7). Gas chromatography data contained peaks with retention times for the ergosterol standard, DDY 4263, and DDY 4261 respectively at 11.46, 11.46, and 11.47 minutes. The mass ions coinciding with these specific retention times gives spectral data with ions 468, 363, 337, and 253 m/z as previously described as ergosterol mass ions by Griffiths et al [36]. These experiments demonstrate that expression of ERG3 from C. reinhardtii can restore ergosterol biosynthesis in erg3Δ yeast.
Figure 7. GC/MS data.
Total ion chromatographs were analyzed to look specifically for the 100% ion of ergosterol. Mass ion 363 gives the best indication of the presence or absence of ergosterol in the samples. (A) Ergosterol standard, (B) Lipid extract from DDY4259 vector control, (C) DDY4263 expressing C.reinhardtii ERG3, (D) DDY4261 expressing S. cerevisiae ERG3.
Discussion
Ergosterol is the major sterol found in membranes of C. reinhardtii. However, the specific genes involved in sterol biosynthesis in Chlamydomonas have not yet been fully characterized. Several sterol mutant strains were previously identified in C. reinhardtii by selecting for resistance to the polyene antibiotic, nystatin [13], [37]. Polyene antibiotics function by forming complexes with the sterols in the membrane and decreasing membrane selective permeability, thereby causing cell death [38]. This method of selection was used to identify ergosterol mutants and allow researchers to delineate the final steps of ergosterol biosynthesis in C. reinhardtii, namely from ergosta- 7, 24(28)- dienol or 5- dehydroepisterol to ergosterol (Fig. 1). However, until recently very little was known about the steps from squalene epoxide to ergosta- 7, 24(28)-dienol. With the publication of the Chlamydomonas genome [12], we are now able to identify genes that may play a role in sterol biosynthesis in C. reinhardtii. Analysis of the genome indicates that the C. reinhardtii sterol biosynthetic pathway from squalene epoxide to episterol most closely follows the higher plant pathway. This means that C. reinhardtii is synthesizing ergosterol differently than yeast although many of the individual steps and enzymes are homologous. Yeast genetics allows not only the study of the function of yeast genes, but is also useful for cross-species complementation studies, and unlike C. reinhardtii, several ergosterol mutants have been identified in yeast, and their phenotypes have been well documented [16], [39]. We chose ERG3 for further study, as specific phenotypes previously characterized in yeast erg3 mutants are readily complemented.
ERG3 encodes a sterol C-5 desaturase, and belongs to the fatty acid hydroxylase superfamily. This superfamily of proteins includes C-5 sterol desaturases as well as C-4 sterol methyl oxidases [32], [40], [41]. This family of proteins possesses four putative iron-binding domains [28]. In yeast, loss of ERG3 function leads to an apparent loss of ergosterol in the membrane and an increase in the closely related precursor, episterol. The replacement of ergosterol with episterol in yeast leads to increased sensitivity to cycloheximide and an inability to grow with acetate as a sole carbon source [24]. The efficient gene replacement techniques used in Sacchromyces cerevisiae, has allowed for the creation of mutant strains in which the ERG3 gene has been replaced with URA3 [42], [43], providing an experimental tool for complementation studies.
Complementation of yeast knockout strains has been used successfully to identify genes involved in sterol biosynthesis in Arabidopsis [20]. The yeast Erg25 (sterol 4α-methyloxidase) [44], Erg24 (sterol C-14 reductase) [45], Erg2 (Δ8-Δ7 sterol isomerase) [46] and Erg3 [47] mutants have all been complemented by the corresponding Arabidopsis genes, even though the homology between the yeast and Arabidopsis proteins was fairly low and the natural substrate in Arabidopsis differs somewhat from that of the yeast pathway. These previous results with Arabidopsis suggested that Chlamydomonas genes encoding homologous proteins might also be identified by complementation in yeast. In the work described here we were able to confirm the annotation of the C. reinhardtii ERG3 ortholog as a C-5 sterol desaturase by both phenotypic complementation and direct biochemical analysis.
While the C. reinhardtii ERG3 ORF does function in yeast, it must be noted that it does not complement as fully as the yeast ERG3 gene. There are several explanations for this. Traditionally, 2 micron plasmids have been used in yeast for heterologous complementation, in order to increase the copy number of the plasmid and possibly circumvent issues regarding variations in protein stability, codon usage, and enzyme activity across species [48]. This study uses a moderate copy number plasmid. Secondly, yeast and C. reinhardtii undergo two very different biochemical pathways for the production of sterols. Yeast use the traditional MVA (IPP) pathway for the synthesis of sterols, while C. reinhardtii utilizes the DOXP pathway for the synthesis of sterols [49]. Evolutionary differences across species allow for several variations in the ergosterol biosynthetic pathway. For example, C. reinhardtii and S. cerevisiae produce precursors to ergosterol by two independent pathways so therefore similar enzymes can often catalyze somewhat different reactions [27], [49]. Finally, ergosterol biosynthesis is a very metabolically taxing process that requires molecular oxygen, sources of energy, and optimal temperatures [50]. Despite these potential complications, complementing the yeast ERG3 null mutation with the ERG3 gene from C. reinhardtii results in the production of ergosterol, and increased survival of the cells during exposure to cycloheximide and growth on acetate.
Sterol biosynthesis is very intricate in nature, and while several ERG3 orthologs from different eukaryotic species have been identified and characterized, ERG3 mutants in Chlamydomonas have not been created. Previous studies using random mutagenesis in Chlamydomonas identified mutants in the last two steps of ergosterol biosynthesis but not in the earlier steps of sterol biosynthesis, which includes ERG3. Presently, there are no consistently reliable methods to knock-out specific genes in Chlamydomonas although new methods are being developed. While bioinformatics as well as functional complementation gives insight into the function of a gene and its corresponding proteins, knockout experiments in the future will allow for a more complete characterization of the gene, as a targeted mutation of the gene we have annotated as ERG3 would allow confirmation of the C-5 sterol desaturase activity observed by complementation in yeast.
Elucidation of the ergosterol biosynthetic pathway in C. reinhardtii is of considerable interest and importance, as plant and algal sterols are used as dietary supplements, and as components of cosmetics and pharmaceuticals [51]. For example, experimental evidence has shown that uptake of plant sterols can help to reduce cholesterol levels in humans [52], [53]. The similarity of the C. reinhardtii ERG3 gene with that of higher plants indicates that the Chlamydomonas system may provide an excellent model system in which to study plant sterol biosynthesis. Better understanding of this pathway will lead to the development of genetically modified strains in Chlamydomonas that may allow for overproduction of specific ergosterol precursors.
Materials and Methods
Yeast Competent Cell Preparation and Transformation
For complementation experiments, yeast were transformed with plasmids expressing ERG3 from Chlamydomonas and ERG3 from yeast using the protocol of Geitz et al [54]. For the construction of erg3Δ strains, single yeast colonies were inoculated and grown overnight. In the morning, cultures were diluted to an optical density of 0.2 (A600), and cells were grown until reaching an approximate optical density of 0.7. Cells were harvested by centrifugation (2000×g for 5 minutes) at room temperature, and the cell pellet was resuspended in 1 mL of 1xTEL (10 mM Tris-HCl pH 7.5, 1 mM EDTA pH 8.0, 100 mM LiAc) per 10 mL culture volume and rocked overnight at room temperature.
Yeast Strains and Growth
Yeast strains were constructed by homologous recombination using primers 5′- TGCATTTGTAAAAAAAGATAAAAGAAAAATATTCGTCTAGATTTGAGA TGCAG ATTGTACTGAGAGTGC- 3′ (forward) and 5′- TCTTGAACGTGAAAGAAAGAA AAAAGATGAGACAAACAAGGCAACCGTA TCTCCTTACGCATCTGTGCGG-3′ (reverse) to amplify the yeast URA3 gene flanked by ERG3 sequence. This PCR product was transformed into yeast as described above, Ura+ colonies were isolated, and erg3Δ isolates verified by PCR analysis. Deletion of ERG3 was done in DDY2, a diploid version of S. cerevisiae W303), which was then sporulated to haploid (see below). Yeast were cultured in YPD (1% yeast extract, 2% peptone, 2% dextrose) or YMD (synthetic minimal medium, 2% dextrose, U. S. Biologicals catalog # Y2025) supplemented with required amino acids and nucleotides [55]. Unless otherwise stated, all yeast operations were carried out according to standard procedures [56]. To construct an ADH1 promoter vector, the plasmid pOAD (gift from Dr. Stan Fields, University of Washington) was modified as follows. The GAL4 coding sequence was removed by digestion with Hind III, and re-ligated to produce the ADH1 promoter control vector pDD1193, and ERG3 genes were coned into the resulting unique Hind III site. Expression of S. cerevisiae and C. reinhardtii ERG3 cDNA from the ADH1 promoter of yeast expression vector pDD1193 was carried out by culture of the transformed yeast strain in minimal media lacking leucine.
Yeast Genomic DNA Extraction
Yeast genomic DNA was extracted by the Winston protocol [57].
Tetrad Dissection
Heterozygous diploid erg3Δ::URA3 cells were cultured on YPD plates, then transferred to spore plates. A colony-sized mass of cells was re-suspended in 6 µL Zymolase 100T (1 mg/mL, Sigma) for each sporulation and incubated for 2 minutes at room temperature. After incubation, 300 µL of water was added to each sample. Each sample solution was then spread along the upper portion of a petri dish and tetrads were separated with an Olympus B201 Dissecting scope. Each plate was incubated at 30°C, and cells were re-patched to a master plate. Each master plate was then replica plated to YMD dropout plates and mating tester lawns to determine genotype.
cDNA Cloning
Chlamydomonas Core Library. The Chlamydomonas cDNA core library was purchased from the Chlamydomonas Center (http://www.chlamy.org/). The library was amplified using the host strain XL1 Blue MRF' (Stratagene catalog number 200301) according to the manufacturer's instructions at http://www.stratagene.com/lit/manuals/aspx (catalog number 236201).
The S. cerevisiae ERG3 protein coding sequence (accession number NP_013157) was used to identify the C. reinhardtii EST with the best homology using the BLAST server (http://genomeportal.jgi-psf.org/Chlre4/Chlre4.home.html). Primers were designed based on the EST sequence in order to clone the coding region of cDNA from the core library described above. The primers used to clone the cDNA by PCR included the forward primer: 5′-GCGGCCGCCGATCGAAGCTTAATGTCAACCTCGCTCAAAATGA-3′ and the reverse primer: 5′-GCGGCCGCCGATCGAAGCTTTACTGCGCCTTGACGGCCT-3′.
Sequence Analysis
Multiple sequence alignment programs from the European Bioinfomatics Institute EMBL-EBI server (http://www.ebi.ac.uk/clustalw/) were used for sequence analysis and alignment data. The Prosite database from EBI server (http://ca.expasy.org/prosite/) was used to identify putative consensus motifs for specific domains in ERG3. The C. reinhardtii genomic database provided information about the genomic sequence, intron-exon structure of ERG3, as well as information regarding potential membrane spanning regions of the protein coding sequence (http://genomeportal.jgi-psf.org/Chlre4/Chlre4.home.html).
Drug Resistance Screen
Cycloheximide was purchased from Sigma Aldrich (catalog# C- 7698-1G). S. cerevisiae minimal media was prepared as previously described with the addition of cycloheximide [34] at a final concentration of 0.13 µg/mL from a stock solution of 2 mg/ml in 100% ethanol.
Lipid Extraction
Yeast cells were grown to an optical density (A600) of 0.7-0.8. The cells were pelleted at 2000 rpm for 5 minutes and resuspended in 4 mL hot isopropanol (70°C) for 30 minutes. The cell wall was disrupted by vortexing with 0.5 mm glass beads for 2 minutes. Cell pellets were then resuspended in 4 mL hot isopropanol and lipids were extracted at 70°C for 2 hours. Next, 4 mL of chloroform/methanol (1∶2) and 2 mL of 1 M KCL were added to the lipid extracts, which were vortexed and centrifuged at 5000 rpm on a benchtop centrifuge for 5 minutes. The top aqueous phase was discarded and the previous extraction step was repeated three times. Then, 2 mL of water were then added to the samples. The samples were vortexed and centrifuged at 5000 rpm for 5 minutes, and the aqueous phase was discarded again. Lipids were dried under nitrogen and stored at −80°C until analysis. The lipid extraction protocol was derived from the Bligh and Dyer method [58].
GC/MS Analysis
Ergosterol standard was purchased from Fluka. N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) was used as the silylating reagent and was purchased from Sigma Aldrich (catalog #33084). 100 µL of BSTFA was added to dried lipid samples and heated to 60°C for 30 minutes. Derivatized samples were resuspended in 25 µL of dichloromethane and prepared for analysis. GC/MS was carried out on an Agilent 6890 gas chromatograph with an autosampler and Agilent 5973 mass selective detector. Then, 1 µL of sample was injected into a splitless system with a flow rate of 1 mL/min for the carrier gas, helium. The front inlet temperature was maintained at 250°C, while the mass selective detector line heater was at 280°C. The oven temperature was programmed to a final temperature of 300°C with an initial temperature of 100°C. The GC column, DB-5 ms, was 30 meters by 250 µm of internal diameter with a film thickness of 0.25 µm. Data was acquired by selective ion monitoring to detect ions at m/z 143, 211,337, 363, 468.
Acknowledgments
We would like to thank Dr. Steven Barker at the Louisiana State University School of Veterinary Medicine Analytical Systems Laboratory for analysis of lipids as well as Dr. Debbie Grimm at the Tulane Coordinated Instrumentation Facility for helpful advice.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Science Foundation (Grants to MCB- 0817823 to D.D. and IOS-0816957 to J.V.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bard M, Lees ND, Burrows LS, Kleinhans FW. Differences in crystal violet uptake and cation-induced death among yeast sterol mutants. J Bacteriol. 1978;135:1146–1148. doi: 10.1128/jb.135.3.1146-1148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleinhans FW, Lees ND, Bard M, Haak RA, Woods RA. ESR determinations of membrane permeability in a yeast sterol mutant. Chem Phys Lipids. 1979;23:143–154. doi: 10.1016/0009-3084(79)90042-2. [DOI] [PubMed] [Google Scholar]
- 3.Lees ND, Bard M, Kemple MD, Haak RA, Kleinhans FW. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979;553:469–475. doi: 10.1016/0005-2736(79)90302-x. [DOI] [PubMed] [Google Scholar]
- 4.Lees ND, Kleinhans FW, Broughton MC, Pennington DE, Ricker VA, et al. Membrane fluidity alterations in a cytochrome P-450-deficient mutant of Candida albicans. Steroids. 1989;53:567–578. doi: 10.1016/0039-128x(89)90032-9. [DOI] [PubMed] [Google Scholar]
- 5.Summons RE, Bradley AS, Jahnke LL, Waldbauer JR. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc Lond B Biol Sci. 2006;361:951–968. doi: 10.1098/rstb.2006.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benveniste P. Biosynthesis and Accumulation of Sterols. Annual Review of Plant Biology. 2004;55:429–457. doi: 10.1146/annurev.arplant.55.031903.141616. [DOI] [PubMed] [Google Scholar]
- 7.Jayasimha P BC, Pedroza J, Nes WD. Engineering pathway enzymes to understand the function and evolution of sterol structure and activity. Recent Advances in Phytochemistry. 2006;40 [Google Scholar]
- 8.Nes WD, Norton RA, Crumley FG, Madigan SJ, Katz ER. Sterol phylogenesis and algal evolution. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:7565–7569. doi: 10.1073/pnas.87.19.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker B, Kerr R. Biosynthesis of marine sterols. 1993. pp. 1–31. Marine Natural Products — Diversity and Biosynthesis.
- 10.Giner JL. Biosynthesis of marine sterol side chains. Chemical Reviews. 1993;93:1735–1752. [Google Scholar]
- 11.Gealt M, Adler J, Nes W. The sterols and fatty acids from purified flagella of Chlamydomonas reinhardtii. Lipids. 1981;16:133–136. [Google Scholar]
- 12.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bard M, Wilson K, Thompson R. Isolation of sterol mutants inChlamydomonas reinhardi: Chromatographic analyses. Lipids. 1978;13:533–539. doi: 10.1007/BF02533592. [DOI] [PubMed] [Google Scholar]
- 14.Salimova E, Boschetti A, Eichenberger W, Lutova L. Sterol mutants of Chlamydomonas reinhardtii: Characterisation of three strains deficient in C24(28) reductase. Plant Physiology and Biochemistry. 1999;37:241–249. [Google Scholar]
- 15.Arthington-Skaggs BA, Crowell DN, Yang H, Sturley SL, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. doi: 10.1016/0014-5793(96)00807-1. [DOI] [PubMed] [Google Scholar]
- 16.Osumi T, Taketani S, Katsuki H, Kuhara T, Matsumoto I. Ergosterol biosynthesis in yeast. Pathways in the late stages and their variation under various conditions. J Biochem. 1978;83:681–691. doi: 10.1093/oxfordjournals.jbchem.a131961. [DOI] [PubMed] [Google Scholar]
- 17.Cowen LE, Steinbach WJ. Stress, Drugs, and Evolution: the Role of Cellular Signaling in Fungal Drug Resistance. Eukaryotic Cell. 2008;7:747–764. doi: 10.1128/EC.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White TC, Marr KA, Bowden RA. Clinical, Cellular, and Molecular Factors That Contribute to Antifungal Drug Resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Z, Nes W. Sterol Biosynthesis Inhibitors: Potential for Transition State Analogs and Mechanism-Based Inactivators Targeted at Sterol Methyltransferase. Lipids. 2007;42:15–33. doi: 10.1007/s11745-006-3017-1. [DOI] [PubMed] [Google Scholar]
- 20.Benveniste P. Biosynthesis and Accumulation of Sterols. Annual Review of Plant Biology. 2004;55:429–457. doi: 10.1146/annurev.arplant.55.031903.141616. [DOI] [PubMed] [Google Scholar]
- 21.Yang W, Moroney JV, Moore TS. Membrane lipid biosynthesis in Chlamydomonas reinhardtii: ethanolaminephosphotransferase is capable of synthesizing both phosphatidylcholine and phosphatidylethanolamine. Arch Biochem Biophys. 2004;430:198–209. doi: 10.1016/j.abb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Yang W, Mason CB, Pollock SV, Lavezzi T, Moroney JV, et al. Membrane lipid biosynthesis in Chlamydomonas reinhardtii: expression and characterization of CTP:phosphoethanolamine cytidylyltransferase. Biochem J. 2004;382:51–57. doi: 10.1042/BJ20040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osumi T, Nishino T, Katsuki H. Studies on the delta 5-desaturation in ergosterol biosynthesis in yeast. J Biochem. 1979;85:819–826. [PubMed] [Google Scholar]
- 24.Smith SJ, Parks LW. The ERG3 gene in Saccharomyces cerevisiae is required for the utilization of respiratory substrates and in heme-deficient cells. Yeast. 1993;9:1177–1187. doi: 10.1002/yea.320091104. [DOI] [PubMed] [Google Scholar]
- 25.Parks LW, McLean-Bowen C, Bottema CK, Taylor FR, Gonzales R, et al. Aspects of sterol metabolism in the yeast Saccharomyces cerevisiae and in Phytophthora. Lipids. 1982;17:187–196. doi: 10.1007/BF02535102. [DOI] [PubMed] [Google Scholar]
- 26.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 27.Gachotte D, Husselstein T, Bard M, Lacroute F, Benveniste P. Isolation and characterization of an Arabidopsis thaliana cDNA encoding a delta 7-sterol-C-5-desaturase by functional complementation of a defective yeast mutant. Plant J. 1996;9:391–398. doi: 10.1046/j.1365-313x.1996.09030391.x. [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki Y, Geber A, Miyazaki H, Falconer D, Parkinson T, et al. Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene. 1999;236:43–51. doi: 10.1016/s0378-1119(99)00263-2. [DOI] [PubMed] [Google Scholar]
- 29.Nes WR, Dhanuka IC. Inhibition of sterol synthesis by delta 5-sterols in a sterol auxotroph of yeast defective in oxidosqualene cyclase and cytochrome P-450. J Biol Chem. 1988;263:11844–11850. [PubMed] [Google Scholar]
- 30.Norton R, David Nes W. Identification of ergosta-6(7),8(14),25(27)-trien-3β-ol and ergosta-5(6),7(8),25(27)-trien-3β-ol, two new steroidal trienes synthesized by Prototheca wickerhamii. Lipids. 1991;26:247–249. [Google Scholar]
- 31.Broach JR, Pringle JR, Jones EW. Cold Spring Harbor, N.Y.: Cole Spring Harbor Laboratory Press; 1991. The Molecular and cellular biology of the yeast Saccharomyces. [Google Scholar]
- 32.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arthington BA, Bennett LG, Skatrud PL, Guynn CJ, Barbuch RJ, et al. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene. 1991;102:39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- 34.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:2005 0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemmi K, Julmanop C, Hirata D, Tsuchiya E, Takemoto JY, et al. The physiological roles of membrane ergosterol as revealed by the phenotypes of syr1/erg3 null mutant of Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1995;59:482–486. doi: 10.1271/bbb.59.482. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths KM, Bacic A, Howlett BJ. Sterol composition of mycelia of the plant pathogenic ascomycete Leptosphaeria maculans. Phytochemistry. 2003;62:147–153. doi: 10.1016/s0031-9422(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 37.Salimova E, Boschetti A, Eichenberger W, Lutova L. Sterol mutants of Chlamydomonas reinhardtii: Characterisation of three strains deficient in C24(28) reductase. Plant Physiology and Biochemistry. 1999;37:241–249. [Google Scholar]
- 38.Lampen JO. Interference by polyenic antifungal antibiotics (especially nystatin and Filipin) with specific membrane functions. In: functions. IbpaaenaFwsm, editor. A Symposium of the Society for General Microbiology. Cambridge: Published for the Society for General Microbiology at the University Press; 1966. 111 [Google Scholar]
- 39.Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 43.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 44.Darnet S, Bard M, Rahier A. Functional identification of sterol-4alpha-methyl oxidase cDNAs from Arabidopsis thaliana by complementation of a yeast erg25 mutant lacking sterol-4alpha-methyl oxidation. FEBS Lett. 2001;508:39–43. doi: 10.1016/s0014-5793(01)03002-2. [DOI] [PubMed] [Google Scholar]
- 45.Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, et al. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- 46.Souter M, Topping J, Pullen M, Friml J, Palme K, et al. hydra Mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell. 2002;14:1017–1031. doi: 10.1105/tpc.001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taton M, Husselstein T, Benveniste P, Rahier A. Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase studied by site-directed mutagenesis. Biochemistry. 2000;39:701–711. doi: 10.1021/bi991467t. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig DL, Ugolini S, Bruschi CV. High-level heterologous gene expression in Saccharomyces cerevisiae from a stable 2[mu]m plasmid system. Gene. 1993;132:33–40. doi: 10.1016/0378-1119(93)90511-z. [DOI] [PubMed] [Google Scholar]
- 49.Lichtenthaler HK. The 1-Deoxy-D-Xylulose-5-Phosphate Pathway of Isoprenoid Biosynthesis in Plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- 50.Parks LW, Casey WM. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 51.Fernandes P, Cabral JM. Phytosterols: applications and recovery methods. Bioresour Technol. 2007;98:2335–2350. doi: 10.1016/j.biortech.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Moghadasian MH, Frohlich JJ. Effects of dietary phytosterols on cholesterol metabolism and atherosclerosis: clinical and experimental evidence. Am J Med. 1999;107:588–594. doi: 10.1016/s0002-9343(99)00285-5. [DOI] [PubMed] [Google Scholar]
- 53.Tapiero H, Townsend DM, Tew KD. Phytosterols in the prevention of human pathologies. Biomed Pharmacother. 2003;57:321–325. doi: 10.1016/s0753-3322(03)00104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 55.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 56.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 57.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 58.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]