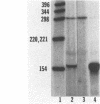
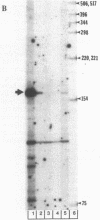
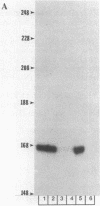
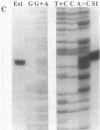
Abstract
We used molecular cloning to isolate a functional gene for mouse ornithine decarboxylase (OrnDCase; L-ornithine carboxy-lyase, EC 4.1.1.17) from a cell line in which that gene had been selectively amplified. The position of the 5' terminus of the mRNA was identified, and the coding sequence was shown to be preceded by a 312- or 313-nucleotide (nt) untranslated leader. The latter is highly G + C rich, particularly in its 5'-most portion. The leader can be anticipated to have extensive and stable secondary structure. The transcription unit of the gene is of relatively small size, approximately equal to 6.2 kilobases (kb) from the start site to the proximal site of polyadenylylation. Sequence analysis of DNA near the transcription start position demonstrated the presence of a "TATA" box, but no "CAAT" box. Functional properties of the cloned gene were tested by transfecting it into cultured cells. Expression of the putative full-length gene efficiently conferred ornithine decarboxylase activity on recipient mutant cells deficient in that activity. To assess the function and strength of the OrnDCase promoter region and to delimit its boundaries, we used a transient expression assay. Upstream of a bacterial chloramphenicol acetyltransferase gene was placed a portion of the OrnDCase gene, including the presumed promoter region, spanning a region from approximately equal to 3.0 kb 5' of the site of transcription initiation to the first 250 nt of the transcript. When expressed in mouse NIH 3T3 cells, this OrnDCase genomic element was comparable in strength to the Rous sarcoma virus long terminal repeat promoter. A similar construct, truncated so as to retain only 264 base pairs of the OrnDCase gene 5' to the site of transcription start, yielded undiminished levels of expression.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Daar I. O., Krug J. R., Maquat L. E. Characterization of the functional gene and several processed pseudogenes in the human triosephosphate isomerase gene family. Mol Cell Biol. 1985 Jul;5(7):1694–1706. doi: 10.1128/mcb.5.7.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Deng T. L., Li D. W., Jenh C. H., Johnson L. F. Structure of the gene for mouse thymidylate synthase. Locations of introns and multiple transcriptional start sites. J Biol Chem. 1986 Dec 5;261(34):16000–16005. [PubMed] [Google Scholar]
- Dircks L., Grens A., Slezynger T. C., Scheffler I. E. Posttranscriptional regulation of ornithine decarboxylase activity. J Cell Physiol. 1986 Mar;126(3):371–378. doi: 10.1002/jcp.1041260307. [DOI] [PubMed] [Google Scholar]
- Dush M. K., Sikela J. M., Khan S. A., Tischfield J. A., Stambrook P. J. Nucleotide sequence and organization of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that has a variable intron/exon arrangement. Proc Natl Acad Sci U S A. 1985 May;82(9):2731–2735. doi: 10.1073/pnas.82.9.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Edlund T., Walker M. D., Barr P. J., Rutter W. J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5' flanking elements. Science. 1985 Nov 22;230(4728):912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Feinstein S. C., Dana S. L., McConlogue L., Shooter E. M., Coffino P. Nerve growth factor rapidly induces ornithine decarboxylase mRNA in PC12 rat pheochromocytoma cells. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5761–5765. doi: 10.1073/pnas.82.17.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W. A., Sypherd P. S. Expression of the gene for ornithine decarboxylase of Saccharomyces cerevisiae in Escherichia coli. Mol Cell Biol. 1985 Jan;5(1):161–166. doi: 10.1128/mcb.5.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. K., Avdalovic N., Madara T., O'Brien T. G. Induction of ornithine decarboxylase by 12-O-tetradecanoylphorbol 13-acetate in hamster fibroblasts. Relationship between levels of enzyme activity, immunoreactive protein, and RNA during the induction process. J Biol Chem. 1985 Dec 25;260(30):16439–16444. [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gupta M., Coffino P. Mouse ornithine decarboxylase. Complete amino acid sequence deduced from cDNA. J Biol Chem. 1985 Mar 10;260(5):2941–2944. [PubMed] [Google Scholar]
- Heby O. Role of polyamines in the control of cell proliferation and differentiation. Differentiation. 1981;19(1):1–20. doi: 10.1111/j.1432-0436.1981.tb01123.x. [DOI] [PubMed] [Google Scholar]
- Hickok N. J., Seppänen P. J., Gunsalus G. L., Jänne O. A. Complete amino acid sequence of human ornithine decarboxylase deduced from complementary DNA. DNA. 1987 Jun;6(3):179–187. doi: 10.1089/dna.1987.6.179. [DOI] [PubMed] [Google Scholar]
- Hunt T. False starts in translational control of gene expression. Nature. 1985 Aug 15;316(6029):580–581. doi: 10.1038/316580a0. [DOI] [PubMed] [Google Scholar]
- Hölttä E., Pohjanpelto P. Control of ornithine decarboxylase in Chinese hamster ovary cells by polyamines. Translational inhibition of synthesis and acceleration of degradation of the enzyme by putrescine, spermidine, and spermine. J Biol Chem. 1986 Jul 15;261(20):9502–9508. [PubMed] [Google Scholar]
- Ingolia D. E., Al-Ubaidi M. R., Yeung C. Y., Bigo H. A., Wright D., Kellems R. E. Molecular cloning of the murine adenosine deaminase gene from a genetically enriched source: identification and characterization of the promoter region. Mol Cell Biol. 1986 Dec;6(12):4458–4466. doi: 10.1128/mcb.6.12.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomaa V. V., Pajunen A. E., Bardin C. W., Jänne O. A. Ornithine decarboxylase in mouse kidney. Purification, characterization, and radioimmunological determination of the enzyme protein. J Biol Chem. 1983 Jun 10;258(11):6735–6740. [PubMed] [Google Scholar]
- Kahana C., Nathans D. Isolation of cloned cDNA encoding mammalian ornithine decarboxylase. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3645–3649. doi: 10.1073/pnas.81.12.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana C., Nathans D. Nucleotide sequence of murine ornithine decarboxylase mRNA. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1673–1677. doi: 10.1073/pnas.82.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana C., Nathans D. Translational regulation of mammalian ornithine decarboxylase by polyamines. J Biol Chem. 1985 Dec 15;260(29):15390–15393. [PubMed] [Google Scholar]
- Kameji T., Pegg A. E. Inhibition of translation of mRNAs for ornithine decarboxylase and S-adenosylmethionine decarboxylase by polyamines. J Biol Chem. 1987 Feb 25;262(6):2427–2430. [PubMed] [Google Scholar]
- Kim S. H., Moores J. C., David D., Respess J. G., Jolly D. J., Friedmann T. The organization of the human HPRT gene. Nucleic Acids Res. 1986 Apr 11;14(7):3103–3118. doi: 10.1093/nar/14.7.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontula K. K., Torkkeli T. K., Bardin C. W., Jänne O. A. Androgen induction of ornithine decarboxylase mRNA in mouse kidney as studied by complementary DNA. Proc Natl Acad Sci U S A. 1984 Feb;81(3):731–735. doi: 10.1073/pnas.81.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters J. N., Attardi G. Discrete human dihydrofolate reductase gene transcripts present in polysomal RNA map with their 5' ends several hundred nucleotides upstream of the main mRNA start site. Mol Cell Biol. 1985 Mar;5(3):493–500. doi: 10.1128/mcb.5.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McConlogue L., Coffino P. A mouse lymphoma cell mutant whose major protein product is ornithine decarboxylase. J Biol Chem. 1983 Oct 25;258(20):12083–12086. [PubMed] [Google Scholar]
- McConlogue L., Coffino P. Ornithine decarboxylase in difluoromethylornithine-resistant mouse lymphoma cells. Two-dimensional gel analysis of synthesis and turnover. J Biol Chem. 1983 Jul 10;258(13):8384–8388. [PubMed] [Google Scholar]
- McConlogue L., Dana S. L., Coffino P. Multiple mechanisms are responsible for altered expression of ornithine decarboxylase in overproducing variant cells. Mol Cell Biol. 1986 Aug;6(8):2865–2871. doi: 10.1128/mcb.6.8.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConlogue L., Gupta M., Wu L., Coffino P. Molecular cloning and expression of the mouse ornithine decarboxylase gene. Proc Natl Acad Sci U S A. 1984 Jan;81(2):540–544. doi: 10.1073/pnas.81.2.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. W., Konecki D. S., Brennand J., Caskey C. T. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2147–2151. doi: 10.1073/pnas.81.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. I., Framson P. E., Caskey C. T., Chinault A. C. Fine structure of the human hypoxanthine phosphoribosyltransferase gene. Mol Cell Biol. 1986 Feb;6(2):393–403. doi: 10.1128/mcb.6.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. A., Coffino P., Wang C. C. Cloning and sequencing of the ornithine decarboxylase gene from Trypanosoma brucei. Implications for enzyme turnover and selective difluoromethylornithine inhibition. J Biol Chem. 1987 Jun 25;262(18):8721–8727. [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Seely J. E., Pegg A. E. Changes in mouse kidney ornithine decarboxylase activity are brought about by changes in the amount of enzyme protein as measured by radioimmunoassay. J Biol Chem. 1983 Feb 25;258(4):2496–2500. [PubMed] [Google Scholar]
- Singer-Sam J., Keith D. H., Tani K., Simmer R. L., Shively L., Lindsay S., Yoshida A., Riggs A. D. Sequence of the promoter region of the gene for human X-linked 3-phosphoglycerate kinase. Gene. 1984 Dec;32(3):409–417. doi: 10.1016/0378-1119(84)90016-7. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Steglich C., Scheffler I. E. An ornithine decarboxylase-deficient mutant of Chinese hamster ovary cells. J Biol Chem. 1982 Apr 25;257(8):4603–4609. [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Valerio D., Duyvesteyn M. G., Dekker B. M., Weeda G., Berkvens T. M., van der Voorn L., van Ormondt H., van der Eb A. J. Adenosine deaminase: characterization and expression of a gene with a remarkable promoter. EMBO J. 1985 Feb;4(2):437–443. doi: 10.1002/j.1460-2075.1985.tb03648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. K., Masters J. N., Attardi G. Human dihydrofolate reductase gene organization. Extensive conservation of the G + C-rich 5' non-coding sequence and strong intron size divergence from homologous mammalian genes. J Mol Biol. 1984 Jun 25;176(2):169–187. doi: 10.1016/0022-2836(84)90419-4. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]