Abstract
Using transcript profile analysis, we explored the nature of the stem cell niche in roots of maize (Zea mays). Toward assessing a role for specific genes in the establishment and maintenance of the niche, we perturbed the niche and simultaneously monitored the spatial expression patterns of genes hypothesized as essential. Our results allow us to quantify and localize gene activities to specific portions of the niche: to the quiescent center (QC) or the proximal meristem (PM), or to both. The data point to molecular, biochemical and physiological processes associated with the specification and maintenance of the niche, and include reduced expression of metabolism-, redox- and certain cell cycle-associated transcripts in the QC, enrichment of auxin-associated transcripts within the entire niche, controls for the state of differentiation of QC cells, a role for cytokinins specifically in the PM portion of the niche, processes (repair machinery) for maintaining DNA integrity and a role for gene silencing in niche stabilization. To provide additional support for the hypothesized roles of the above-mentioned and other transcripts in niche specification, we overexpressed, in Arabidopsis, homologs of representative genes (eight) identified as highly enriched or reduced in the maize root QC. We conclude that the coordinated changes in expression of auxin-, redox-, cell cycle- and metabolism-associated genes suggest the linkage of gene networks at the level of transcription, thereby providing additional insights into events likely associated with root stem cell niche establishment and maintenance.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-009-1059-3) contains supplementary material, which is available to authorized users.
Keywords: Quiescent center, Root, Stem cell, Stem cell niche, Zea mays
Introduction
Plants maintain special microenvironments (stem cell niches [SCN]) at both the root and the shoot tips (apical meristems). As a consequence of their location within the niche, cells may display varying degrees of “stemness”, including variations in their ability to divide (or not), as well as in their states of differentiation and capacity for self-renewal (Cai J et al. 2004). Based on these features, the root SCN can be divided into two stem cell subpopulations; one characterized by cells with long cell cycles (120–200 h), and a second population characterized by more rapidly dividing cells that may also show varying degrees of differentiation. The former population comprises the quiescent center (QC) (Clowes 1961), whereas the latter, more rapidly cycling cells, populate the proximal meristem (PM). In roots of the model plant Arabidopsis, the mitotically active initials of the PM overlay the less active QC cells, which number 4–7 (Campilho et al. 2006; Xu et al. 2006). The root SCN in Arabidopsis can be contrasted with that of larger roots, such as those of corn (maize), in which the QC is composed of many more cells, perhaps 500–1,000, which show a gradation in cell activities and states of differentiation as one moves basally toward and the PM/QC border.
In addition to information on the state of differentiation of the cells populating the root SCN, and on their capacity for apparently unlimited proliferation, self-maintenance and self-renewal (Clowes 1961), it has recently been discovered that signals move between the two sets of initials (van den Berg et al. 1995; Sarkar et al. 2007), though the identities of the signals, which may include the SHR protein (Nakajima and Benfey 2002), are not known. The expression of WOX5, a member of the WUSCHEL-related Homeobox gene family, suggests also the possible involvement of CLAVATA-like signaling (Schoof et al. 2000). The demonstration that the plant growth regulator auxin plays a central role in niche specification (Xu et al. 2006; Sena et al. 2009) points to possible physiological events underlying niche development and maintenance. Indeed, the involvement of auxin in SCN specification likely begins early in embryogenesis (Friml et al. 2003) and is most probably mediated by a number of auxin-regulated transcription factors (TF), including members of the PLETHORA protein family and the auxin response factor, MONOPTEROS (Hardtke and Berleth 1998; Sabatini et al. 1999). Evidence also points to an involvement of members of both the PIN and RETINOBLASTOMA protein families as having inputs in the designation and/or maintenance of the SCN (Aida et al. 2002; Wildwater et al. 2005; Galinha et al. 2007).
Building on what has already been established or suggested about the nature of the root SCN (Vernoux and Benfey 2005; Iyer-Pascuzzi and Benfey 2008; Sena et al. 2009), our goal in this report was to elucidate additional characteristics of plant stem cells, with particular attention directed to heretofore unreported characteristics of the stem cells that form the QC and the PM in roots of maize (Feldman and Torrey 1976). We begin this report with a consideration of the global patterns of gene expression in the maize QC and PM. These data were generated as part of a microarray data set previously used for profiling the maize root cap (RC) (Jiang et al. 2006), but these data have not been previously published. We group these QC/PM genes into categories based on putative function and then select for further analysis by real-time quantitative PCR (qPCR) specific genes hypothesized by us or others, (Blau et al. 2001; Baharvand et al. 2006) to affect “stemness”.
Central to this work is an assessment of the likelihood of a linkage between the expression levels of these genes with the specification, elaboration and maintenance of the SCN. For this effort, we reasoned that if we could cause QC cells to lose their typical stem cell characteristics (i.e., stereotypically undifferentiated, with cells accumulating at the G1/S checkpoint of the cell cycle), we may also lose or at least change that gene expression characteristic of cells occupying this portion of the niche. Toward altering the status of the SCN, we surgically excised the RC (decapping) (Fig. 1a, b), which stimulates (activates) QC cells to transit the G1/S checkpoint, to divide relatively rapidly and to begin differentiating into new RC cells (Feldman 1975). Eventually, the QC reforms and QC/PM stem cells return to their pre-decapping status. Using this technique, we identified genes, the activities of which changed (increased/decreased) coincident with an alteration in SCN size and activity. To investigate further whether specific genes with changed activities could be associated with the development/maintenance of the root SCN, we overexpressed the homologs of maize candidate genes in Arabidopsis. We consider results from both maize and Arabidopsis in terms of providing insights into the molecular and physiological basis underlying the development and maintenance of the root SCN. Additionally, we consider whether these data point to a “signature” gene expression pattern for plant (root) stem cells.
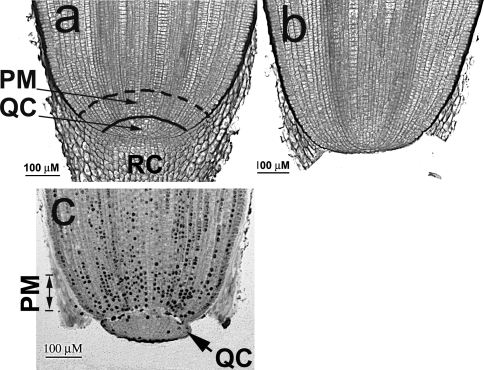
Fig. 1.
Maize root apical meristems as seen in longitudinal section. a Intact primary root tip with the quiescent center (QC), proximal meristem (PM) and root cap (RC) (still attached). b A root from which the root cap has been excised leaving the QC intact. c Autoradiograph showing the excised (QC) and adjacent (PM)
Materials and methods
Isolation of root tissues
Zea mays cv. Merit seeds were imbibed for 2 h in rapidly running water and germinated in the dark at 24°C for 48 h. For QC activation experiments, RCs were excised and QC and PM tissues either collected immediately (time 0) (as described in detail in Feldman and Torrey 1976) (Fig. 1a–c), or roots were returned to the dark at 24°C for varying periods of time (6, 20 or 32 h), and then both the QC and PM collected. As tissues were excised, they were immediately frozen in liquid N2, pooled, and stored at −80°C until the RNA was extracted (Jiang et al. 2006). For time course experiments, root tissues (100–160 QCs or 20–25 PMs) were collected into 500 μl of RNAwiz (Ambion, Austin, TX), and stored at −80°C until use.
Microarray data analysis
Using the rice GeneChip (Zhu et al. 2003), representing 23,779 genes [approximately half of the rice genome (Bennetzen et al. 2004)], we previously obtained gene expression profiles for the RC, QC and PM of maize (but to date have published only the data on RC) (Jiang et al. 2006). Previous comparative sequence analysis showed that more than 93% of full-length, nonredundant, complete coding sequences (696 in total) of maize were highly conserved with the rice genes (E ≤ −25) (Goff et al. 2002). Zhu et al. (2003) have shown that over 90% of the maize transcripts cross-hybridize with the probes on the rice GeneChip. QC and PM microarray data were analyzed using the ANOVA one-factor F-test, followed by Fisher’s LSD and PCA. A total of 316 genes were found to be statistically enriched/reduced in their expression in the QC. Specifics for the RNA isolation, cDNA preparation and hybridization and the data transformation and statistics analysis are as described by Jiang et al. (2006). Of the 316 genes differentially expressed in the QC, 125 were reduced and 191 enriched. However, because of the cross-species nature of this methodology, we have taken a conservative approach and further filtered (using a 1.2-fold change) these two sets of genes, resulting in a reduction in the number of down- or enriched-QC genes to 70 (Supplemental Table 1) and 119 (Supplemental Table 2), respectively. Using a 1.2-fold change allowed us to capture approximately 59.8% (189) of the statistically differentially expressed QC genes, of which ten were later analyzed in more detail, and their expression levels confirmed using real-time quantitative RT-PCR (RT-qPCR) with primers selected from maize (Supplemental Table 3).
To identify the putative maize orthologs, we first searched for rice locus IDs using rice GeneChip probe region sequences in the TIGR database. Then, using the rice locus sequences, we identified the maize orthologs using the Inparanoid algorithm (O’Brien et al. 2005) (Supplemental Table 1 and 2). In addition, we also used the GeneChip rice probe region sequences for blast searches employing various maize ESTs databases (http://tigrblast.tigr.org/tgi, http://www.ncbi.nlm.nih.gov/blast), which provided either the tentative consensus (TC) number or GenBank access number for the best matching maize sequence. These identifying numbers are listed in Supplemental Tables 1 and 2. To validate the expression data, the top matching TC sequences were re-assessed, and the TC sequence(s) that contained the EST from the root tissue cDNA library was used for PCR primer design.
Real-time quantitative RT-PCR
For real-time quantitative RT-PCR, total RNA was extracted from either 400–600 RCs, 800–1,000 QCs, 300–400 PMs or 100–150 elongation zones (EZ) (a 1 mm-long segment 10 mm from the root tip), using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. For each RNA sample, a 1.2-kb kanamycin RNA control was added, as previously described by Jiang et al. (2006), and then 2 μl of this mixture was used for the cDNA synthesis.
For time course quantitative RT-PCR, RNA was extracted using 500 μl of RNAwiz (Ambion, Austin, TX); 100 μl of chloroform was added and the mixture shaken vigorously for 20 s. The upper aqueous phase was transferred to a new tube and 250 μl of DEPC-treated H2O was added. After adding 500 μl of isopropanol, the RNA was placed at −20°C for 30 min, collected by centrifuging at 10,000 rpm for 15 min at 4°C and then solubilized in 10 μl of DEPC-treated H2O. At 37°C for 30 min, 4 μg of the RNA was digested with 2 μl of RQ1 RNase-free DNase (Promega, Madison, WI) in a 20 μl reaction volume. The DNase was then inactivated by adding 2 μl of RQ1 stop solution and incubated at 65°C for 10 min. After DNase digestion, 100 μl of DEPC-treated H2O was added to the RNA, and the concentration was measured at 260 nm. For each sample, an equal amount of RNA was used and sufficient DEPC H2O was added to attain the same final concentration, and then 65 μl of an approximately 10−7-fold dilution of the kanamycin 1.2-kb control RNA was added to each sample as an internal standard for normalization. The RNA sample was then loaded onto a Microcon-100 column and spun 8 min at 2,500 rpm. After washing with 200 μl of DEPC H2O, the RNA was eluted from the column by adding 15 μl of DEPC H2O to the column, waiting for 5 min, and then the column was spun and inverted for 3 min at 1000 rpm. As much as 8 μl of RNA was used for cDNA synthesis.
The cDNA synthesis and qPCR were preformed as described by Jiang et al. (2006). Target cDNAs were normalized to the kanamycin 1.2-kb control. For comparisons between tissues, the differences in transcript levels were further expressed as a ratio after setting the PM value as 1. The results are presented as the means of at least two biological replicates and are listed in Supplemental Tables 3 and 4.
To determine whether there were coherent, dominant expression patterns in either the QC or PM transcripts, we performed a tight clustering analysis of the qPCR data using the software tightClust (Tseng and Wong 2005). Tight clustering is a re-sampling-based approach for identifying the most informative, tight and stable clusters from the data. As it does not force all genes into clusters, it helps avoid the contamination of tight expression patterns of biologically relevant genes from other genes with only loosely compatible patterns. In our study, we slightly modified the tight clustering algorithm by using a more reliable and sensitive gene co-expression measure PoissonC (Cai L et al. 2004; Kim et al. 2007) to generate the results.
Primers used for this work are listed in Supplemental Table 5. The identity of the PCR product was confirmed either by sequencing or by sizing on an agarose gel.
Transgenic Arabidopsis lines
Eight genes (AHL1 [At4g12080], GATA2 [At2g45050], RHC1a [At2g40830], At1g75710, At1g76510, At2g34710, At2g41310 and At3g58120), all encoding TFs, were selected for overexpression in Arabidopsis Col-0 under the 35S promoter. These putative maize homologs were selected based on the similarity of their protein sequences and their expression patterns in the root tissue. Briefly, the maize gene coding sequences were queried against the Arabidopsis database (http://www.arabidopsis.org/) employing the BLASTX program, and the top three matching genes were screened by comparing their expression patterns in the root tissue [using, http://bbc.botany.utoronto.ca/efp/ (Birnbaum et al. 2003; Nawy et al. 2005), (Supplemental Table 1&2)]. Those genes with QC expression pattern in Arabidopsis closest to the pattern in maize were selected.
For the 35S::GATA2 construct, the entire coding sequence of At2g45050 was amplified from a full-length cDNA clone (accession NO. BT000921) and cloned into the pCAMBIA1304 (Cambia, Canberra) at the Nco I/BstE II site, downstream from the CaMV 35S promoter, replacing the mGFP5-GUS gene. The original pCAMBIA1304 was used as a control. Transgenic plants were generated by transforming Col-0 wild type (Clough and Bent 1998).
The AHL1, RHC1a, At2g34710 and At3g58120 overexpression lines are the gifts from Mendel Biotechnology Inc., Hayward, CA. Briefly, for the 35S::AHL1 construct, At4g12080 cDNA was cloned into the Sal I/Not I site of pMEN65 downstream from the CaMV35S promoter. For construction of the 35S::RHC1a, At2g40830 genomic DNA was introduced into pMEN65 downstream from the CaMV35S promoter using a Gateway genomic clone. Control lines were produced by transforming with empty pMEN65.
For the At1g76510, At1g75710 and At2g41310 overexpression lines, the CaMV 35S promoter was inserted into EcorV/SacI sites of pGII0179 and the entire coding sequences from each gene were cloned into the downstream CaMV 35S promoter. The coding sequences of At1g76510, At1g75710 and At2g41310 were amplified from cDNA that was reverse transcribed from mRNA of Col-0 wild type using RT-PCR. Subsequently, the coding sequences of At1g76510 and At2g41310 were inserted between the XbaI/SacI sites, and the coding sequence of At1g75710 was inserted between XbaI/BamHI sites.
Overexpression for GATA2, AHL1, RHC1a transgenic lines was confirmed on individual plants by qPCR (Supplemental Table 6). To characterize root phenotypes, ten individual lines from each transgenic line were screened against the empty vector transgenic lines. Three lines with the short root phenotype were then selected for further analysis. T2 plants were used from either the 35S::GATA2 or 35S::RHC1a lines. T3 plants were used for 35S::AHL1 lines.
T-DNA insertion lines for At2g45050 (Salk_008743, Salk_008794 and Salk_008845), At4g12080 (Salk_024324, Salk_026823, Salk_052728 and Salk_086713, Salk_092100) and At2g40830 (Salk_127503, Salk_143110, Salk_137684, Salk_037121) were obtained from the ABRC. Lines showing the short root phenotype were genotyped by PCR using the primers listed in Supplemental Table 5.
Phenotype analysis
Arabidopsis transgenic seeds were germinated in a 1× Murashige–Skoog medium (minus sucrose) under long-day conditions (16-h light/8-h dark) at 22°C, and at an age of 2 weeks, fixed in 1 N HCl, followed by staining with Acriflavin neutral according to Ruzin (1999) and then examined by confocal microscopy.
GenBank accession numbers
See Supplemental Table 5.
Results
Global gene expression patterns in the maize root SCN
There is little difference in the number of genes expressed among the three root tissues (RC, QC, PM), irrespective of the cut-off level (data not shown). No genes were found expressed exclusively in the QC, nor did we detect any genes lacking expression in either the QC or PM. These results have been confirmed using RT-qPCR, as noted subsequently, and point to the transcriptionally active status of the QC, even though chromatin in these cells is reported to be highly condensed (Clowes 1961). Using the microarray data set (Jiang et al. 2006), we identified 316 genes with specific changed expression in the QC, and 176 with changed expression in the PM. Of the 316 genes differentially expressed in the QC, the expression levels of 125 were reduced and of 191 enriched. Further filtering of these two sets of genes using a 1.2-fold change cut-off (genes with expression either 1.2-fold greater or lesser in the QC, as compared to both the PM and RC) reduced the number of genes with changed expression (either reduced or enriched) in the QC to 70 (Supplemental Table 1) and 119 (Supplemental Table 2), respectively.
Next, we consider the specifics of the genes in the QC showing changed expression, drawing particular attention to both differences and similarities with Arabidopsis, the only other root system for which the root SCN has been profiled (Nawy et al. 2005). These results form the basis of the subsequent experimental manipulations and activation of the SCN.
Major themes shown by genes with reduced levels of expression
Of the transcripts showing reduced expression, we note that the largest number (20 of 70) are from genes encoding enzymes that function in various metabolic pathways, including proteins involved in sugar metabolism/the energy production pathway (glycolysis) (Supplemental Table 1). As well, genes encoding proteins for early steps of secondary metabolism pathways are also reduced (Supplemental Table 1). Of particular interest, and not reported for Arabidopsis, is the detection of eight TFs with lower expression levels, of which five encode GATA types of zinc finger proteins (Supplemental Table 1).
Major themes shown by genes with enriched levels of expression
Significantly enriched transcripts in the QC, and not reported for Arabidopsis, include several encoding proteins influencing membrane lipid composition, and vesicle-trafficking-associated activities (Supplemental Table 2). Proteins encoded by these genes are particularly important because they generate and regulate asymmetries, integrate multiple signals, coordinate diverse cellular pathways (Xu and Scheres 2005; Nibau et al. 2006; Teale et al. 2006; Cockcroft and Carvou 2007), affect auxin-mediated AUX/IAA proteolysis (Scherer et al. 2007), alter the auxin response (Bloch et al. 2005) and may affect mitosis and/or the cell cycle (McElver et al. 2000). We also report that six genes associated with DNA replication/repair/chromatin modifications are enriched in the QC (Supplemental Table 2), including a homolog (OS017994.1) of Arabidopsis FAS2, which is a subunit of the CHROMATIN ASSEMBLY FACTOR-1 [CAF-1] complex (Supplemental Table 2). CAF-1 is associated with the ability of stem cells to maintain genomic integrity (stemness) (Ridgway and Almouzni 2000; Kaya et al. 2001; Park and Gerson 2005; Houlard et al. 2006; Nabatiyan et al. 2006; Nijnik et al. 2007). In fas2 mutants, the aberrant phenotype is associated with a misexpression of WUSCHEL (WUS) in the shoot and of SCARECROW (SCR) in root meristems (Kaya et al. 2001).
From this global survey, we conclude that differences between the PM and QC are quantitative (enriched or reduced) and not qualitative (transcripts for all genes are detected), and that the QC is relatively active at the level of gene expression, with much of this activity directed at maintaining underlying metabolic machinery and the integrity of the DNA, thereby keeping the system primed to transit the G1/S checkpoint on receipt of the appropriate signal/stimulus. Nevertheless, unlike what has been reported for Arabidopsis (Nawy et al. 2005), we detect a reduction in the expression of a significant number of genes, many of which encode TFs.
Exploring the activities of genes hypothesized to have a role in the stem cell niche
Based on the fact that the SCN consists of populations with transcription profiles that appear to be quantitatively but not qualitatively different, we next employed qPCR, allowing greater resolution of these differences. For this effort, we selected 35 genes (10 from Supplemental Tables 1, 2) hypothesized by us, or others, as having a role in SCN establishment and/or maintenance. We compared and contrasted their activities in the QC and PM to the activities of these same genes in the root cap (RC) and elongation zone (EZ), both in the intact root and in roots in which the QC had been activated by decapping (monitored at time points 6, 20 and 32 h after decapping). Here, we report and emphasize the most significant results (Tables 1, 2, 3, 4, 5 and Figs. 2, 3, 4, 5, 6).
Table 1.
Regulators of transcription in the maize root tip
| Gene | RC | QC | PM | EZ |
|---|---|---|---|---|
| ZmAGO4 | 0.42 | 1.47 | 1 | 0.08 |
| ZmRR | 0.71 | 0.18 | 1 | 0.63 |
| ZmATH | 1.28 | 1.35 | 1 | 0.15 |
| ZmGATA | 1.84 | 0.37 | 1 | 0.06 |
Table 2.
Auxin-associated gene expression in the maize root tip
| Gene | RC | QC | PM | EZ |
|---|---|---|---|---|
| ZmARF2 | 4.20 | 1.73 | 1 | 2.38 |
| ZmARF5/MP | 0.24 | 2.32 | 1 | 0.02 |
| ZmARF6 | 1.24 | 1.34 | 1 | 0.19 |
| ZmAFR7a | 0.96 | 2.34 | 1 | 0.35 |
| ZmARF7b | 2.57 | 0.80 | 1 | 2.68 |
| ZmARF7_NHP4 | 21.8 | 0.94 | 1 | 0.64 |
| ZmARF8 | 3.34 | 1.32 | 1 | 0.42 |
| ZmARF10 | 5.35 | 0.43 | 1 | 0.23 |
| ZmAUX1 | 2.14 | 2.94 | 1 | 0.10 |
| ZmPIN1a | 2.00 | 0.58 | 1 | 0.23 |
| ZmPIN1b | 3.04 | 1.92 | 1 | 0.48 |
| ZmPIN2 | 2.72 | 0.32 | 1 | 0.08 |
| ZmPLT | 0.83 | 3.8 | 1 | 0.02 |
Table 3.
Cell cycle-associated gene expression in the maize root tip
| Gene | RC | QC | PM | EZ |
|---|---|---|---|---|
| ZmCYCC | 1.11 | 0.82 | 1 | 0.38 |
| ZmCYCD2;1 | 1.63 | 0.63 | 1 | 0.21 |
| ZmCYCL1;1 | 3.12 | 0.92 | 1 | 0.42 |
| ZmCDKA | 0.65 | 0.49 | 1 | 0.40 |
| ZmKRP4 | 1.24 | 1.64 | 1 | 0.12 |
| ZmRBR2;1 | 3.25 | 0.63 | 1 | 0.91 |
| ZmCKS1 | 0.42 | 1.47 | 1 | 0.14 |
Table 4.
Redox-associated gene expression in the maize root tip
| Gene | RC | QC | PM | EZ |
|---|---|---|---|---|
| ZmGR | 2.11 | 0.49 | 1 | 1.27 |
| ZmMDAR | 2.86 | 0.45 | 1 | 1.84 |
| ZmDHAR | 3.11 | 0.79 | 1 | 1.35 |
| Zmγ-GCS | 0.43 | 0.33 | 1 | –a |
| ZmGS | 1.43 | 0.69 | 1 | 0.59 |
| ZmGLDH | 0.95 | 0.78 | 1 | 0.21 |
| ZmAPX | 1.53 | 0.91 | 1 | –a |
| ZmAAO | 0.96 | 1.7 | 1 | 6.8 |
aNot measured by qRT-PCR
Table 5.
Metabolism-related gene expression in the maize root tip
| Gene | RC | QC | PM | EZ |
|---|---|---|---|---|
| ZmGAPDH | 3.09 | 0.33 | 1 | 0.31 |
| ZmV-ATPAPE | 2.23 | 0.46 | 1 | 1.18 |
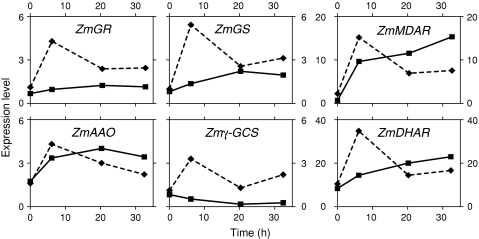
Fig. 2.
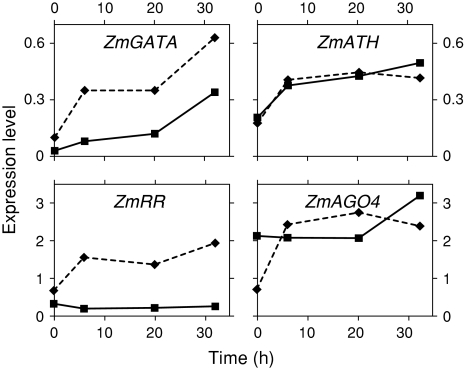
Relative levels of transcription factors in the QC (filled squares on continuous line) and PM (filled diamonds on broken line) at various times following decapping (activation of the stem cell niche). Values from Supplemental Table 4
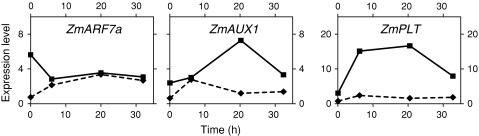
Fig. 3.
Relative levels of auxin-associated transcripts in the QC (filled squares on continuous line) and PM (filled diamonds on broken line) at various times following decapping (activation of the SCN). Values from Supplemental Table 4
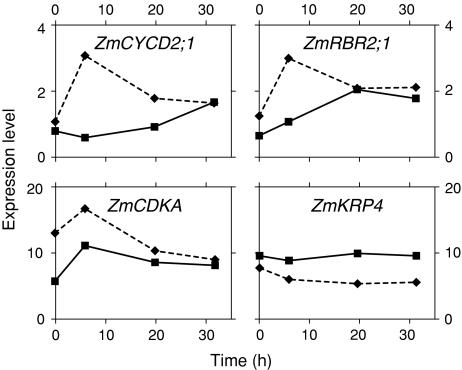
Fig. 4.
Relative levels of cell cycle-related transcripts in the QC (filled squares on continuous line) and PM (filled diamonds on broken line) at various times following decapping (activation of the SCN). Values from Supplemental Table 4
Fig. 5.
Relative levels of redox-associated transcripts in the QC (filled squares on continuous line) and PM (filled diamonds on broken line) at various times following decapping (activation of the SCN). Values from Supplemental Table 4
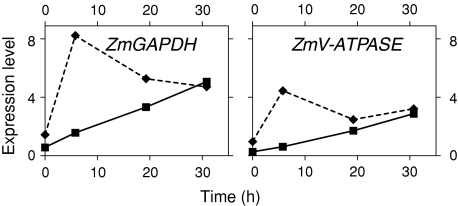
Fig. 6.
Relative levels of metabolism-associated transcripts in the QC (filled squares on continuous line) and PM (filled diamonds on broken line) at various times following decapping (activation of the stem cell niche). Values from Supplemental Table 4
Regulators of transcription
The expression of a GATA-type zinc finger transcription factor (ZmGATA) and the A-type response regulator (ZmRR) are both reduced in the QC, compared to their status in the PM and RC (Table 1, Supplemental Table 3), whereas the expression of an Argonaute-related gene (ZmAGO4) and an AT-hook motif gene (ZmATH) are both enriched in the QC. With niche activation, ZmGATA activity increases threefold in the QC and nearly sixfold in the PM (Fig. 2 and Supplemental Table 4).
Auxin related
We quantified the spatial expression of three genes in the auxin efflux carrier family (ZmPINs), one gene in the auxin influx carrier family (ZmAUX1s), eight genes in the AUXIN RESPONSE FACTOR (ZmARFs) family and one gene in the PLETHORA (ZmPLTs) family. Genes with increased expression in the QC, and thus overlapping in their spatial expression patterns, include ZmARF5/MP, ZmARF7a, ZmAUX1 and ZmPLT (Table 2, Supplemental Table 3). This is the first report that quantitatively measures, localizes and compares the levels of these important auxin-associated transcripts in the root SCN (QC and PM).
Activation of the QC by decapping resulted in a substantial increase in the expression in both the QC (4–8-fold) and PM (2–4-fold) of two auxin-regulated genes (ZmAUX1 and ZmPLT), whereas the expression of a third gene, ZmARF7a, is reduced in the QC portion of the niche, but increased substantially in the PM portion (Fig. 3 and Supplemental Table 4).
Cell cycle regulators
We quantified seven genes, the level of transcription of which is often associated with regulation of the cell cycle (Table 3, Supplemental Table 3). In general, the levels of these transcripts are lower in the QC compared to the PM and RC, but with the two prominent exceptions: CYCLIN-DEPENDENT KINASE REGULATORY SUBUNIT (ZmCKS1) and cyclin-dependent kinase inhibitor KIP-RELATED PROTEIN (ZmKRP4) genes, both of which show enriched expression in the QC. Significantly, the transcript levels of both of these genes in the non-dividing cells of the EZ are at levels tenfold less than in the QC (Table 3). In this regard, transcripts of a RETINOBLASTOMA (Rb) homolog (ZmRBR2;1) (Rymen et al. 2007) can be detected in the QC, PM and RC (Table 3, Supplemental Table 3), and its level is significantly greater in the RC (5-fold> than in the QC, and 3-fold>than in the PM) (Table 3).
Activation of the QC resulted in an equalizing of the transcript levels in the PM and the QC, so that by 32 h after decapping transcript levels for each gene (D-type cyclin [ZmCYCD2;1], ZmRBR2;1, and an A-type cyclin-dependent kinase [ZmCDKA]) changed from a ratio of approximately 2:1 (PM:QC) to nearly 1:1, mostly as a consequence of an increased expression in the QC (Fig. 4 and Supplemental Table 4). Changes in expression of ZmRBR2;1 are especially noteworthy (3-fold increase in the QC) (Fig. 4).
Redox related (ascorbate–glutathione cycle)
We observed reduced expression in both the QC and, with some exceptions, in the PM of the suite of genes encoding redox-regulating enzymes (Table 4, Supplemental Table 3). Zmγ-GCS and ZmGS, which encode proteins associated with the second to the last, and the last step, respectively, in the synthesis of glutathione, are expressed two to threefold higher in the PM than in other root tissues.
Decapping leads to an increase in the expression of six genes encoding proteins central to regulating redox status (glutathione reductase [ZmGR], glutathione synthetase [ZmGSH], γ-glutamylcysteine synthetase [Zmγ-GCS], monodehydroascorbate reductase [ZmMDAR], ascorbate oxidase [ZmAAO] and dehydroascorbate reductase [ZmDHAR]) (Fig. 5 and Supplemental Table 4). For both the QC and PM, the expression of these genes peaks 6–20 h after decapping, with levels, in general, at 32 h (the last time point) 2–22-fold higher than at time 0, immediately after RC removal.
Energy/metabolism related
The expression of two metabolism-/energy-associated transcripts encoding glyceraldehyde-3-phosphate dehydrogenase (ZmGAPDH) and a vacuolar ATP synthase (ZmV-ATPASE) is reduced in the QC (5–9-fold less) and in the PM (2–3-fold less), in comparison to the adjacent RC (Table 5 and Supplemental Table 3). Compared to the QC, cells of the EZ have similar levels of expression of ZmGAPDH, but more than twofold the level of expression of the ZmV-ATPASE.
Following decapping, the expression of ZmV-ATPASE and ZmGAPDH increases markedly (5–10-fold) with QC activation (Fig. 6 and Supplemental Table 4).
Tight clustering results
By performing the tight clustering analysis on the pooled qPCR data of the 19 selected genes from PM and QC (30 expression profiles in total), we obtained one tight cluster consisting of 12 expression profiles (Supplemental Fig. 1). This is also the only significant tight cluster we found in the data. In this cluster, the identified 12 expression profiles, showing a very strong coherent expression pattern, correspond to 12 different genes in the PM.
Overexpression in Arabidopsis of GATA2, AHL1, RHC1a and other TF genes
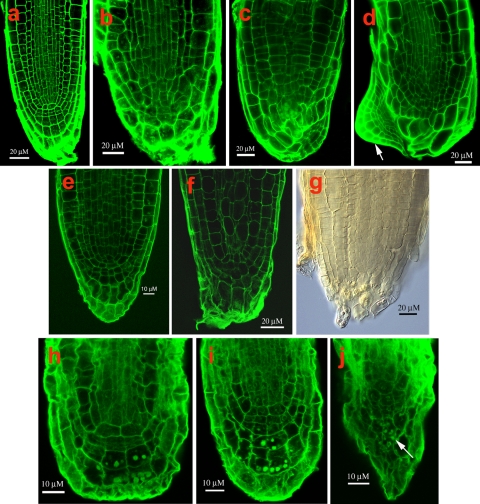
Because of a central role for TFs in linking transcriptional networks (Iyer-Pascuzzi and Benfey 2008), we selected for further analysis four TF genes showing reduced expression, and four TF genes showing enriched expression in the maize QC (each from different TF gene families), and overexpressed their sequence homologs in Arabidopsis (Supplemental Table 1 and 2). Overexpression in Arabidopsis of the GATA2 (At2g45050), AHL1 (At4g12080) (AT-hook protein) and RHC1a (At2g40830) (RING-H2) TF genes under the control of the 35S-promoter (overexpression confirmed by qPCR, Supplemental Table 6) resulted in alterations ranging from subtle to profound in the development of the root SCN and of the adjacent root cap (RC). Apical meristems of roots overexpressing the GATA2 TF gene were disorganized with no distinct boundary between the RC and the root proper. Cap cells were often enlarged and their shapes distorted (Fig. 7b–d). In one overexpressing line, a new root meristem formed adjacent to the enlarged cap cells (Fig. 7d). Roots overexpressing the AHL1gene displayed the most variability in apical meristem architecture, varying in apical patterning from relatively typical to grossly abnormal (Fig. 7e–g). The region occupied by the QC often penetrated into the RC, destroying the RC initials, resulting in a decrease in size and often disappearance of the RC (Fig. 7f, g). In roots overexpressing RHC1a, apical architecture is also altered, first in the cells of the QC that become atypically enlarged, followed by changes in the shapes and activities of the RC initials, resulting in a highly disorganized RC and in the root becoming determinate (Fig. 7h–j). No root phenotypes were observed in the T-DNA insertion lines for GATA2 (Salk_008743, Salk_008794 and Salk_008845), for AHL1 (Salk_024324, Salk_026823, Salk_052728, and Salk_086713, Salk_092100), and for RHC1a (Salk_127503, Salk_143110, Salk_137684, Salk_037121), suggesting redundancy of gene function.
Fig. 7.
Confocal microscope views of Arabidopsis root apical meristems from plants overexpressing various transcription factor genes (TF). a Wild type. b–d GATA2 TF; arrow in d points to a displaced meristem. e–g AHL1 TF; h–j, RHC1a TF; arrow in j points to amyloplasts
Discussion
The objectives for this work are to understand the factors underlying SCN specification and elaboration in maize roots, and to determine whether a molecular “signature” can be discerned for the root SCN. Toward addressing these questions, we first developed a baseline for gene expression in the QC and PM using a global microarray-based approach. We found that differences in gene expression between the QC and PM were quantitative and not qualitative, and using qPCR we ascertained how gene activity changed (or did not) in both SCN populations following activation and perturbation of the niche. Our working hypothesis is that genes essential to and/or linked to niche specification will have their activities altered when the niche is perturbed (Xu et al. 2006). Genes that we have confirmed, or have newly hypothesized as important in SCN formation and maintenance, fall into the broad categories discussed below.
A role for ascorbate/monodehydroascorbate/dehydroascorbate cycle genes in SCN specification and maintenance
One of the emerging themes in biology is that stem cells have attributes of cells under stress (Ramalho-Santos et al. 2002; Baharvand et al. 2006). In maize, as a consequence of a highly oxidized microenvironment at the root tip, the cells comprising the QC are under oxidative stress, a condition that has been linked to shifts in the ratios of the oxidized and reduced forms of GSH and AA in the QC, favoring an oxidized redox status (Jiang et al. 2003). Thus, for roots, as for other organisms [e.g., yeast, C. elegans (Essers et al. 2005)], a strong correlation exists between oxidative stress and the development and maintenance of the SCN. Evidence supporting a role for redox status in niche establishment/maintenance comes from work on the Arabidopsis mutant, ROOTMERISTEMLESS, characterized by a defect in GSH synthesis (Vernoux et al. 2000), resulting in disorganization of the root meristem shortly after germination. This phenotype can be rescued by supplying GSH. Similarly, our earlier work showing niche disorganization following decapping is interpreted as occurring as a consequence of the changes in the redox status in both the QC and PM (Jiang et al. 2003). Here, we show that the oxidized status in the QC is, in part, paralleled by the relatively low levels in the QC of transcripts encoding proteins that regulate the ascorbate/monodehydroascorbate/dehydroascorbate cycle, including ZmGR, ZmMDAR, ZmDHAR and other associated enzymes (Table 4). Based on the kinetics and the magnitude of transcript changes following decapping (Fig. 5), we would hypothesize that the development of the less oxidizing environment is mainly a consequence of an increase in the activity (3–30-fold) of the ascorbate/monodehydroascorbate/dehydroascorbate cycle and to a lesser degree due to the synthesis of more glutathione (GSH), the biosynthesis of which in the activated QC is probably low, judging by the low levels of expression of transcripts encoding 2 key GSH-biosynthesis enzymes (γ-glutamyl-cysteine synthetase [γ-GCS] and glutathione synthetase [GS], Fig. 5).
Regulation of cell cycle genes’ transcription is associated with SCN maintenance
Stereotypically, adult stem cells divide relatively slowly, often accumulating at cell cycle checkpoints (Robert 2004). QC cells typify this pattern (Clowes 1961). The maintenance of these characteristics is essential for meristem organization (Willemsen et al. 1998; Riou-Khamlichi et al. 1999; Wildwater et al. 2005). In contrast, PM cells that occupy the basal portion of the SCN divide relatively more frequently, about once every 20–30 h. Toward an understanding of differences in the regulation of the cell cycle between the two stem cell populations, and other non-stem cells, we focused on specific transcripts associated with core cell cycle regulators, including transcripts encoding ZmCYCD2;1, ZmCYCL1;1, ZmCDKA, ZmKRP4, ZmCKS1 and ZmRBR2;1 (Table 3). Here, we provide the first spatial measurements of the levels of cell cycle-associated gene activities in the root SCN. Low ZmRBR2;1 transcription in the non-activated maize QC accords well with reports linking a reduction of RBR function with the maintenance of the stem cell state in Arabidopsis (Wildwater et al. 2005). In maize, we show that there is at least a threefold difference (increase) in RBR expression when the QC is activated (Fig. 4). Thus, for maize our results suggest the possibility that re-patterning of the root SCN is associated with an increase of ZmRBR2;1 expression in the QC.
G1-to-S transition is also governed by an increased expression of cyclin D (Menges et al. 2005), which Wildwater et al. (2005) suggests influences RBR control of root stem cell pool size. Our results with maize roots support this possibility, since activation of the niche leads to differences (increases) in ZmCYCD2;1 expression (Fig. 4), reinforcing the notion that maintenance of the stem cell cycle hinges on a balance of the CYCD/RBR pathway.
Although the levels of most cell cycle-associated transcripts are reduced in the QC relative to the RC and PM, we found enhanced expression of two transcripts encoding negative regulators of the cell cycle, ZmCKS1 and ZmKRP4 (De Veylder et al. 2001; Wang et al. 2006) (Table 3, Supplemental Table 3). ZmCKS1 proteins associate with cyclin-dependent kinases (CDKs) and play an essential role in the regulation of the cell cycle. ZmCKS1 is abundant in non-dividing cells, and its level is reduced during G1/S transition (Menges et al. 2005). Overexpression of CKS1 in Arabidopsis causes a reduction in root growth rates, which is primarily a consequence of an increase in the duration of the cell cycle and a shortening of the meristem (De Veylder et al. 2001). The increased levels of expression of both of these genes in the QC may point to an underlying mechanisms contributing to the reduced rate of cell division in the QC.
Other regulators of transcription related to SCN establishment and maintenance
We identified several different types of transcription factors (TFs) that are enriched or reduced in the QC (Supplemental Tables 1, 2). Because of the central role for TFs in linking gene networks, we selected for further study four genes representing different classes of regulators of transcription, including ZmRR, the homolog of ARABIDOPSIS RESPONSE REGULATOR (ARR) genes. Ioio et al. (2007) hypothesize that members of this family interact with cytokinins to control the rate of cell differentiation in the meristem by affecting the position of the transition zone (TZ) where cells pass from being meristematic to elongating and differentiating. Our data point to a role for ZmRR in the PM (site of the transition zone), but not in the QC. In support of this view, we note that for the one member of the RR gene family that we investigated in detail (OS009399, an Arabidopsis homolog of ARR8/ATRR3), the expression in the QC is exceedingly low and unchanged by decapping and activation (Table 1 and Fig. 2), whereas decapping caused a threefold increase in ZmRR expression in the PM (Fig. 2 and Supplemental Table 4). Thus, we suggest that development and maintenance of the QC portion of the SCN may involve a repression of interactions between cytokinins and members of the RR gene family.
A second gene studied in more detail is ZmAGO4, the homolog in Arabidopsis which has been linked to endogenous siRNA, suggesting the possibility of epigenetic silencing (Baumberger and Baulcombe 2005). In maize, we report that this homolog is expressed most highly in the QC portion of the SCN and is barely detectable in the EZ (Table 1). Previous work which suggests that AtAGO4 and long siRNAs direct chromatin modifications, including histone methylation and non-CpG DNA methylation (Zilberman et al. 2003), may point to other important mechanisms underlying development and maintenance of the SCN. Because decapping results in an increase in ZmAGO4 expression (Fig. 2 and Supplemental Table 4), this suggests that AGO4-mediated processes may be associated with specification of the niche.
Finally, to obtain additional evidence for a role for TFs and their associated networks in niche specification and/or maintenance, we focused on three additional TFs, an AT-hook protein and two Zn finger proteins, (a RING-H2 type and a GATA type). For each, we overexpressed their homologs (AHL1, RHC1a and GATA2) in Arabidopsis, which resulted in mild to severe alterations in root meristem architecture (Fig. 7a–j). Of the three TF types that we overexpressed, GATAs have received the most attention. Recent studies also point to an important role for GATAs in stem cell maintenance (Zhao et al. 2004; Liu et al. 2005; Manfield et al. 2007; Naito et al. 2007). HAN, which encodes a GATA-3-like protein in Arabidopsis, controls the number and position of cells expressing WUS and thereby affects cell proliferation and differentiation. As for GATA2, Manfield et al. (2007) report that it is strong transcriptionally and co-expresses with a significant number of genes with roles in cell wall assembly, suggesting that GATA2 could be a possible link to proliferation/differentiation processes.
Transcript profiles from the maize and Arabidopsis QCs show similarities, but also differences
In 2005, Nawy et al. reported the transcriptional profile for the Arabidopsis QC. Using a technical approach that involved protoplasting and sorting of GFP-expressing QC cells, obtained from many roots, they found enrichment in the QC of 290 genes, which they grouped into various functional categories (their Table I). Satisfyingly, we find similarities in the expression profiles of Arabidopsis (a eudicot with a small QC) to the QC of maize (a monocot with a much larger QC), including a number of transcripts associated with transcriptional regulators, DNA replication/repair and auxin. Two specific groupings not reported for the Arabidopsis QC, but detected in maize, include a relatively large number of enriched transcripts for mitochondrial/plastid genes, and with the exception of thioredoxin reductase (Nawy et al. 2005), transcripts associated with the redox status (Table 4 and Supplemental Table 2). Additionally, we report a large number of genes with significantly reduced expression in the maize QC (Supplemental Table 1) and which were not reported in the profile data for the Arabidopsis QC (Nawy et al. 2005), thereby providing additional insights into the possible molecular controls of the SCN.
Activation of the maize SCN provides additional support for a role for redox-, metabolism- and cell cycle-associated transcripts in SCN specification
Xu et al. (2006) perturbed the stem cell status in Arabidopsis by laser ablating one or more QC cells, resulting in a loss of expression of typical QC auxin-associated markers (e.g., PLT1, WOX5). Sena et al. (2009) have likewise explored genes in relation to niche specification during meristem root regeneration in Arabidopsis. They excised 130 μm of the root tip (which includes the QC and PM), and then followed gene expression in the 70 μm stump region distal to the excision. Of the gene families (auxin-, redox-, metabolism- and cell cycle-associated) for which we find increased expression in the reforming maize SCN, the only overlap with the early expressed genes in the reforming Arabidopsis SCN is for members of the PLT gene family (auxin-related genes).
Activation and re-establishment of the SCN reveals similarities and differences in transcript expression in the QC and PM
While the SCN activation experiments point to similarities in the underlying molecular machinery that may be important in the PM and QC in defining stemness, the experiments also suggest differences between these two zones. These differences are most evident from a consideration of the kinetics of changes in gene expression patterns following decapping. Taken as a group, PM transcripts generally peak at 6 h and then decrease and stabilize 20–32 h after decapping, while the expression of QC transcripts does not show a coherent pattern. When we performed a cluster analysis for all the PM and QC transcripts, we found a high expression at the second time point and a low expression at the first time point for most PM transcripts in the tightest cluster. But no QC transcripts are found to have this tightest cluster expression pattern. From this analysis, we conclude that differences in the expression patterns/levels of genes in the QC versus PM may be important in defining distinct regions of the niche.
Conclusions
We conclude that there is no specific gene expression “signature”, which distinguishes one stem cell sub-population from the other. Further, we suggest that the pattern of increased expression of auxin-regulated genes, coupled with the reduced expression of metabolism-, redox- and some cell cycle-associated genes, comes closest to a root SCN “signature” and supports the notion that “stemness” represents a state rather than a quantifiable entity (Zipori 2004; Mikkers and Frisén 2005).
Our findings not only extend the recent work on Arabidopsis to the specification of the maize root SCN, but also, as importantly, we have been able to add significantly to the number of genes enriched and reduced in both the QC and PM. We thereby are able to suggest additional steps and processes possibly associated with the specification/maintenance of the root SCN, including; controls of the state of differentiation of QC cells, processes for maintaining DNA integrity (repair machinery), a possible role for cytokinins specifically in the PM portion of the niche, a role for gene silencing in niche stabilization and an underlying and central role for redox. Finally, with these new results, we demonstrate that plant and animal stem cells share an increased number of molecular characteristics, and in many ways, therefore, are more alike than they are different.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Dr. Peter Repetti of Mendel Biotechnology, Hayward, CA for kindly providing seeds of Arabidopsis overexpressing lines. We thank Dr. Shibo Zhang for assisting with the analysis of the microarray data.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AA
Ascorbic acid
- AAO
ASCORBATE OXIDASE
- ARF
AUXIN RESPONSE FACTOR
- AGO4
Argonaute-related gene 4
- AHL1
AT-hook motif nuclear localized protein 1
- ATH
AT-hook protein gene
- APX
ASCORBATE PEROXIDASE
- ARR
ARABIDOPSIS RESPONSE REGULATOR (ARR)
- AUX1
Auxin influx transporter
- CAF-1
CHROMATIN ASSEMBLY FACTOR-1
- CDKA
A-type cyclin-dependent kinase
- CKS1
CYCLIN-DEPENDENT KINASE REGULATORY SUBUNIT
- CYCD
D-type cyclin
- CYCC
C-type cyclin
- CYCL
L-type cyclin
- DHAR
DEHYDROASCORBATE REDUCTASE
- EZ
ELONGATION ZONE
- FAS2
FASCIATA 2
- GAPDH
GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE
- GATA2
GATA-type zinc finger protein 2
- GCS
γ-GLUTAMYLCYSTEINE SYNTHETASE
- GLDH
L-GALACTONO-1, 4-LACTONE DEHYDOROGENASE
- GR
GLUTATHIONE REDUCTASE
- GS
GLUTATHIONE SYNTHASE
- GSH
Glutathione
- KRP
KIP-RELATED PROTEIN
- MDAR
MONODEHYDROASCORBATE REDUCTASE
- PIN
PIN-FORMED
- PLT
PLETHORA
- PM
Proximal meristem
- QC
Quiescent center
- Rb
RETINOBLASTOMA
- RbR
RETINOBLASTOMA RELATED
- RC
Root cap
- RHC1a
RINGH-H2 group C protein 1a
- SCR
SCARECROW
- SCN
Stem cell niche
- TF
Transcription factor
- V-ATPASE
Vacuolar ATP synthase
- WUS
WUSCHEL
- Zm
Zea mays
References
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development. 2002;129:3965–3974. doi: 10.1242/dev.129.17.3965. [DOI] [PubMed] [Google Scholar]
- Baharvand H, Hajheidari M, Ashtiani SK, Salekdeh GH. Proteomic signature of human embryonic stem cells. Proteomics. 2006;6:3544–3549. doi: 10.1002/pmic.200500844. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W. Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol. 2004;7:732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung J, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Blau HM, Brazelton TR, Weimann JM. The evolving concept of a stem cell: entity or function? Cell. 2001;105:829–841. doi: 10.1016/S0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, Sadot E, Yalovsky S. Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol Biol Cell. 2005;16:1913–1927. doi: 10.1091/mbc.E04-07-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Weiss ML, Rao MS. In search of “stemness”. Exp Hematol. 2004;32:585–598. doi: 10.1016/j.exphem.2004.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Huang H, Blackshaw S, Liu JS, Cepko C, Wong WH. Clustering analysis of SAGE data using a Poisson approach. Genome Biol. 2004;5:R51. doi: 10.1186/gb-2004-5-7-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campilho A, Garcia B, Toorn HVD, Wijk HV, Campilho A, Scheres B (2006) Time-lapse analysis of stem-cell divisions in the Arabidopsis thaliana root meristem. Plant J 48:619–627 [DOI] [PubMed]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Clowes FAL. Apical meristems. Oxford: Blackwell; 1961. [Google Scholar]
- Cockcroft S, Carvou N. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beemster GTS, Beeckman T, Inzé D. CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 2001;25:617–626. doi: 10.1046/j.1365-313x.2001.00996.x. [DOI] [PubMed] [Google Scholar]
- Essers MAG, De Vries-Smits LMM, Barker N, Polderman PE, Burgering BMT. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Feldman LJ. Cytokinins and quiescent center activity in roots of Zea. In: Torrey JG, Clarkson D, editors. The development and function of roots. London: Academic Press; 1975. pp. 55–72. [Google Scholar]
- Feldman LJ, Torrey JG. The isolation and culture in vitro of the quiescent center of Zea mays. Am J Bot. 1976;63:345–355. doi: 10.2307/2441580. [DOI] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luiten M, Wilmsen V, Bilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lang BM, Moughamer T, Xia Y, Budworth P, Zong J, Miguel T, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:1686–1696. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioio RD, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Benfey PN. Transcriptional networks in root cell fate specification. Biochim Biophys Acta. 2008;1789:315–325. doi: 10.1016/j.bbagrm.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Meng YL, Feldman LJ. Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development. 2003;130:1429–1438. doi: 10.1242/dev.00359. [DOI] [PubMed] [Google Scholar]
- Jiang K, Zhang S, Lee S, Tsai G, Kim K, Huang H, Chilcott C, Zhu T, Feldman LJ. Transcription profile analyses identify genes and pathways central to root cap functions in maize. Plant Mol Biol. 2006;60:343–363. doi: 10.1007/s11103-005-4209-4. [DOI] [PubMed] [Google Scholar]
- Kaya H, Shibahara K-i, Taoka K-i, Iwabuchi M, Bruce Stillman B, Takashi Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- Kim K, Zhang S, Jiang K, Cai L, Lee IB, Feldman LJ, Huang H. Measuring similarities between gene expression profiles through data transformations. BMC Bioinform. 2007;8:29–43. doi: 10.1186/1471-2105-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Koizuka N, Martin RC, Nonogaki H. The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 2005;44:960–971. doi: 10.1111/j.1365-313X.2005.02588.x. [DOI] [PubMed] [Google Scholar]
- Manfield IW, Devlin PF, Jen C-H, Westhead DR, Gilmartin PM. Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 2007;143:941–958. doi: 10.1104/pp.106.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C, Yang LJ, Meinke D. The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell. 2000;12:1379–1392. doi: 10.1105/tpc.12.8.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- Mikkers H, Frisén J. Deconstructing stemness. EMBO J. 2005;24:2715–2719. doi: 10.1038/sj.emboj.7600749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatiyan A, Szüts D, Krude T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Mol Cell Biol. 2006;26:1839–1849. doi: 10.1128/MCB.26.5.1839-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T. Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2007;71:1557–1560. doi: 10.1271/bbb.60692. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Benfey PN (2002) Signaling in and out: control of cell division and differentiation in the shoot and root. Plant Cell 14 Suppl:S265–S276 [DOI] [PMC free article] [PubMed]
- Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcription profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Wu H-M, Cheung AY. RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci. 2006;11:309–315. doi: 10.1016/j.tplants.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- O’Brien KP, Remm M, Sonnhammer ELL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33 (Database Issue):D476–D480 [DOI] [PMC free article] [PubMed]
- Park Y, Gerson SL. DNA repair defects in stem cell function and aging. Annu Rev Med. 2005;56:495–508. doi: 10.1146/annurev.med.56.082103.104546. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Soonsang Y, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Ridgway P, Almouzni G. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J Cell Sci. 2000;113:2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- Robert JS. Model systems in stem cell biology. BioEssays. 2004;26:1005–1012. doi: 10.1002/bies.20100. [DOI] [PubMed] [Google Scholar]
- Ruzin SE. Plant microtechnique and microscopy. New York: Oxford University Press; 1999. [Google Scholar]
- Rymen B, Fiorani F, Kartal F, Vandepoele K, Inzé D, Beemster GTS. Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol. 2007;143:1429–1438. doi: 10.1104/pp.106.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoye T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/S0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Scherer GF, Zahn M, Callis J, Jones AM. A role for phospholipase A in auxin-regulated gene expression. FEBS Lett. 2007;581:4205–4211. doi: 10.1016/j.febslet.2007.07.059. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu H-Y, Hofhuis H, Birnbaum KD. Organ regeneration does not require a functional stem cell niche in plants. Nature. 2009;457:1150–1153. doi: 10.1038/nature07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Tseng GC, Wong WH. Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics. 2005;61:10–16. doi: 10.1111/j.0006-341X.2005.031032.x. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signaling. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Benfey P. Signals that regulate stem cell activity during plant development. Curr Opin Genet Dev. 2005;15:388–394. doi: 10.1016/j.gde.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld J-P, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al. The root meristemless1/cadmium sensitive 2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell. 2000;12:97–110. doi: 10.1105/tpc.12.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Fowke LC. The emerging importance of cyclin-dependent kinase inhibitors in the regulation of the plant cell cycle and related processes. Can J Bot. 2006;84:640–650. doi: 10.1139/B06-043. [DOI] [Google Scholar]
- Wildwater M, Campiho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Wolkenfelt H, De Vrieze G, Weisbeek P, Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development. 1998;125:521–531. doi: 10.1242/dev.125.3.521. [DOI] [PubMed] [Google Scholar]
- Xu J, Scheres B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell. 2005;17:525–536. doi: 10.1105/tpc.104.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, J Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell. 2004;16:2586–2600. doi: 10.1105/tpc.104.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen W, Provart N, Chang H-S, Guimil S, Su WP, Zou G, Wang X. Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J. 2003;1:59–70. doi: 10.1046/j.1467-7652.2003.00006.x. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Zipori D. The nature of stem cells: state rather than entity. Nat Rev Genet. 2004;5:873–878. doi: 10.1038/nrg1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.