Abstract
The molecular chaperone, heat shock protein 70 (Hsp70), acts at multiple steps in a protein’s life cycle, including during the processes of folding, trafficking, remodeling and degradation. To accomplish these various tasks, the activity of Hsp70 is shaped by a host of co-chaperones, which bind to the core chaperone and influence its functions. Genetic studies have strongly linked Hsp70 and its co-chaperones to numerous diseases, including cancer, neurodegeneration and microbial pathogenesis, yet the potential of this chaperone as a therapeutic target remains largely underexplored. Here, we review the current state of Hsp70 as a drug target, with a special emphasis on the important challenges and opportunities imposed by its co-chaperones, protein-protein interactions and allostery.
Keywords: proteostasis, flavonoids, dihydropyrimidines, spergualin, sulfoglycolipids, geranylgeranyl acetone, protein folding, ATPase, protein-protein interactions
1. Introduction
Heat-shock protein 70 (Hsp70) is a molecular chaperone that interacts with exposed hydrophobic amino acid in polypeptides [1–4]. This interaction assists in the folding of newly synthesized proteins and minimizes their aggregation, but, in addition to these activities, Hsp70 also binds partially folded substrates and regulates their functions [5–8]. For example, this chaperone is required for clathrin re-modeling during endocytosis, it stabilizes proteins associated with apoptotic and nuclear hormone signaling and it assists in protein turnover [9–12]. Thus, Hsp70 is able to intersect with a protein during virtually every stage of its life cycle: from primary folding to function and degradation. Moreover, Hsp70 is thought to bind promiscuously, so it is able to serve as a core chaperone for the proteome [13, 14]. Because of this diversity and breadth of functions, Hsp70 is considered a central mediator of protein homeostasis and, not surprisingly, it has been implicated in many diseases as a potential therapeutic target.
Despite the promise of Hsp70 as a pharmacological objective, its targeting is complicated by a number of factors. For example, Hsp70 is abundant (~ 1–2% of total cellular protein) and highly conserved, with approximately 50% sequence identity between prokaryotic and mammalian family members. In addition, many organisms express multiple Hsp70s (e.g. 13 in humans) and members are found in all the major subcellular compartments. The complexity of Hsp70’s functions (e.g. folding, degradation, trafficking and remodeling) and its ubiquitous expression patterns create numerous challenges in designing safe and effective therapeutics [15]. How can specific Hsp70 functions (e.g. folding) be disrupted to achieve desired therapeutic outcomes? Can subsets of Hsp70 substrates be preferentially disrupted, despite the broad activity of the chaperone? Can prokaryotic Hsp70s be selectively targeted for anti-bacterial applications, despite the high sequence homology? The field of Hsp70 therapeutics is in its infancy, so many of these questions remains unanswered. However, in this review, we will discuss Hsp70’s roles in disease and specifically focus on how structure and function studies might assist identification of therapeutic leads.
2. Structure and Function of Hsp70
Domain architecture
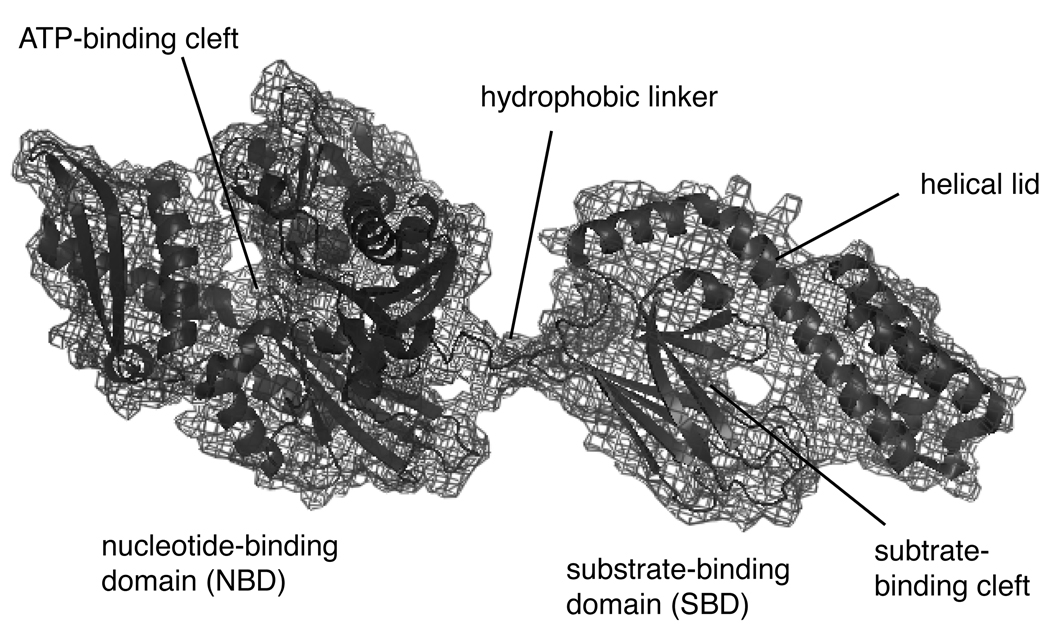
Hsp70 is a 70 kDa molecular machine that binds hydrophobic peptide sequences, hydrolyzes ATP and directs its substrates into a variety of distinct fates. These tasks are accomplished by a relatively minimal structure composed of three major domains: a ~44 kDa N-terminal nucleotide binding domain (NBD), a ~15 kDa substrate binding domain (SBD) and a ~10 kDa C-terminal alpha helical, “lid” domain (Figure 1). The NBD contains the important site of ATP binding and hydrolysis. When ATP is bound, the SBD and NBD exhibit coupled motion, suggestive of their tight association [16, 17]. Also in this ATP-bound form, the lid domain remains open, which facilitates transient interactions with substrates (Figure 2). Following ATP hydrolysis, a conformational change releases the SBD, resulting in closure of the lid and a ~10-fold increase in the affinity for substrate [18, 19]. The conformation change associated with ATP hydrolysis is communicated through a key proline switch and involves the conserved, hydrophobic linker that connects the NBD to the SBD [20]. Together, these structural and biochemical studies have begun to reveal the dynamic changes in Hsp70 that accompany nucleotide hydrolysis and substrate binding [21]. However, the intrinsic ATPase rate of Hsp70 is remarkably slow (on the order of 0.2 nmol/µg/min) [22], so one question in chaperone biology is to understand how this enzyme is regulated and stimulated in vivo. Towards that end, multiple physiological factors, including substrates and co-chaperone proteins, bind Hsp70 and modify its nucleotide turnover. Because the ATPase activity of Hsp70 is central to controlling its structure and function, these regulatory interactions are likely key to understanding chaperone biology.
Fig. 1.
Domain architecture of heat shock protein 70 (Hsp70). An N-terminal nucleotide-binding domain (NBD) contains the nucleotide binding and hydrolysis center. The substrate-binding domain (SBD), which binds to exposed hydrophobic polypeptides, is attached through a short linker. The C-terminus of the SBD is a mostly helical subdomain, which is termed the “lid”. The prokaryotic Hsp70, DnaK, is shown (PDB code #2HKO), but the general architecture appears to be conserved amongst prokaryotic and eukayrotic family members.
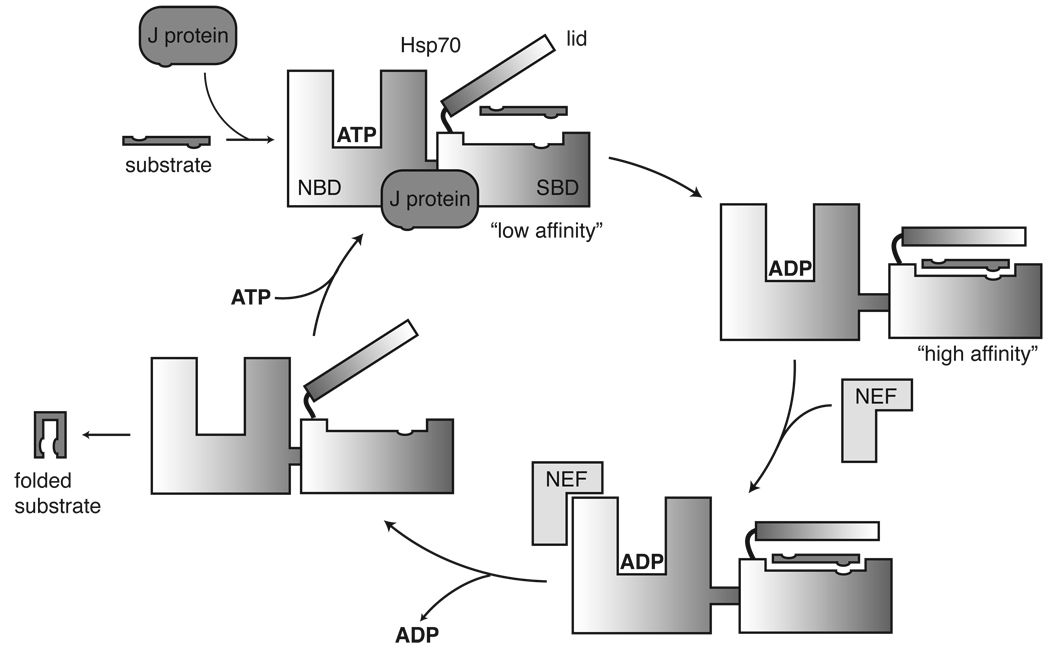
Fig. 2.
Nucleotide hydrolysis, allostery and substrate binding in the Hsp70 complex. Substrate binding in the SBD, coupled with J-domain co-chaperone interactions in the NBD, promote ATP hydrolysis. Conformational changes associated with ATP conversion close the lid and enhance affinity for the substrate. To complete the cycle, nucleotide exchange factor interacts with the NBD and assists with ADP release. The released substrate can adopt one of many fates, including proper folding (as shown).
J-domain co-chaperones
The J-domain proteins are a large class of Hsp70-associated co-chaperones; there are six of these proteins in Escherichia coli, ~20 in Saccharomyces cerevisiae and ~40 in humans [23, 24]. These factors are characterized by a conserved ~70 amino acid J-domain, which is named after the founding member of the class, E. coli DnaJ. The main role of this domain is to stimulate the intrinsically slow ATPase activity of Hsp70 [25, 26] and the key region required for this process is an invariant histidine-proline-aspartic acid (HPD) motif, which resides in a loop between helix 2 and 3 of the J-domain [27–29]. Interactions between the J-domain and Hsp70’s NBD stimulate ATPase activity by approximately 5- to 10-fold [22, 30], resulting in enhanced substrate affinity. In addition to the J-domain, which is typicaly at their N-termini, members of this co-chaperone family contain a wide variety of distinct domains at their C-termini. The identity of this C-terminal domain is used for classification; briefly, proteins in class I and II contain domains involved in dimerization and substrate binding [31, 32], while the class III members have domains with a variety of predicted functions [33]. Consistent with this diversity of functions, deletion studies have suggested that individual J-protein co-chaperones play distinct cellular roles. For example, complementation studies involving thirteen cytosolic J-domain proteins revealed that at least four examples (Sis1, Jjj1, Jjj3, Cwc23) fulfill unique functions in yeast [34]. For Sis1, its C-terminal region was responsible for its specificity because fusing it to the J-domain of Ydj1 was sufficient to suppress the loss-of-function phenotype [35]. Similarly, the mammalian J-domain protein, DJA1, is competent for refolding of denatured proteins in vitro, but the related co-chaperone, DJA2, is not [36]. These and other studies have repeatedly shown non-overlapping roles for the J-proteins [24, 37, 38]; however, how these factors assemble and interchange on Hsp70 in vivo remains unclear.
Nucleotide exchange factor co-chaperones
Completion of Hsp70’s ATPase cycle requires release of ADP, which is a process that is catalyzed by another class of co-chaperones, the nucleotide exchange factors (NEFs). These co-chaperones accelerate ADP release through interactions with the NBD and they can be categorized into four distinct, evolutionarily unrelated families named after representative members: GrpE, Bag1, Hsp110 and HspBP1. Prokaryotic GrpE was shown to increase the rate of release of nucleotide from the bacterial Hsp70, DnaK, without affecting the rate of hydrolysis [39]. Similar findings were shown with the mammalian GrpE and mtHsp70 counterparts [40]. Together with DnaJ, GrpE stimulates the overall ATPase activity of DnaK by ~50 fold [41], which suggests that the catalytically active chaperone is minimally composed of this multi-protein complex [42, 43]. Co-crystallization revealed that a dimer of GrpE binds asymmetrically to a single molecule of DnaK and forces the two lobes of the NBD open by 14° [44]. In addition to its roles as a NEF, GrpE has also been shown to function as a thermosensor; it reversibly unfolds in response to heat shock, thus decreasing the rate of nucleotide exchange and favoring substrate retention until conditions improve [43, 45, 46]. The second class of NEFs is named after Bag-1 (Bcl-2-associated athanogene-1), which was the first eukaryotic exchange factor to be identified [47]. This family contains 6 members in humans, characterized by a C-terminal BAG domain that interacts with Hsp70. Like GrpE, the BAG domain induces a 14° outward rotation in the NBD lobes [48, 49]. The conservation of this mechanism is interesting because BAG domains are structurally and evolutionarily unrelated to GrpE. Importantly, the BAG family members typically possess a variety of additional protein-protein interaction motifs and, like the J-domain co-chaperones, these regions are thought to bring unique capabilities into the Hsp70 machinery. For example, Bag1 has an ubiquitin-like domain through which it interacts with the proteasome, stimulating degradation of Hsp70 substrates [50]. Although GrpE and Bag-1 share mechanistic features, the other major classes, HspBP1 and Hsp110, are both structurally and functionally distinct. For example, the co-crystal structure of HspBP1 revealed that this NEF reduces the affinity for nucleotide by binding to the NBD and displacing one lobe through a steric conflict [51]. Members of the Hsp110 family are structurally similar to Hsp70 and, consistent with this observation, they protect against aggregation of heat-denatured proteins in addition to their poorly understood NEF functions [52–56]. Importantly, the substrate-binding activity of Hsp110 is strictly associated with ‘holdase’ outcomes and these factors are not competent in the active refolding of denatured substrates. Together, these four classes of NEFs comprise a diverse array of important co-chaperones that interact in only partially overlapping regions on the Hsp70 surface. Moreover, like J-domain proteins, they possess separate, intrinsic capabilities and/or interact with other cellular pathways to recruit Hsp70 into a variety of tasks.
TPR-domain co-chaperones
The tetratricopeptide repeat (TPR) is a 34 amino acid motif found in a class of co-chaperones that bind at the extreme C-terminus of Hsp70’s lid domain, a region characterized by four conserved amino acids, EEVD. Interestingly, the C-terminal EEVD motif is found in both Hsp70 and Hsp90, but not the prokaryotic DnaK or the mitochondrial or ER-resident Hsp70 isoforms. The structure of a representative TPR reveals that it interacts with the negatively charged side chains of the linear EEVD peptide [57]. In addition, residues upstream of the conserved EEVD are also recognized and these sites have been hypothesized to provide specificity [58, 59]. Binding of some TPR domains appears to be dependent on the nucleotide state of Hsp70, for example, the TPR-domain protein, Hop, binds tighter to the ADP-bound form [60]. Moreover, yeast Sti1 can actively stimulate the ATPase activity of the yeast Hsp70 Ssa1 [61], suggesting crosstalk between the TPR co-chaperones and nucleotide turnover. In addition to their ability to bind Hsp70 and regulate ATP hydrolysis, the various TPR-containing proteins exhibit an array of additional activities, provided via their other domains. For example, Hop has two TPR domains and it mediates the association between Hsp70 and Hsp90 [62]. Hop-mediated substrate transfer between these major chaperone networks is thought to be critical to some functions, such as nuclear hormone receptor signaling [10, 63]. Conversely, CHIP (carboxyl terminus of Hsc70 interacting protein) is a TPR-domain protein that both inhibits J-domain-stimulated ATPase activity and also contains a U-box domain associated with E3 ubiquitin ligase activity. Through this modality, CHIP has been shown to ubiquitinate Hsp70 substrates, diverting them to the proteasome [64–67]. Thus, CHIP functions as a chaperone-associated, quality control monitor that can tag substrates for degradation. Knockouts of CHIP are viable and show distinct phenotypes [68], suggesting that this co-chaperone plays roles that are not entirely redundant with other TPR family members.
Combinatorial assembly of Hsp70 complexes
Mammalian cells contain approximately 13 Hsp70s, over 40 J-domain proteins, at least 4 distinct types of NEFs and (conservatively) dozens of proteins with TPR-domains. At any given time, an individual Hsp70 molecule can only interact with a single representative of each major class; therefore, in theory, tens of thousands of possible complexes might be formed (Figure 3). Thus, a major question in chaperone biology is how these various components are assembled and whether different combinations always lead to specific outcomes. The answer to this question might lead to a better understanding of how Hsp70 can engage in its dizzying array of biological process, from folding to trafficking, endocytosis and protein turnover. Moreover, understanding the emergent properties of each complex might provide insight into how Hsp70 can act on such a wide range of substrates. Most important for this discussion, the ‘combinatorial assembly’ hypothesis creates a number of important challenges and opportunities for drug discovery. For example, can compounds selectively block contact between Hsp70 and individual co-chaperones, thus shaping chaperone outcomes? Although chemical perturbation of protein-protein interactions has become increasingly common [69–71], the path to these compounds remains less straightforward. Given that Hsp70 is central to many cellular processes, effective and safe compounds might be required to act on specific Hsp70 complexes in order to avoid toxicity associated with global disruption of proteostasis.
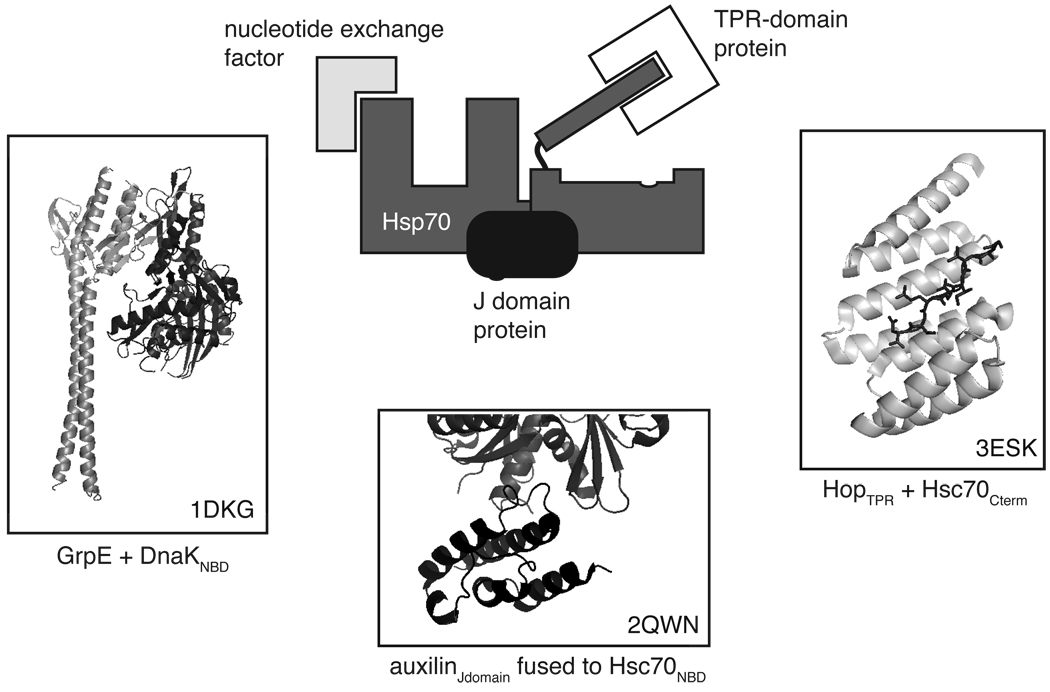
Fig. 3.
Hsp70 forms the core of a multi-protein machine. A least three distinct classes of co-chaperones interact with Hsp70 and regulate its activity, its localization and the fate of its associated substrates. Representative structures are shown to highlight the macro-molecular interactions and protein-protein surfaces involved in these regulatory conduits. The J domain co-chaperones and NEFs interact with the NBD, while the TPR-domain proteins bind the C-terminal region in the lid. In addition, each cell is thought to harbor multiple co-chaperones from each class, which suggests that combinatorial assembly can occur at these distinct surfaces. The identity of the specific structures and the corresponding PDB code numbers are included. As noted in the text, there are multiple classes of NEFs and only the GrpE interaction is pictured. All images were prepared in PyMol.
3. Roles of Hsp70 in Disease
Consistent with Hsp70’s many roles in the maintenance of the proteome, genetic studies have implicated this chaperone in numerous diseases. In some of these instances, disease pathology is associated with “too much” Hsp70 activity (e.g. aberrant stabilization of a specific substrate). In other examples, defects appear to arise from a failure of Hsp70 to properly recognize and remove misfolded substrates. Thus, disease might arise from disruption of the proteostasis network in either direction. These observations are certainly consistent with the complexity of hsp70s functions in protein processing. In this section, we will briefly outline some of the molecular roles for Hsp70 in disease and point out some of the remarkably diverse opportunities for therapeutic intervention.
Cancer
Hsp72, a stress inducible Hsp70, is known to directly inhibit several steps in apoptotic signaling, including both intrinsic and extrinsic pathways [72]. Hsp72 inhibits lysosomal membrane permeabilization [73], activation and translocation of pro-apoptotic factors such as JNK, BID, BAX, or AIF, and release of cytochrome c from mitochondria. In addition, Hsp72 has been shown to suppress cellular senescence pathways [74, 75]. Consequently, its levels are elevated in several cancer cell lines and overexpression of this chaperone is correlated with poor prognosis [76] and resistance to chemotherapies [77]. Important roles have also been attributed to the mitochondrial Hsp70 isoform, mtHsp70 or Grp75, which was identified as a mortality factor in cancer and thus named "mortalin". mtHsp is over-expressed in breast, colon and colorectal cancer cells and its upregulation has been shown to induce malignant transformation in a mouse model [78, 79]. Moreover, RNAi knockdown of the chaperone causes growth arrest in human immortalized cells [80]. Finally, the ER-localized BiP has also been implicated in cancer [81]. It is highly expressed in glioblastomas and its expression exhibited a negative relationship with patient survival [82]. Recently, BiP was found to promote angiogenesis and assist proliferation of tumor cells [83]. Together, these studies illustrate Hsp70’s anti-apoptotic roles, consistent with a general capacity as a stress-inducible, pro-survival factor.
Based on these studies, Hsp70 has been proposed to be a potential drug target in cancer. In part, this model is based on parallel studies on Hsp90. Briefly, Hsp90 is thought to both enhance the cellular capacity to accommodate otherwise toxic proteins, while also preventing the onset of apoptosis by stabilizing pro-survival substrates, such as Akt [84–89]. Inhibitors of Hsp90 show selective toxicity against cancer cells in vitro and they are currently in multiple clinical trials [90]. Interestingly, Hsp90 in tumor cells is predominantly complexed with its co-chaperones, whereas, normal cells contain higher levels of free (or latent) Hsp90 [91]. Consistent with this concept, the Hsp90 inhibitor, 17-AAG, binds with higher affinity to the Hsp90 complex than to the latent chaperone, suggesting a mechanism for specific toxicity in cancer cells. The analogous studies have not been thoroughly performed for Hsp70, but, as discussed above, this chaperone is also known to interact with numerous substrates and co-chaperones. Moreover, Hsp90 and Hsp70 appear to share common substrates in anti-apoptotic signaling and both chaperones are over-expressed in tumor cells, presumably because of the higher demand for protein folding and the stressful microenvironments that these cells encounter. Thus, Hsp70 may also be a good target for anti-cancer strategies in much the same way as Hsp90.
One interesting aspect of Hsp90-based treatments, which has yet to be thoroughly explored for Hsp70, is the ability of some Hsp90 inhibitors to induce a compensatory stress response. Hsp90 inhibitors lead to over-expression of Hsp72 and many other heat shock proteins, through release of HSF1. Briefly, the transcription factor, HSF1, is held in an inactive state by the Hsp90 complex [92, 93]. Accumulation of misfolded substrates diverts the chaperone and triggers nuclear localization of HSF1, which results in upregulation of targets involved in protection from cellular stress. This feedback loop has been hypothesized to decrease the overall effectiveness of Hsp90 inhibition by elevating the overall chaperone pool [94, 95]. Moreover, these observations suggest that the efficacy of Hsp90 inhibitors may be improved by co-administration with Hsp70 inhibitors. Consistent with this idea, Guo et al. used siRNA against Hsp70 to increase the efficacy of 17-AAG in cancer cells [96]. Recently, Powers et al. showed that simultaneous suppression of two cytosolic Hsp70s, Hsc70 and Hsp72, also sensitized cancer cells to 17-AAG [97]. Together, these findings highlight the inter-connectivity of the chaperone networks and the ability of the basic biological knowledge to assist in design of therapeutic strategies.
Neurodegenerative and protein misfolding diseases
Many neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases, are characterized by abnormal protein misfolding and accumulation [98–100]. In these disorders, misfolded proteins are not properly cleared by the chaperone / quality control system and, subsequently, they self-associate into cytotoxic oligomers. Conversely, other indications such as cystic fibrosis and Gaucher disease involve premature protein degradation [101]. For example, certain point mutations in the cystic fibrosis transmembrane receptor (CFTR) cause aberrant degradation in the secretory pathway, leading to pathology consistent with a loss-of-function. Thus, many important diseases involve alterations in protein homeostasis, but some involve seemingly insufficient chaperone capacity while others appear to arise from overactive chaperone decisions. Recently, the “proteostasis boundary” hypothesis has been put forward to describe this concept and its relationship with disease (for a review of this concept: [102]). Because of its central role in protein processing, it isn’t surprising that Hsp70 has been linked to both types of disorders [103, 104]. For example, Hsp70 has been over-expressed in S. cerevisiae [105], mouse [106, 107], Caenorhabditis elegans [108] and Drosophila melanogaster [109–111] models of neurodegenerative disease. In each of these experiments, high levels of Hsp70 were found to partially restore normal physiology or block protein aggregation. Similarly, pharmacological induction of Hsp70 has been found to be beneficial effect in some models; compounds such as 17-AAG or celasterol, which stimulate a stress response and enhance expression of heat shock proteins, are sometimes protective [112, 113]. This recovery is thought to result, in part, from enhanced cellular capacity to process proteins, combined with enhanced pro-survival signaling. Conversely, reduction of chaperone function has been found to be beneficial in cases involving premature degradation; or example, mutation of either Hsp70 or the J-domain co-chaperones, YDJ1 and HLJ1, reduces degradation of CFTR in models of cystic fibrosis [114, 115]. Thus, the relationship between Hsp70 and disease is complex, and the desired properties of a therapeutic are expected to be dependent on the specific damage to the proteostasis boundary.
Consistent with this idea, co-chaperones that regulate Hsp70 are also thought to be important in disease. For example, the interaction of Hsp70 with the TPR-domain proteins CHIP [116–118] and BAG2 [119] are important for clearance of tau, a protein implicated in Alzheimer’s disease. Similarly, over-expression of the J-domain co-chaperone, Hsp40, or the Hsp70-interacting protein, E6-AP, has been found to be sufficient to inhibit polyglutamine aggregation [105, 120] and RNA interference of the Hsp90 co-chaperone, Aha1, partially recovers CFTR stability [121]. Together, these genetic findings point to the Hsp70 complex (e.g. the core chaperone and its associated factors) as potential targets for pharmacologic intervention in neurodegenerative and protein misfolding disorders.
Microbial pathogenesis
The prokaryotic Hsp70, DnaK, has been strongly linked to bacterial survival under stress [122–125]. In addition, Hsp70s have also been linked to malarial infection [126] and host-pathogen interactions [127]. Consistent with these roles, knockouts of E. coli DnaK are viable under normal laboratory conditions, but they are sensitive to elevated temperature [128] or addition of antibiotics [124]. Moreover, null mutations in DnaK make Staphalococcus aureus less efficient in mouse infection models [125] and Streptococcus mutans dnakΔ strains have impaired biofilm formation [123]. Consistent with the entire Hsp70 chaperone system being important in disease, knockouts of the regulatory co-chaperone, DnaJ, also show growth defects under stress conditions [124, 129]. Together, these observations have led to the hypothesis that inhibitors of DnaK and its co-chaperones might have antibiotic applications. Importantly, the expected mechanism would be to sensitize pathogens towards additional stresses.
Other diseases
In addition to being implicated in cancer, protein misfolding disorders and infection, Hsp70 and its co-chaperones have been associated with numerous other diseases, including Crohn’s disease [130], Behcet’s disease [131], ischemic stroke [132] and ALS [133], and this chaperone also plays roles in normal aging [134, 135]. The breadth of these indications highlights the chaperone’s central role in protein homeostasis. Moreover, these observations suggest that chemical modulators of Hsp70 might find use in a wide variety of indications, but that therapeutic selectivity might be challenging to achieve.
4. Targeting the Nucleotide Binding Domain
As mentioned above, Hsp70 is composed of multiple domains, including the NBD and SBD. These domains each harbor specific activities and interact with key co-chaperones. Thus, Hsp70 is a dynamic, multi-component machine, with numerous potential locations for chemical manipulation. Accordingly, the known modulators of Hsp70 share little structural similarity and they bind multiple regions of the Hsp70 protein. Here, we will separate the compounds based on their chemical scaffold and their proposed binding location in either the NBD or the SBD. In addition, there are a few compounds whose binding sites have not been explored [136].
4.1 Nucleotide mimetics
Compounds that compete with ATP for binding to Hsp70 would be expected to have significant effects on chaperone function, as this turnover is required for many of its activities. By analogy, many of the successful Hsp90 inhibitors, which have been informative in mechanistic and therapeutic studies [137, 138], are ATP-competitive. Thus, compounds that displace ATP from Hsp70 are also expected to be powerful tools [72]. Unlike Hsp90, the Hsp70 chaperone binds to nucleotide using the actin-like fold, so compounds that bind Hsp90’s ATP-binding sites are not cross-reactive with Hsp70. To solve this problem, Williamson et al. recently designed and synthesized adenosine-derived molecules based on the X-ray crystal structure of a commercially available ATP analog in complex with Hsc70/Bag1 [139]. Several rounds of SAR studies afforded molecules with affinity for Hsp70 that are comparable to that of ATP; for example, compound 12 showed an IC50 of 0.5 µM by a fluorescent polarization assay and a KD of 0.3 µM by surface plasmon resonance (Figure 4). Consistent with a role for Hsp70 in cancer cell viability, this compound also exhibited a GI50 of 5 µM against human colon tumor (HCT)-116 cells and caused degradation of the Hsp90 clients, Her2 and Raf-1. Following the logic that Hsp90 inhibitors have been useful probes, it seems likely that these Hsp70-directed compounds will also be important reagents in the arsenal. Of course, further studies on their selectivity and potency are needed.
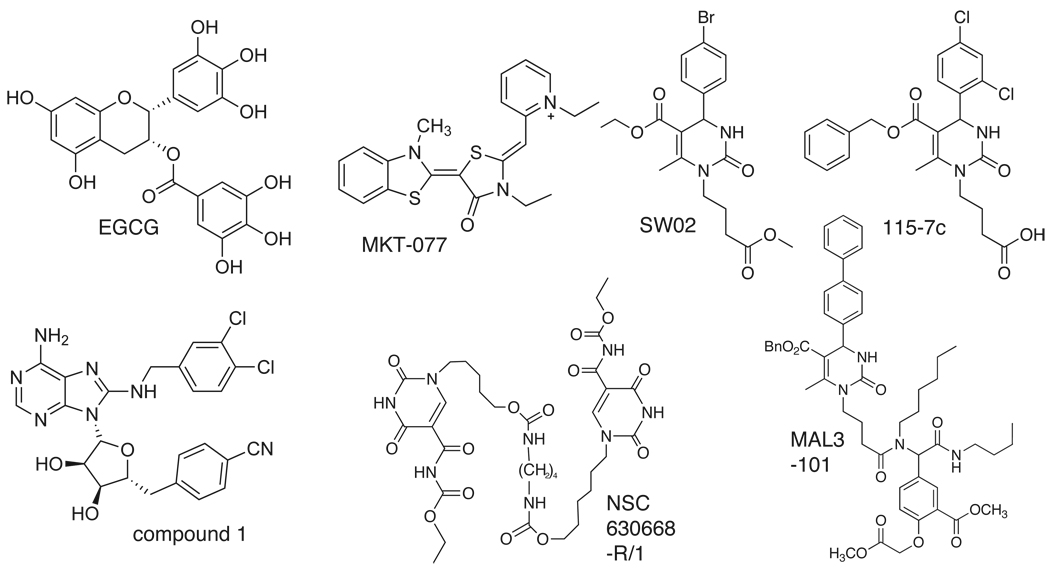
Fig. 4.
Structures of Hsp70 modulators that are thought to interact with the nucleotide-binding domain (NBD). In some cases, representative structures from a series are shown.
4.2 (−)-epigallocatechin gallate (EGCG)
EGCG is a naturally occurring polyphenolic flavonoid that has various biological activities, including its functions as an antioxidant. Among these functions, it has also been shown to interact with the ER-resident Hsp70, BiP, by affinity chromatography [140]. Subsequently, the binding site was localized to the NBD using deletion constructs and it was shown to compete with ATP for binding. Although the selectivity of this compound is uncertain, it was shown to enhance the apoptotic activity of the chemotherapeutic, etoposide, consistent with a role for BiP in pro-survival signaling. Thus, one of the activities of EGCG in cancer cells might involve partial inactivation of Hsp70 function. In support of this model, another flavonoid, myrectin, has recently been reported as an inhibitor of this chaperone (Jinwal et al. (in press)). This compound inhibits the ATPase activity of Hsp70 (IC50 ~ 10 µM) and initiates rapid, Hsp70-mediated degradation of the Alzheimer’s disease-related substrate, tau. Importantly, over-expression of Hsp70 significantly enhanced the potency of myrectin in cell-based models, consistent with this chaperone being a cellular target. It remains to be seen whether more selective compounds can be developed, based on these studies and the polyphenol structure.
4.3 MKT-077
The NBD is also thought to be the site of action of MKT-077, a cationic, rhodacyanine dye with selective toxicity against cancer cells. This compound killed CX-1 colon carcinoma cells (IC50 ~ 7 µM) but had no effect on normal epithelial cells [141] and it has been explored in phase I clinical trials as an anti-tumor agent [142]. Using a pull down assay, Wadhwa, et al. first showed that the molecule binds to mtHsp70 [143]. In support of the mtHsp70 binding, MKT-077 is known to localize to mitochondria, likely because it is cationic [144]. The same group also localized the binding site to the NBD of mtHsp70 using deletion mutants [145]. Interestingly, the binding site of MKT-077 overlapped with that of p53, suggesting that it might disrupt p53-mtHsp70 interactions and influence apoptotic signaling [146]. Selectivity wasn’t reported and the compound also appears to bind actin [147], but the ‘drug-like’ nature of this scaffold warrants further investigation.
4.4 Sulfoglycolipids
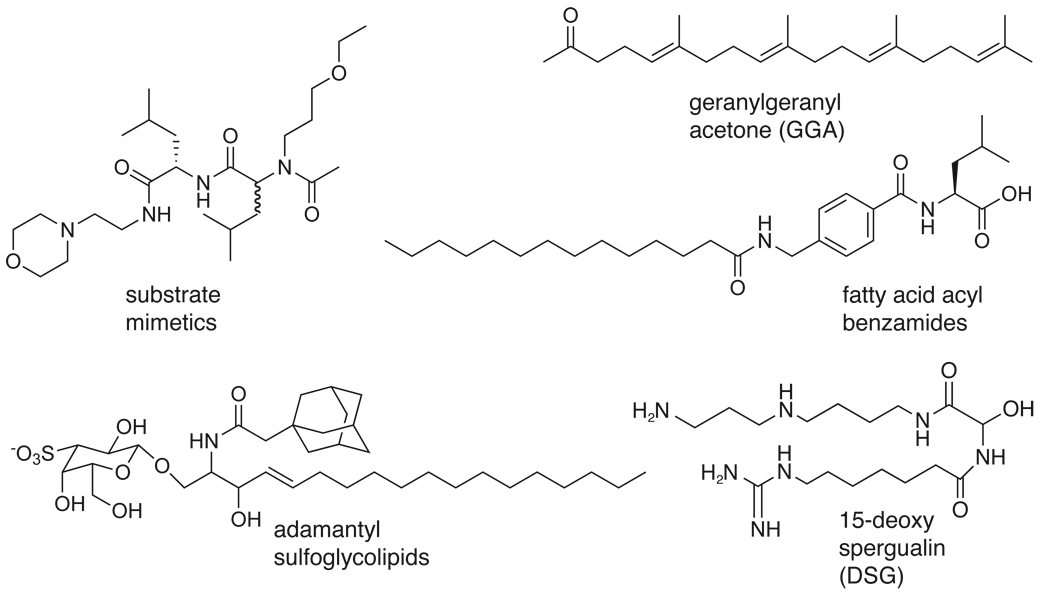
In binding assays, sulfogalactoglycerolipid (SGG) and sulfoglactosylceramide (SGC) were identified as having affinity for a testis-specific Hsp70 [148]. Using domain deletions and site-directed mutagenesis, the binding site was determined to reside in the NBD [149]. These compounds also bind Hsc70s from multiple organisms, highlighting the potential problem of selectivity in this highly conserved family. Interestingly, these compounds did not affect the Km of ATP turnover but they did decrease Vmax, suggesting a noncompetitive mechanism. Further structural studies will be required to understand whether these compounds operate by an allosteric binding site. However, such studies seem worthwhile, as a derivative of SGC, adamantylSGC, inhibited the ATPase activity of bovine brain Hsc70, a potential target in neurodegenerative disease [150]. Because of the similarity of these compounds to endogenous glycolipids, it is also interesting to hypothesize that they might be taking advantage of intrinsic regulatory mechanisms (e.g. mimicking interactions between Hsp70 and signaling lipids) [151].
4.5 Dihydropyrimidines
Because ATPase activity appears to be important for many chaperone functions, screens for this activity have been useful tools for identifying new inhibitors and activators [15]. Using this approach and a subset of the NCI drug collection, NSC 630668-R/1 (R/1) was identified as an inhibitor of both endogenous and J-domain stimulated ATPase activity against two different Hsp70s, Ssa1p and BiP. Moreover, this compound blocked Hsp70-dependent protein translocation in cells [152]. R/1 contains a keto-functionalized pyridine core and, based this general heteroaromatic structure, a more focused cheminformatic search was conducted. These efforts led to the testing of 31 small molecules, mostly peptoid-functionalized 2-dihydropyrimidinones generated by sequential Biginelli and Ugi reactions. This pilot library was screened for the ability to modulate the endogenous and J-protein stimulated ATPase activity of recombinant Ssa1p. From these efforts, compounds such as MAL3–101 were identified as inhibitors of J-domain protein stimulated Hsp70 activity. Conversely, MAL3–90 and related compounds enhance the endogenous ATPase activity but inhibit J-domain co-chaperone activity [153]. Thus, compounds from this class are able to modulate ATP turnover in either direction and their functions depend on the co-chaperone context. Given the complexity of Hsp70 biology, access to these different classes of compounds may be fortuitous. In support of this idea, dihydropyrimidines are able to tune Hsp70-mediated luciferase folding activity in either direction [154]. Following these initial studies, a focused library of functionalized dihydropyrimidines was assembled by microwave-assisted, in situ Biginelli cyclocondensation at the terminus of a resin-bound β-peptide. A series of di and tripeptides with a range of hydrophobic and polar side chains were utilized to explore SAR in an ATPase assay against DnaK [22, 154]. These results confirmed that dihydropyrimidines operate as either stimulators or inhibitors of nucleotide hydrolysis. Members of this collection also showed some selectivity for Hsp70 isoforms, as one of these substituted dihydropyrimidines inhibited bovine Hsc70 but not E. coli DnaK [155]. Finally, hydrophobic groups pendant to the dihydropyrimidinone core were found to be important for activity.
Although the potency of these first-generation compounds is not yet optimized and they have modest IC50 values ranging from approximately 10 to over 200 µM, representatives of the class have been shown to possess interesting biological activities. Compound SW02, which stimulates ATPase activity, was shown to enhance the ability of Hsp70 to block aggregation of amyloid beta, an important target in Alzheimer’s disease target [156]. In another model, analogs of MAL3–101 were shown to inhibit proliferation of SK-BR-3 cell cancer cells, presumably by modulating the anti-apoptotic functions of Hsp70 [157]. A similar derivative, MAL2-11B, was shown to inhibit the activity of a viral J-domain protein, T antigen. This activity was associated with a block in viral replication in a plaque assay, suggesting that MAL2–11B may represent a new class of polyomavirus inhibitors [158]. Select pyrimidinone-peptoid hybrid compounds also exhibit potent effects on the malaria parasite, Plasmodium falciparum, and were shown to inhibit its replication in human red blood cells [159]. Recently, compounds from this class have also been shown to alter processing of the microtubule-associated protein, tau. Specifically, the dihydropyrimidines, SW02 and 115-7c, stimulate the ATPase activity of Hsp70 in vitro and these compounds also promote tau accumulation in models of Alzheimer’s disease (Jinwal et al. (in press)). These results highlight the interesting relationship between Hsp70 and the stability of its substrates. While more potent derivatives will likely arise from further studies, the first-generation probes have been successfully used in a diverse collection of applications.
5. Targeting the Substrate Binding Domain
5.1 Substrate mimetics
A number of short, antibacterial peptides, including drosocin, pyrrhocoricin and apidaecin, have been shown to bind E. coli DnaK [160]. These peptides are interesting because an analog of pyrrhocoricin exhibits broad-spectrum antibacterial activity against both gram-positive and gram-negative species. Competition experiments suggest that pyrrhocoricin (Kd ~ 50 µM) binds to DnaK at two different sites, including the substrate-binding cleft of the SBD. Using fragments of the chaperone, another binding site was identified within the C-terminal “lid” domain. The interaction between this peptide and DnaK inhibits ATP turnover in vitro and folding of model substrates in vivo [161], but it isn’t clear if these particular activities are responsible for the observed anti-bacterial function. Interestingly, pyrrhocoricin, failed to bind DnaK from S. aureus and it also had no anti-bacterial activity against this species, consistent with this chaperone as a target. In addition to these natural antibacterial peptides, Haney et al. synthesized peptides that were patterned after DnaK’s model substrate, NRLLLTG [162]. One of these peptides activated the ATPase activity of yeast Hsp70 Ssa1p by 10 % at 300 µM and modeling confirmed that this compound could occupy the SBD cleft. The stereochemistry of this peptide was important for function, as the opposite diasteromer was inactive. Together, these studies indicate that targeting the substrate-binding pocket is an effective strategy for manipulating chaperone function. Another interesting observation in these studies was that pyrrhocoricin had no activity against human Hsc70, suggesting that this binding site might be leveraged to gain selectivity.
5.2 Geranylgeranyl acetone (GGA)
GGA induces the expression of heat shock proteins and this compound has been used to activate the stress response [163, 164]. For many of the pharmacological inducers of the stress response, such as celasterol, their molecular mechanisms are only beginning to emerge [165, 166]. In an analogous attempt to explore how GGA operates, this compound was immobilized onto Sepharose and potential cellular targets identified by mass spectrometry. Interestingly, the major protein was found to be Hsp70 itself [167]. Subsequent experiments using truncated domains of Hsp70 suggest that the binding site of GGA is within the SBD. It still isn’t clear why this interaction leads to a stress response. However, the similarity between GGA and the endogenous lipid modifications found on some proteins, such as farnesyl and geranylgeranyl groups, raises the interesting possibility that Hsp70 might natively recognize these lipidated substrates, but this hypothesis has not been thoroughly explored. Moreover, like many of the scaffolds, there has not been much chemistry performed on GGA or its derivatives, so its utility as a drug lead is largely unexplored.
5.3 Acyl benzamides
In addition to their ATPase activity, Hsp70s are known to possess amide peptide bond cis/trans isomerase (APIase) activity [168]. This activity was specifically targeted by Liebscher et al., who designed and synthesized fatty acyl benzamides, consisting of three chemical moieties; a fatty acid, an aromatic linker and an amino acid residue (Figure 5) [169]. One of these compounds inhibited the APIase activity of DnaK with an IC50 value of 2.7 µM and bound with a KD of 2.6 µM. The compound also inhibited the chaperone-mediated refolding of denatured firefly luciferase with an IC50 value of 9.5 µM. This compound was shown to compete with a model peptide for binding to DnaK, suggesting that it binds to the SBD. This compound was also tested for its antibacterial activity against E. coli and it was shown to decrease viability at a nonpermissive temperature (42 °C) with an MIC of ~380 µg/mL. This action mirrors the failure of Δdnak strains to grow at elevated temperatures, consistent with this chaperone as a cellular target. Also consistent with anti-Hsp70 functions, electrophoresis experiments showed a significant increase in the amount of insoluble protein in the treated cells. To explore the potential of the acyl benzamide scaffold as a drug lead, the SAR was explored using both DnaK’s APIase activity and antibacterial potency as indicators. Variations in the fatty acid moiety, including the number of carbons and unsaturations, impacted inhibition of DnaK in vitro, but the SAR did not correlate well with antibacterial activity. Similarly, substitution on the benzoic acid linker increased MIC but had no effect on the activity against DnaK. Together, these SAR studies suggest that antibacterial activity may involve cellular targets in addition to DnaK. However, it is important to note that Hsp70 is a complex machine and the relationships between APIase activity and other in vivo functions are uncertain. To better understand this relationship, the focused collection was examined for effects on chaperone-mediated refolding of firefly luciferase, but again the SAR of antibacterial activity and folding efficiency did not correlate. Despite some mechanistic questions, these acyl benzamides appear to be promising anti-bacterial leads. It will be interesting to see if they have selectivity for prokaryotic Hsp70 isoforms and whether they might be used in other systems to manipulate APIase and chaperone functions.
Fig. 5.
Structures of Hsp70 modulators that are thought to interact with the substrate-binding domain (SBD). In some cases, representative structures from a series are shown.
5.4 Spergualin Derivatives
The natural product spergualin was first identified as an antibiotic from culture filtrates of Bacillus laterosporus BMG162-aF2 [170]. Subsequently its structure was determined to be (15S)-1-amino-19-guanidino-11,15-dihydroxy-4,912-triazanonadecane-10,13-dione and only the (−)-spergualin enantiomer was found to be active. The Umezawa group accomplished the first total synthesis of spergualin and its analogues by the acid-catalyzed condensation of ω-guanidino alkanamides with glyoxyloylspermidine. They showed that both the carbon chain length in the polyamine region and the 15-hydroxyl group affected activity [171, 172]. Using these compounds in T-cell assays, they focused on (−)-15-deoxyspergualin (DSG) as a potent immunosuppressive agent [173]. To identify the cellular targets of DSG, human T-cell lysates were analyzed by affinity chromatography using Sepharose beads covalently coupled to methoxy-DSG [174]. Using Western blotting and peptide sequencing, the interacting protein was identified to be Hsc70. This interaction was further characterized through affinity capillary electrophoresis to obtain binding affinities for purified Hsc70 (KD = 4 µM). Further studies revealed that DSG and its analogs stimulate the ATPase activity of Hsc70 (~2-fold; Km = 3 µM), but had no effect on stimulation by J-domain co-chaperones or on the release of substrate [175]. To further clarify the binding site of the compound, 14C-DSG was cross-linked to purified bovine Hsc70 by addition of EDC (1-ethyl-3-(3-dimethoxyaminopropyl)-carbodiimide). Interestingly, mass spectrometry analysis of the resulting peptide fragments localized the binding site to the C-terminal amino acids, EEVD, which is the same site of binding to the TPR-domain co-chaperones. Consistent with this binding site, DSG was also found to bind Hsp90 with an affinity of 5 µM [176].
DSG is likely the Hsp70 modulator with the best-characterized clinical utility. Although DSG was originally identified as a potential anti-bacterial agent, it also has anti-tumor activity and it has found clinical use in organ transplantation and autoimmune diseases [177–179]. DSG prolongs survival time after allogenic graft and minimizes acute rejection. Unfortunately, its mechanism-of-action is poorly understood and the potential roles played by its Hsp70/90 binding are unclear. One possibility arises from observations that DSG interferes with antigen processing/presentation by monocytes, which is an Hsp70-dependent process [180]. Alternatively, DSG has been found to block B cell development by inhibiting the nuclear translocation of transcription factor NF-κB [181], another activity that may involve Hsp70 and Hsp90. Similarly, it was shown that DSG kills the malaria parasite by interfering with the trafficking of essential proteins [182]. Unlike other immunosuppressive agents, such as FK506 and cyclosporin A, DSG does not suppress cytokine production (IL-1, IL-2) but does inhibit cytotoxic lymphocyte (CTL) induction [183]. Consistent with this model, DSG prophylaxis with FK506 improves long-term graft survival in renal transplant patients [184]. Unfortunately, DSG has a very low bioavailability (<5%) and is very unstable in aqueous solution because of its labile hydroxylglycine group. To address this issue, Renaut et al., developed an alternative synthetic route in which this group was substituted by a malonyl unit. In a graft-versus-host disease (GVHD) model in mice, these derivatives were found to be significantly more stable, although the SAR around the spermidine moiety is restricted [185]. One of the lead compounds, LF 15-0195, was less toxic and more potent than DSG in a renal allograft rejection primate model, suggesting that further chemical optimization might improve the performance of these compounds [186]. Thus, the strong biological data around these compounds make a case for further development. At the same time, there are many uncertainties in their molecular mechanism-of-action and there are pharmacological hurdles to overcome.
6. Targeting the Co-Chaperones
6.1 J-protein substrate mimetics
As discussed above, J-domain co-chaperones deliver substrate peptides to Hsp70 and stimulate ATP turnover. Consistent with the importance of these functions, Bischofberger et al. reported that D-peptides targeting E. coli DnaJ inhibit chaperone-dependent luciferase refolding in vitro [187]. The peptides were further shown to inhibit DnaJ stimulated ATPase activity of DnaK, which genetic studies have shown to be critical under physiological conditions. These compounds are thought to compete with natural substrates for binding to DnaJ, thus preventing its capacity to engage in productive interactions with the chaperone complex. These results highlight one of the interesting aspects of Hsp70 as a target; compounds might impact Hsp70 functions indirectly via the essential co-chaperones. Such approaches might be expected to have lower toxicity and greater selectivity, although this model remains to be tested.
6.2 Inhibitors of the TPR domain
Another strategy that has been reported recently is direct targeting of the TPR-domain. Using an AlphaScreen high-throughput approach, Yi and Regan identified pyrimidotriazinediones that interfere with the Hop-Hsp90 interaction [188]. These compounds were toxic to WST-1 cells in vitro, consistent with an important role for this contact. In theory, a similar route could be used to develop selective inhibitors that decouple this class of co-chaperones from their contact with Hsp70.
7. Prospectus and Future Opportunities
Hsp70 regulates multiple aspects of protein homeostasis. As such, it has been linked to numerous diseases and it has been suggested as a potential therapeutic target for many indications. Genetic and biochemical studies support this model and limited pharmacological findings have also provided intriguing insights. However, the discovery of potent, selective and well-characterized Hsp70 modulators remains an on-going task. Given recent clinical success with other proteins in the proteostasis network, such as Hsp90 or the proteasome, aggressive pursuit of Hsp70 seems worthwhile. The challenge will be to identify potent and selective chemical scaffolds that target the many functions of this chaperone.
In this review, we have focused on the interesting structural biology of Hsp70 and its many co-chaperones and how these complexes might provide unique opportunities for drug discovery. Specifically, Hsp70 operates as part of a combinatorial, multi-protein complex, with protein-protein interfaces, allosteric sites, a catalytic center and a distinct substrate-binding cleft. Partly owing to this structural and regulatory complexity, many different chemical scaffolds have been reported to interact with Hsp70 and their binding sites are spread across the chaperone’s surface. Importantly, many of these first-generation compounds have biological activities and both activators and inhibitors appear to have useful applications. While the complexity of Hsp70 makes it a challenging target, the same complexity also creates opportunities to design molecules with interesting capabilities. Based on the limited evidence collected thus far, we suggest that the toolbox of Hsp70-targeted compounds will include compounds that operate at distinct sites and impact distinct functions. Ultimately, each disease system might require a specific Hsp70 modulator that tunes the proteostasis boundary to the desired and appropriate level. Thus, the goal of this pharmacological intervention will be to restore balance to the system and the number and diversity of chemical probes may need to mirror the complexity of the biology.
Acknowledgements
Our work on Hsp70 is funded by grants from the McKnight Foundation, the Alzheimer’s Association (IIRG-07-60067), NIH (NS059690) and NSF (MCB-0844512). The authors thank Jeffrey Brodsky, Betty Craig, Chad Dickey, Martin Duennwald, Jill Johnson, Jeffrey Kelly, Bill Pratt, and Erik Zuiderweg for particularly helpful conversations.
REFERENCES
- 1.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125(3):443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66(4):840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 4.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 5.Kramer G, et al. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16(6):589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 6.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2(10):885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28(10):541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42(1):1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 9.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. Embo J. 2008;27(2):328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pratt WB, et al. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006;172:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8(6):640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 12.Nollen EA, Morimoto RI. Chaperoning signaling pathways: molecular chaperones as stress-sensing 'heat shock' proteins. J Cell Sci. 2002;115(Pt 14):2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- 13.Erbse A, Mayer MP, Bukau B. Mechanism of substrate recognition by Hsp70 chaperones. Biochem Soc Trans. 2004;32(Pt 4):617–621. doi: 10.1042/BST0320617. [DOI] [PubMed] [Google Scholar]
- 14.Rudiger S, et al. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. Embo J. 1997;16(7):1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6(11):1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 16.Bertelsen EB, et al. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuermann JP, et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31(2):232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittung-Stafshede P, et al. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 2003;42(17):4937–4944. doi: 10.1021/bi027333o. [DOI] [PubMed] [Google Scholar]
- 19.Slepenkov SV, Witt SN. Kinetic analysis of interdomain coupling in a lidless variant of the molecular chaperone DnaK: DnaK's lid inhibits transition to the low affinity state. Biochemistry. 2002;41(40):12224–12235. doi: 10.1021/bi0263208. [DOI] [PubMed] [Google Scholar]
- 20.Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21(3):359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Swain JF, et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26(1):27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang L, et al. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Qiu XB, et al. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vos MJ, et al. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47(27):7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- 25.Russell R, et al. DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry. 1999;38(13):4165–4176. doi: 10.1021/bi9824036. [DOI] [PubMed] [Google Scholar]
- 26.Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95(11):6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genevaux P, et al. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics. 2002;162(3):1045–1053. doi: 10.1093/genetics/162.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269(7):5446–5451. [PubMed] [Google Scholar]
- 29.Huang K, et al. Backbone dynamics of the N-terminal domain in E. coli DnaJ determined by 15N- and 13CO-relaxation measurements. Biochemistry. 1999;38(32):10567–10577. doi: 10.1021/bi990263+. [DOI] [PubMed] [Google Scholar]
- 30.Gassler CS, et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci U S A. 1998;95(26):15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo A, et al. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. Embo J. 1996;15(2):408–417. [PMC free article] [PubMed] [Google Scholar]
- 32.Acebron SP, et al. DnaJ recruits DnaK to protein aggregates. J Biol Chem. 2008;283(3):1381–1390. doi: 10.1074/jbc.M706189200. [DOI] [PubMed] [Google Scholar]
- 33.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3(1):28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A. 2007;104(17):7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19(11):7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzankov S, et al. Functional divergence between co-chaperones of Hsc70. J Biol Chem. 2008;283(40):27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hennessy F, et al. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14(7):1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Braun AP, Braun JE. Biological roles of neural J proteins. Cell Mol Life Sci. 2008;65(15):2385–2396. doi: 10.1007/s00018-008-8089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Packschies L, et al. GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry. 1997;36(12):3417–3422. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- 40.Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. Embo J. 1997;16(20):6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberek K, et al. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88(7):2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierpaoli EV, et al. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J Biol Chem. 1998;273(12):6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- 43.Siegenthaler RK, Christen P. Tuning of DnaK chaperone action by nonnative protein sensor DnaJ and thermosensor GrpE. J Biol Chem. 2006;281(45):34448–34456. doi: 10.1074/jbc.M606382200. [DOI] [PubMed] [Google Scholar]
- 44.Harrison CJ, et al. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276(5311):431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 45.Groemping Y, Reinstein J. Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol. 2001;314(1):167–178. doi: 10.1006/jmbi.2001.5116. [DOI] [PubMed] [Google Scholar]
- 46.Bimston D, et al. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. Embo J. 1998;17(23):6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takayama S, et al. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80(2):279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 48.Sondermann H, et al. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291(5508):1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 49.Briknarova K, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol. 2001;8(4):349–352. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- 50.Demand J, et al. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11(20):1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 51.Shomura Y, et al. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17(3):367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 52.Polier S, et al. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133(6):1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Andreasson C, et al. Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc Natl Acad Sci U S A. 2008;105(43):16519–16524. doi: 10.1073/pnas.0804187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andreasson C, et al. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008;283(14):8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 55.Raviol H, et al. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. Embo J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaner L, et al. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280(50):41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- 57.Scheufler C. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101(2):199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 58.Odunuga OO, et al. Tetratricopeptide repeat motif-mediated Hsc70-mSTI1 interaction. Molecular characterization of the critical contacts for successful binding and specificity. J Biol Chem. 2003;278(9):6896–6904. doi: 10.1074/jbc.M206867200. [DOI] [PubMed] [Google Scholar]
- 59.Brinker A, et al. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 × Hop × Hsp90 complexes. J Biol Chem. 2002;277(22):19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 60.Carrigan PE, et al. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J Biol Chem. 2004;279(16):16185–16193. doi: 10.1074/jbc.M314130200. [DOI] [PubMed] [Google Scholar]
- 61.Wegele H, et al. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278(28):25970–25976. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- 62.Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273(52):35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 63.Wegele H, et al. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356(3):802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 64.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Connell P, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 66.Meacham GC, et al. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 67.Jiang J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 68.Min JN, et al. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28(12):4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 70.Gestwicki JE, Crabtree GR, Graef IA. Harnessing chaperones to generate small-molecule inhibitors of amyloid beta aggregation. Science. 2004;306(5697):865–869. doi: 10.1126/science.1101262. [DOI] [PubMed] [Google Scholar]
- 71.Gestwicki JE, Marinec PS. Chemical control over protein-protein interactions: Beyond inhibitors. Combi. Chem. High Throughput Screen. 2007;10(8):667–675. doi: 10.2174/138620707782507296. [DOI] [PubMed] [Google Scholar]
- 72.Powers MV, Clarke PA, Workman P. Death by chaperone: HSP90, HSP70 or both? Cell Cycle. 2009;8(4):518–526. doi: 10.4161/cc.8.4.7583. [DOI] [PubMed] [Google Scholar]
- 73.Nylandsted J, et al. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200(4):425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yaglom JA, Gabai VL, Sherman MY. High levels of heat shock protein Hsp72 in cancer cells suppress default senescence pathways. Cancer Res. 2007;67(5):2373–2381. doi: 10.1158/0008-5472.CAN-06-3796. [DOI] [PubMed] [Google Scholar]
- 75.Gabai VL, et al. Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol Cell Biol. 2009;29(2):559–569. doi: 10.1128/MCB.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10(2):86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pocaly M, et al. Overexpression of the heat-shock protein 70 is associated to imatinib resistance in chronic myeloid leukemia. Leukemia. 2007;21(1):93–101. doi: 10.1038/sj.leu.2404463. [DOI] [PubMed] [Google Scholar]
- 78.Dundas SR, et al. Mortalin is over-expressed by colorectal adenocarcinomas and correlates with poor survival. J Pathol. 2005;205(1):74–81. doi: 10.1002/path.1672. [DOI] [PubMed] [Google Scholar]
- 79.Wadhwa R, et al. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int J Cancer. 2006;118(12):2973–2980. doi: 10.1002/ijc.21773. [DOI] [PubMed] [Google Scholar]
- 80.Wadhwa R, et al. Reduction in mortalin level by its antisense expression causes senescence-like growth arrest in human immortalized cells. J Gene Med. 2004;6(4):439–444. doi: 10.1002/jgm.530. [DOI] [PubMed] [Google Scholar]
- 81.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67(8):3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 82.Lee HK, et al. GRP78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neuro Oncol. 2008;10(3):236–243. doi: 10.1215/15228517-2008-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dong D, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68(2):498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 84.Taldone T, Sun W, Chiosis G. Discovery and development of heat shock protein 90 inhibitors. Bioorg Med Chem. 2009;17(6):2225–2235. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bishop SC, Burlison JA, Blagg BS. Hsp90: a novel target for the disruption of multiple signaling cascades. Curr Cancer Drug Targets. 2007;7(4):369–388. doi: 10.2174/156800907780809778. [DOI] [PubMed] [Google Scholar]
- 86.Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev. 2006;26(3):310–338. doi: 10.1002/med.20052. [DOI] [PubMed] [Google Scholar]
- 87.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 88.Mahalingam D, et al. Targeting HSP90 for cancer therapy. Br J Cancer. 2009;100(10):1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271(37):22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 90.Banerji U. Heat shock protein 90 as a drug target: some like it hot. Clin Cancer Res. 2009;15(1):9–14. doi: 10.1158/1078-0432.CCR-08-0132. [DOI] [PubMed] [Google Scholar]
- 91.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425(6956):407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 92.Guo Y, et al. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem. 2001;276(49):45791–45796. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- 93.Zou J, et al. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 94.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets. 2009;13(4):469–478. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 95.Bagatell R, et al. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents. Clin Cancer Res. 2000;6(8):3312–3318. [PubMed] [Google Scholar]
- 96.Guo F, et al. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylaminodemethoxy geldanamycin. Cancer Res. 2005;65(22):10536–10544. doi: 10.1158/0008-5472.CAN-05-1799. [DOI] [PubMed] [Google Scholar]
- 97.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell. 2008;14(3):250–262. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 98.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426(6968):900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 99.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 100.Koo EH, Lansbury PT, Jr, Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci U S A. 1999;96(18):9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 102.Powers ET, et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 103.Barral JM, et al. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15(1):17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 104.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6(1):11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 105.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci U S A. 2000;97(4):1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cummings CJ, et al. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet. 2001;10(14):1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 107.Klucken J, et al. Hsp70 Reduces alpha-Synuclein Aggregation and Toxicity. J Biol Chem. 2004;279(24):25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 108.Fonte V, et al. Interaction of intracellular beta amyloid peptide with chaperone proteins. Proc Natl Acad Sci U S A. 2002;99(14):9439–9444. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chan HY, et al. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9(19):2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 110.Auluck PK, et al. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295(5556):865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 111.Warrick JM, et al. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23(4):425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 112.Hay DG, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13(13):1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 113.Waza M, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11(10):1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y, et al. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12(5):1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Youker RT, et al. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15(11):4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dickey CA, et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci U S A. 2008;105(9):3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dickey CA, et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J Neurosci. 2006;26(26):6985–6996. doi: 10.1523/JNEUROSCI.0746-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Petrucelli L, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13(7):703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 119.Carrettiero DC, et al. The cochaperone BAG2 sweeps paired helical filament-insoluble tau from the microtubule. J Neurosci. 2009;29(7):2151–2161. doi: 10.1523/JNEUROSCI.4660-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mishra A, et al. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem. 2008;283(12):7648–7656. doi: 10.1074/jbc.M706620200. [DOI] [PubMed] [Google Scholar]
- 121.Wang X, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127(4):803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 122.Henderson B, Allan E, Coates AR. Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect Immun. 2006;74(7):3693–3706. doi: 10.1128/IAI.01882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lemos JA, Luzardo Y, Burne RA. Physiologic effects of forced downregulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol. 2007;189(5):1582–1588. doi: 10.1128/JB.01655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wolska KI, et al. Antibiotic susceptibility of Escherichia coli dnaK and dnaJ mutants. Microb Drug Resist. 2000;6(2):119–126. doi: 10.1089/107662900419429. [DOI] [PubMed] [Google Scholar]
- 125.Singh VK, et al. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153(Pt 9):3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 126.Acharya P, Kumar R, Tatu U. Chaperoning a cellular upheaval in malaria: heat shock proteins in Plasmodium falciparum. Mol Biochem Parasitol. 2007;153(2):85–94. doi: 10.1016/j.molbiopara.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 127.Stewart GR, Young DB. Heat-shock proteins and the host-pathogen interaction during bacterial infection. Curr Opin Immunol. 2004;16(4):506–510. doi: 10.1016/j.coi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 128.Wild J, et al. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc Natl Acad Sci U S A. 1992;89(15):7139–7143. doi: 10.1073/pnas.89.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sell SM, et al. Isolation and characterization of dnaJ null mutants of Escherichia coli. J Bacteriol. 1990;172(9):4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zouiten-Mekki L, et al. Crohn's disease and polymorphism of heat shock protein gene HSP70-2 in the Tunisian population. Eur J Gastroenterol Hepatol. 2007;19(3):225–228. doi: 10.1097/01.meg.0000252625.65549.29. [DOI] [PubMed] [Google Scholar]
- 131.Birtas-Atesoglu E, et al. Serum levels of free heat shock protein 70 and anti-HSP70 are elevated in Behcet's disease. Clin Exp Rheumatol. 2008;26(4) Suppl 50:S96–S98. [PubMed] [Google Scholar]
- 132.Sharp FR, et al. HSP70 heat shock gene regulation during ischemia. Stroke. 1993;24(12 Suppl):I72–175. [PubMed] [Google Scholar]
- 133.Koyama S, et al. Alteration of familial ALS-linked mutant SOD1 solubility with disease progression: its modulation by the proteasome and Hsp70. Biochem Biophys Res Commun. 2006;343(3):719–730. doi: 10.1016/j.bbrc.2006.02.170. [DOI] [PubMed] [Google Scholar]
- 134.Gamerdinger M, et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. Embo J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kroll J. Chaperones and longevity. Biogerontology. 2005;6(5):357–361. doi: 10.1007/s10522-005-4809-z. [DOI] [PubMed] [Google Scholar]
- 136.Williams DR, et al. An apoptosis-inducing small molecule that binds to heat shock protein 70. Angew Chem Int Ed Engl. 2008;47(39):7466–7469. doi: 10.1002/anie.200802801. [DOI] [PubMed] [Google Scholar]
- 137.Neckers L. Chaperoning oncogenes: Hsp90 as a target of geldanamycin. Handb Exp Pharmacol. 2006;(172):259–277. doi: 10.1007/3-540-29717-0_11. [DOI] [PubMed] [Google Scholar]
- 138.Workman P, et al. Drugging the cancer chaperone HSP90: Combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci. 2007 doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 139.Williamson DS, et al. Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J Med Chem. 2009;52(6):1510–1513. doi: 10.1021/jm801627a. [DOI] [PubMed] [Google Scholar]
- 140.Ermakova SP, et al. (−)-Epigallocatechin gallate overcomes resistance to etoposide-induced cell death by targeting the molecular chaperone glucose-regulated protein 78. Cancer Res. 2006;66(18):9260–9269. doi: 10.1158/0008-5472.CAN-06-1586. [DOI] [PubMed] [Google Scholar]
- 141.Modica-Napolitano JS, et al. Selective damage to carcinoma mitochondria by the rhodacyanine MKT-077. Cancer Res. 1996;56(3):544–550. [PubMed] [Google Scholar]
- 142.Britten CD, et al. A phase I and pharmacokinetic study of the mitochondrial-specific rhodacyanine dye analog MKT 077. Clin Cancer Res. 2000;6(1):42–49. [PubMed] [Google Scholar]
- 143.Wadhwa R, et al. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000;60(24):6818–6821. [PubMed] [Google Scholar]
- 144.Koya K, et al. MKT-077, a novel rhodacyanine dye in clinical trials, exhibits anticarcinoma activity in preclinical studies based on selective mitochondrial accumulation. Cancer Res. 1996;56(3):538–543. [PubMed] [Google Scholar]
- 145.Deocaris CC, et al. Mortalin sensitizes human cancer cells to MKT-077-induced senescence. Cancer Lett. 2007;252(2):259–269. doi: 10.1016/j.canlet.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 146.Kaul SC, et al. An N-terminal region of mot-2 binds to p53 in vitro. Neoplasia. 2001;3(2):110–114. doi: 10.1038/sj.neo.7900139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tikoo A, et al. Treatment of ras-induced cancers by the F-actin-bundling drug MKT-077. Cancer J. 2000;6(3):162–168. [PubMed] [Google Scholar]
- 148.Boulanger J, et al. Members of the 70 kDa heat shock protein family specifically recognize sulfoglycolipids: role in gamete recognition and mycoplasma-related infertility. J Cell Physiol. 1995;165(1):7–17. doi: 10.1002/jcp.1041650103. [DOI] [PubMed] [Google Scholar]
- 149.Mamelak D, Lingwood C. The ATPase domain of hsp70 possesses a unique binding specificity for 3'-sulfogalactolipids. J Biol Chem. 2001;276(1):449–456. doi: 10.1074/jbc.M006732200. [DOI] [PubMed] [Google Scholar]
- 150.Whetstone H, Lingwood C. 3'Sulfogalactolipid binding specifically inhibits Hsp70 ATPase activity in vitro. Biochemistry. 2003;42(6):1611–1617. doi: 10.1021/bi026735t. [DOI] [PubMed] [Google Scholar]
- 151.Harada Y, Sato C, Kitajima K. Complex formation of 70-kDa heat shock protein with acidic glycolipids and phospholipids. Biochem Biophys Res Commun. 2007;353(3):655–660. doi: 10.1016/j.bbrc.2006.12.068. [DOI] [PubMed] [Google Scholar]
- 152.Fewell SW, Day BW, Brodsky JL. Identification of an inhibitor of hsc70-mediated protein translocation and ATP hydrolysis. J Biol Chem. 2001;276(2):910–914. doi: 10.1074/jbc.M008535200. [DOI] [PubMed] [Google Scholar]
- 153.Fewell SW, et al. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279(49):51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 154.Wisen S, Gestwicki JE. Identification of small molecules that modify the protein folding activity of heat shock protein 70. Anal Biochem. 2008;374(2):371–377. doi: 10.1016/j.ab.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 155.Wisen S, et al. Chemical modulators of heat shock protein 70 (Hsp70) by sequential, microwave-accelerated reactions on solid phase. Bioorg Med Chem Lett. 2008;18(1):60–65. doi: 10.1016/j.bmcl.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 156.Evans CG, Wisén S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1–42) aggregation in vitro. J Biol Chem. 2006;281(44):33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 157.Wright CM, et al. Pyrimidinone-peptoid hybrid molecules with distinct effects on molecular chaperone function and cell proliferation. Bioorg Med Chem. 2008;16(6):3291–3301. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wright CM, et al. Inhibition of Simian Virus 40 replication by targeting the molecular chaperone function and ATPase activity of T antigen. Virus Res. 2009;141(1):71–80. doi: 10.1016/j.virusres.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chiang AN, et al. Select pyrimidinones inhibit the propagation of the malarial parasite, Plasmodium falciparum. Bioorg Med Chem. 2009;17(4):1527–1533. doi: 10.1016/j.bmc.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Otvos L, Jr, et al. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;39(46):14150–14159. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]
- 161.Kragol G, et al. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry. 2001;40(10):3016–3026. doi: 10.1021/bi002656a. [DOI] [PubMed] [Google Scholar]
- 162.Haney CM, et al. Identification of Hsp70 modulators through modeling of the substrate binding domain. Bioorg Med Chem Lett. 2009;19(14):3828–3831. doi: 10.1016/j.bmcl.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sinn DI, et al. Pharmacological induction of heat shock protein exerts neuroprotective effects in experimental intracerebral hemorrhage. Brain Res. 2007;1135(1):167–176. doi: 10.1016/j.brainres.2006.11.098. [DOI] [PubMed] [Google Scholar]
- 164.Asano T, et al. HSP70 confers protection against indomethacin-induced lesions of the small intestine. J Pharmacol Exp Ther. 2009;330(2):458–467. doi: 10.1124/jpet.109.152181. [DOI] [PubMed] [Google Scholar]
- 165.Westerheide SD, et al. Celastrols as inducers of the heat shock response and cytoprotection. J Biol Chem. 2004;279(53):56053–56060. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- 166.Trott A, et al. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19(3):1104–1112. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Otaka M, et al. The induction mechanism of the molecular chaperone HSP70 in the gastric mucosa by Geranylgeranylacetone (HSP-inducer) Biochem Biophys Res Commun. 2007;353(2):399–404. doi: 10.1016/j.bbrc.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 168.Schiene-Fischer C, et al. The hsp70 chaperone DnaK is a secondary amide peptide bond cis-trans isomerase. Nat Struct Biol. 2002;9(6):419–424. doi: 10.1038/nsb804. [DOI] [PubMed] [Google Scholar]
- 169.Liebscher M, et al. Fatty acyl benzamido antibacterials based on inhibition of DnaK-catalyzed protein folding. J Biol Chem. 2007;282(7):4437–4446. doi: 10.1074/jbc.M607667200. [DOI] [PubMed] [Google Scholar]
- 170.Takeuchi T, et al. A new antitumor antibiotic, spergualin: isolation and antitumor activity. J Antibiot (Tokyo) 1981;34(12):1619–1621. doi: 10.7164/antibiotics.34.1619. [DOI] [PubMed] [Google Scholar]
- 171.Umeda Y, et al. Synthesis and antitumor activity of spergualin analogues. I. Chemical modification of 7-guanidino-3-hydroxyacyl moiety. J Antibiot (Tokyo) 1985;38(7):886–898. doi: 10.7164/antibiotics.38.886. [DOI] [PubMed] [Google Scholar]
- 172.Umeda Y, et al. Synthesis and antitumor activity of spergualin analogues. III. Novel method for synthesis of optically active 15-deoxyspergualin and 15-deoxy-11-O-methylspergualin. J Antibiot (Tokyo) 1987;40(9):1316–1324. doi: 10.7164/antibiotics.40.1316. [DOI] [PubMed] [Google Scholar]
- 173.Tepper MA, et al. Inhibition of antibody production by the immunosuppressive agent, 15-deoxyspergualin. Transplant Proc. 1991;23(1 Pt 1):328–331. [PubMed] [Google Scholar]
- 174.Nadler SG, et al. Interaction of the immunosuppressant deoxyspergualin with a member of the Hsp70 family of heat shock proteins. Science. 1992;258(5081):484–486. doi: 10.1126/science.1411548. [DOI] [PubMed] [Google Scholar]
- 175.Brodsky JL. Selectivity of the molecular chaperone-specific immunosuppressive agent 15-deoxyspergualin: modulation of Hsc70 ATPase activity without compromising DnaJ chaperone interactions. Biochem Pharmacol. 1999;57(8):877–880. doi: 10.1016/s0006-2952(98)00376-1. [DOI] [PubMed] [Google Scholar]
- 176.Nadeau K, et al. Quantitation of the interaction of the immunosuppressant deoxyspergualin and analogs with Hsc70 and Hsp90. Biochemistry. 1994;33(9):2561–2567. doi: 10.1021/bi00175a027. [DOI] [PubMed] [Google Scholar]
- 177.Kalsch AI, et al. In vivo effects of cyclic administration of 15-deoxyspergualin on leucocyte function in patients with Wegener's granulomatosis. Clin Exp Immunol. 2006;146(3):455–462. doi: 10.1111/j.1365-2249.2006.03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee J, et al. 15-deoxyspergualin prevents mucosal injury by inhibiting production of TNF-alpha and down-regulating expression of MD-1 in a murine model of TNBS-induced colitis. Int Immunopharmacol. 2007;7(8):1003–1012. doi: 10.1016/j.intimp.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 179.Sugawara A, et al. Polyamine compound deoxyspergualin inhibits heat shock protein-induced activation of immature dendritic cells. Cell Stress Chaperones. 2009;14(2):133–139. doi: 10.1007/s12192-008-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hoeger PH, et al. Immunosuppressant deoxyspergualin inhibits antigen processing in monocytes. J Immunol. 1994;153(9):3908–3916. [PubMed] [Google Scholar]
- 181.Nadler SG, et al. Elucidating the mechanism of action of the immunosuppressant 15-deoxyspergualin. Ther Drug Monit. 1995;17(6):700–703. doi: 10.1097/00007691-199512000-00026. [DOI] [PubMed] [Google Scholar]
- 182.Ramya TN, et al. 15-deoxyspergualin primarily targets the trafficking of apicoplast proteins in Plasmodium falciparum. J Biol Chem. 2007;282(9):6388–6397. doi: 10.1074/jbc.M610251200. [DOI] [PubMed] [Google Scholar]
- 183.Amemiya H Japanese Collaborative Transplant Study Group of NKT-01. Immunosuppressive mechanisms and action of deoxyspergualin in experimental and clinical studies. Transplant Proc. 1995;27(1):31–32. [PubMed] [Google Scholar]
- 184.Amada N, et al. Deoxyspergualin prophylaxis with tacrolimus further improves long-term graft survival in living-related renal-transplant recipients transfused with donor-specific blood. Transplant Proc. 2005;37(2):927–929. doi: 10.1016/j.transproceed.2004.12.289. [DOI] [PubMed] [Google Scholar]
- 185.Lebreton L, et al. Structure-immunosuppressive activity relationships of new analogues of 15-deoxyspergualin. 1. Structural modifications of the hydroxyglycine moiety. J Med Chem. 1999;42(2):277–290. doi: 10.1021/jm980431g. [DOI] [PubMed] [Google Scholar]
- 186.Yang H, et al. Monotherapy with LF 15-0195, an analogue of 15-deoxyspergualin, significantly prolongs renal allograft survival in monkeys. Transplantation. 2003;75(8):1166–1171. doi: 10.1097/01.TP.0000062841.89728.CF. [DOI] [PubMed] [Google Scholar]
- 187.Bischofberger P, et al. D-Peptides as inhibitors of the DnaK/DnaJ/GrpE chaperone system. J Biol Chem. 2003;278(21):19044–19047. doi: 10.1074/jbc.M300922200. [DOI] [PubMed] [Google Scholar]
- 188.Yi F, Regan L. A novel class of small molecule inhibitors of Hsp90. ACS Chem Biol. 2008;3(10):645–654. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]