Abstract
To initiate class switch recombination (CSR) activation-induced cytidine deaminase (AID) induces staggered nick cleavage in the S region, which lies 5′ to each Ig constant region gene and is rich in palindromic sequences. Topoisomerase 1 (Top1) controls the supercoiling of DNA by nicking, rotating, and religating one strand of DNA. Curiously, Top1 reduction or AID overexpression causes the genomic instability. Here, we report that the inactivation of Top1 by its specific inhibitor camptothecin drastically blocked both the S region cleavage and CSR, indicating that Top1 is responsible for the S region cleavage in CSR. Surprisingly, AID expression suppressed Top1 mRNA translation and reduced its protein level. In addition, the decrease in the Top1 protein by RNA-mediated knockdown augmented the AID-dependent S region cleavage, as well as CSR. Furthermore, Top1 reduction altered DNA structure of the Sμ region. Taken together, AID-induced Top1 reduction alters S region DNA structure probably to non-B form, on which Top1 can introduce nicks but cannot religate, resulting in S region cleavage.
Keywords: camptothecin, non-B DNA, translation suppression
Class switching is one of the critical features of antibody memory that is required for vaccination. Class switching is accomplished by region-specific recombination between two switch (S) regions located upstream of the Ig (Ig) heavy-chain constant region (CH) gene (1, 2). Class switch recombination (CSR) is a unique type of recombination distinct from site-specific, homologous, or illegitimate recombination because the S region consists of tandem arrays of repetitive sequences rich in inverted repeats or palindromes. Such sequences facilitate formation of stem-loop, cruciform, and other non-B DNA structures (3).
Transcription of the S region is initiated from the I promoter that is specific to each S region, and is essential to trigger CSR (4, 5). The S region transcripts seem to remain on the DNA template for a short period, thus facilitating the formation of R-loop. Lieber and colleagues reported that the bisulfite treatment of nuclear DNA from switch-induced spleen cells converted dC to dU in the Sμ region, indicating the presence of single stranded DNA in the Sμ region in activated B cells (6, 7). R-loop may not survive long but lead to the formation of non-B structures including stem-loop, cruciform, and triplex in the S region because of abundant inverted repeats (3, 8–10).
Frequent and long deletions and duplications are observed at the CSR junctions in artificial switch substrates designed to select inversion-type products (11). The results suggest that the double strand breaks (DSBs) in the S regions leading to CSR, are generated by the staggered nick-type cleavages because deletions and duplications are most likely to be generated during the repair of staggered nicked ends with single-strand overhangs by exonucleases or DNA polymerases.
AP endonuclease 1 and 2 (Apex 1 and 2) and topoisomerase 1 (Top1) represent evolutionally conserved nicking enzymes. Apex1 and 2 cleave abasic sites generated by base excision enzymes, such as uracil DNA glycosylase, which are proposed to be involved in CSR (12, 13). However, we have shown that both Apex1 and 2 are not essential for CSR (14). Top1 reduces excessive supercoiling in the DNA of mammalian cells (15). It binds to DNA, cleaves one strand and fixes the 3′ phosphate of the cleaved end by forming a transient covalent bond with its tyrosine residue. The transient complex is quickly resolved by religation after rotating the nonfixed end of the DNA strand around the helical axis. A specific inhibitor of Top1, camptothecin (CPT) can be intercalated into this transient cleavage complex of Top1 and DNA to block Top1's catalytic function while CPT does not affect free Top1 (16). Curiously, the reduction of Top1 by RNA mediated knockdown in cultured cells causes the genome-wide instability (17). Activation-induced cytidine deaminase (AID) is essential for CSR and involved in S region cleavage, but its molecular mechanism is unknown (12, 13, 18, 19). AID has two functions in CSR: one for S region cleavage and the other for the end joining step (20). Interestingly, AID introduces DNA cleavage in multiple genetic loci, resulting in the genome instability (20–23).
We found that CPT almost completely inhibited CSR and DNA cleavage in the S region induced by AID, indicating that Top1 is the major, if not a sole, nicking enzyme in CSR. We then found that AID expression reduced the Top1 protein amount primarily by suppression of Top1 mRNA translation. In addition, Top1 reduction altered the Sμ region structure, which probably prevents the rotation and religation steps after cleavage by Top1. From these observations, we propose a model that S region cleavage is due to suppression of religation by Top1 on non-B DNA, which is formed by inefficient relaxation of transcription-coupled supercoiling because of Top1 reduction by AID.
Results
Top1 Inhibition by CPT Blocks CSR.
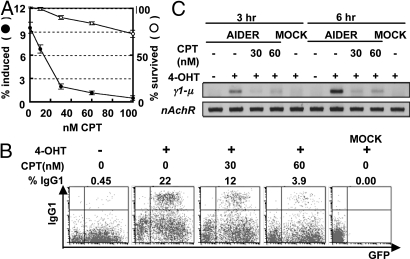
Because Apex1 and 2 are not required for CSR (14), we suspected involvement of Top1. We thus tested the effect on CSR of treating cells with CPT that specifically stabilizes the Top1-DNA intermediate complex and inhibits the catalytic function of Top1. The IgA switching of CH12F3–2 cells 24 h after stimulation with a mixture of CD40L, IL-4, TGFβ (CIT) was reduced in the presence of CPT in a dose dependent manner (Fig. 1A). CPT (60 nM) inhibited CSR to 13% while 80% of cells remained alive even after 24 h. The inhibition of CSR was more directly demonstrated by a drop in α circle transcripts (αCT), which are transcribed from looped-out circular DNA (24) (Fig. S1A). The drop in αCT by increment of CPT paralleled the reduction of IgA expression. We also confirmed that this strong blockade of CSR by CPT was not due to the suppression of either AID expression or germline transcription of the Sμ and Sα regions in CH12F3–2 cells (Fig. S1B).
Fig. 1.
CPT blocks CSR in CH12F3–2 cells and splenic B cells. (A) The percentages of the IgA+ and propidium iodide (PI)− alive cells are presented as closed and open circles, respectively. Each circle represents average with S.D. (n = 3). The backgrounds (subtracted) of IgA+ and PI+ cells at 0 nM CPT were 1.26 and 11.5%, respectively. (B) AID−/− spleen cells were infected by AIDER-IRES-GFP retrovirus. Twenty-four hours later, the cells were treated with 1 μM 4-OHT or EtOH in the presence of 30 to 60 nM CPT. After an additional 24-h incubation, surface IgG1 and GFP expression were analyzed by flow cytometry with biotinylated anti-IgG1 antibody and streptavidin-APC. The percentage of IgG1 switch in the GFP positive cells is shown above each plot. Mock, no retrovirus control. (C) Sγ1-Sμ DC-PCR. Before the flow cytometry in panel B, part of cells were collected 3 and 6 h after 4-OHT and CPT addition. Extracted genomic DNA was then subjected to DC-PCR for the Sγ1-Sμ recombination (γ1-μ) and for the nicotinic acetylcholine receptor (nAchR) locus as control. PCR products were run on a 2% agarose gel and visualized by ethidium bromide.
The inhibition of CSR by CPT was confirmed in splenic B cells. AID deficient splenic B cells were stimulated by LPS and IL-4, and infected with the retrovirus expressing AID fused with the hormone binding domain of the estrogen receptor (ER) (AIDER) (25). AIDER was then activated by the addition of tamoxifen (4-OHT) 24 h after the infection. The surface expression of IgG1 in splenic B cells was inhibited by 30 or 60 nM CPT (Fig. 1B). To examine the effect of CPT more quickly, we performed the digestion circularization (DC) PCR method (26). As early as 3 or 6 h after the activation of AIDER with 4-OHT, we could already detect Sγ1-Sμ DC-PCR products of Sγ1-Sμ recombination, which results from recombination between the Sγ1 and Sμ regions (Fig. 1C and Fig. S1C). The amount of Sγ1-Sμ DC-PCR products was drastically reduced by 3-h incubation with 30 or 60 nM CPT, indicating that the blockage of CSR by CPT is unlikely to be the secondary effect of cell death. None of Top2α inhibitors blocked CSR as assessed by αCT synthesis (Table S1).
CPT Inhibits S Region DNA Cleavage.
To assess whether CPT inhibits DNA cleavage in the S region, we first examined the γH2AX focus formation in the S region. A chromatin immunoprecipitation (CHIP) assay using anti-γH2AX antibodies clearly showed that γH2AX accumulated in the Sμ region and Cμ exons but not in the Gapdh or Icos loci in response to DNA cleavage in AER cells i.e., CH12F3–2 cell expressing AIDER after the addition of 4-OHT, as reported previously (Fig. 2A) (27). γH2AX focus formation at the Sμ region and Cμ exons was severely blocked by 50 nM CPT, indicating that CPT suppresses AID-induced DSBs in the Sμ region.
Fig. 2.
CPT inhibits the DNA cleavage in CSR. (A) AER cells were cultured with 50 nM CPT for 12 h then stimulated with CIT and 1 μM 4-OHT. Cells were harvested 8 h after the stimulation and γH2AX accumulation at the indicated gene loci was assessed by CHIP. Cells treated with 0.5 μM staurosporin were shown as a positive control. A numeric value representing the nonspecific IgG precipitation was subtracted as a background. The graph represents the average of 4 independent experiments with a standard deviation. (B–E) DNA break assay was carried out as described in Materials and Methods. Indicated concentrations of CPT were added at the same time as 4-OHT (1 μM). (B) CPT effect on DNA break induced by AID. AER cells were stimulated for 16 h with 4-OHT. (C) Top1 knockdown was carried out for 24 h with siRNA oligo #33 or #35 in AER cells. Cells were harvested 24 h later from the start of 4-OHT. (D) Knockdown of Top1 in AER cells was carried out by miR expression with 50 ng/mL tetracycline (Tet). AER cells were activated for 24 h by 1 μM 4-OHT. AER cells with microRNA-negative control (miR-neg) were used as control. (E) AER cells with Top1 knockdown by siRNA oligo #33 for 24 h and subsequently stimulated.
Using semiquantification of DSBs ends by ligation of a biotinylated linker, we confirmed that the number of DSBs of the Sμ and Sα region was clearly reduced in AER cells by 10 and 30 nM CPT (Fig. 2B). On the other hand, the Sγ1 region cleavage was not affected by CPT. Also, although 150 nM CPT induced numerous nonspecific DNA cleavage, it did not induce a significant number of such cleavage at <50 nM used in the present study (Fig. S2). Finally, because CPT, a substrate-enzyme intercalating inhibitor of Top1, blocks the AID-induced DSBs of the S region at concentrations far below CPT's IC50 (ca 7 μM) (28), the S region might be preferred target to Top1. These results demonstrate that Top1 is the enzyme responsible for DNA cleavage during AID-induced CSR.
AID Expression Reduces Top1 Protein.
We then explored a possible relationship between Top1 and AID. We compared the Top1 protein level between wild-type and AID deficient splenic B cells stimulated with LPS and IL-4 (Fig. 3A). Surprisingly, 24 h after stimulation the wild-type splenic B cells showed a decreased level of the Top1 protein that was ≈60% of the level in the nonstimulated or AID deficient spleen cells. When we introduced retrovirus carrying AIDER into the AID deficient splenic B cells, wild-type AIDER expression reduced the Top1 protein to 50% 3 h after 4-OHT stimulation compared with expression of a catalytically inactive mutant of AIDER (KSS) (Fig. 3B). This faster reduction of the Top1 protein is probably because of the rapid activation of the accumulated AIDER by 4-OHT.
Fig. 3.
Top1 protein is decreased by AID expression. (A–C) The protein amounts were measured by Western blot analysis using appropriate antibodies. (A) Wild-type (W) and AID−/− (A−/−) splenic B cells were stimulated for the indicated periods by LPS and IL-4. The relative Top1 protein amounts normalized by tubulin are plotted at the right. (B) AID−/− mouse splenic B cells were infected by retroviruses carrying AIDER (W) or its KSS-mutant (K). 4-OHT (1 μM) was added 24 h after infection. Relative Top1 protein levels at indicated time points after 4-OHT addition are plotted as above. (C) Top1 protein levels in the nucleoplasm fraction of AER cells at indicated time points after 4-OHT addition were measured, normalized by lamin, and are plotted as above.
When AER cells were stimulated with 4-OHT or CIT, the Top1 protein was also reduced but more slowly (Fig. S3A), probably because cell lines proliferate much faster and thus contain a much larger amount of the Top1 protein than splenic B cells. The majority of the Top1 protein is localized in nucleoli and the turnover of the nucleolar Top1 protein is much slower than the nucleoplasmic Top1 (29, 30). We therefore quantified the Top1 protein level in the nucleoplasmic fraction of AER cells after the addition of 4-OHT and found that the Top1 amount was reduced quickly (Fig. 3C). Similarly, NIH 3T3 fibroblasts expressing AIDER but not its KSS mutant decreased the Top1 protein amount in the nucleoplasm fraction after 4-OHT stimulation (Fig. S3B). We also found that C-terminally truncated AID (Jp8Bdel) also reduced Top1 protein (Fig. S3C) (20). The mRNA level of Top1 was decreased by ≈20% 48 h after stimulation of splenic B or CH12F3–2 cells (Fig. S3D). We therefore suspected that the reduction in the Top1 protein was mostly due to either the suppression of Top1 mRNA translation or to enhanced degradation of the Top1 protein.
AID Suppresses the Translation of Top1 mRNA.
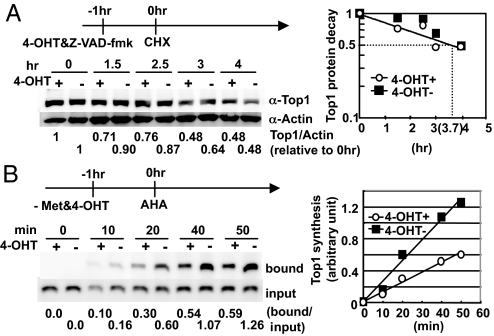
We first determined the half life of the Top1 protein in AER cells to be 3.7 h by the addition of cycloheximide (CHX) (Fig. 4A). The half-life of the Top1 protein was not shortened significantly by AID activation, indicating that AID does not accelerate the degradation of Top1. We then directly examined whether Top1 protein synthesis is reduced by AID induction. AID activation clearly suppressed the rate of the Top1 protein synthesis to a half, while the total protein synthesis is not affected (Fig. 4B and Fig. S4). We sequenced the Top1 cDNA in AID expressing B cells and found no mutations in either the coding or 3′ UTR sequence (Fig. S5). Therefore, Top1 mRNA does not seem to be a direct editing target of AID. The half life (3.7 h) and translation reduction rate (0.5) explains experimental reduction of Top1 protein (see Discussion). Collectively, these results suggest that the Top1 reduction induced by AID activation was caused primarily by the suppression of Top1 mRNA translation and less significantly by Top1 mRNA degradation.
Fig. 4.
AID reduces the Top1 protein level by inhibiting its translation. (A) Half-life of Top1. AER cells were treated with 50 μM Z-VAD-FMK, 1 μM of 4-OHT, and 10 μg/mL of CHX at the indicated time points. Whole cell extract was collected serially, and Top1 was measured by Western blot and plotted. (B) Translation rate of Top1. AER cells were cultured in methionine free medium with or without 1 μM 4-OHT for 1 h before l-azidohomoalanine (AHA) addition at time 0. Cells were harvested at different time points, and AHA incorporated protein was biotinylated as described in Materials and Methods. The Top1 synthesis ratio in the presence vs. absence of 4-OHT was calculated to be 0.53 ± 0.044. Met, methionine; bound, Streptavidin beads-bound fraction.
Top1 Knockdown Enhances CSR.
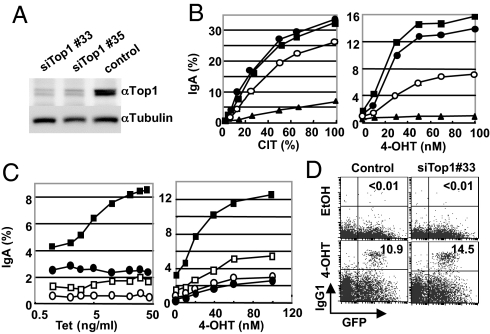
We then examined whether a Top1 reduction induced by AID is functionally relevant to CSR. Small interfering (si)RNA oligos to Top1 mRNA were introduced into AER cells to knock down the Top1 protein, and their effects on CSR were examined. Two Top1 siRNA oligos (Nos. 33 and 35) knocked down the Top1 protein strongly in AER cells although measurable amounts of Top1 always remained (Fig. 5A). Surprisingly, the Top1 protein knockdown by either of the Top1 siRNA oligos clearly augmented CSR induced by various levels of AID activation, which was controlled by changing the concentrations of either CIT or 4-OHT (Fig. 5B). The magnitude of CSR augmentation by Top1 reduction was greater in the 4-OHT activated AER cells than in the CIT-activated cells probably because 4-OHT caused lower AID activation than did CIT as evidenced by a lower efficiency of IgA switching and stronger inhibition by AID knockdown. The augmentation of CSR by the Top1 knockdown was not due to an increase in Iμ and Iα germline transcription, or endogenous AID expression in AER cells with or without 4-OHT stimulation (Fig. S6A).
Fig. 5.
Top1 knockdown enhances CSR in AER and splenic B cells. (A) The Top1 protein level in AER cells was measured by Western blot 48 h after electroporation with the indicated siRNA. (B) Top1 knocked-down AER cells were stimulated with various concentrations of CIT (Left) or 4-OHT (Right) for 24 h, and the surface IgA expression was quantified by flow cytometry. siRNAs used are as follows; closed circles, siTop1#33; squares, siTop#35; triangles, siAID; open circles, control. (C) Knockdown of Top1 was performed in AER cells carrying the Tet-inducible microRNA (miR)-Top1 construct as described in Materials and Methods. The surface IgA expression was quantified by flow cytometry 24 h after 4-OHT addition. Squares, miR-Top1; circles, miR-neg. (Left) Cells were exposed to indicated concentrations of Tet for 24 h, and then incubated with 1 μM of 4-OHT. Closed and open symbols, with and without 4-OHT, respectively. (Right) Cells were initially exposed to 1 μg/mL doxycycline for 24 h and then CSR was induced by indicated concentrations of 4-OHT. Closed and open symbols, with and without doxycycline, respectively. (D) Top1 knockdown augmented CSR in splenic B cells. AIDER-induced CSR in the control siRNA- and Top1 siRNA(#33)-treated splenic B cells. AID−/− splenic cells were cultured for 24 h after siRNA introduction and then infected by the AIDER-IRES-GFP retrovirus. The surface IgG1 and GFP levels were analyzed by flow cytometry 24 h after 4-OHT addition. Numbers in the quadrants are the percentages of switched cells among the GFP positive population.
We also generated AER cell lines containing plasmids carrying tetracycline (Tet) - inducible microRNA directed to Top1 mRNA or a randomized negative control. In this cell line, CSR was gradually augmented in parallel with Top1 knockdown by increasing expression of the Top1-directed microRNA but not the negative control (Fig. 5C, Left and Fig. S6B). The augmentation of CSR by Top1 knockdown was also dependent on AIDER activation by 4-OHT (Fig. 5C, Right). A slight increase in CSR by Top1 knockdown was observed without 4-OHT, which might have been due to the basal level of AID expression in CH12F3–2 cells. CSR enhancement by Top1 knockdown was also obvious in splenic AID−/− B cells that were electroporated to incorporate Top1 siRNA and then infected by AIDER retrovirus, followed by 4-OHT activation (Fig. 5D and Fig. S6C).
Top1 Knockdown Enhances S Region Cleavage.
We then asked whether Top1 reduction stimulated DNA cleavage in the S region. When the Top1 protein was decreased by either Top1 siRNA No. 33 or No. 35, the DSBs were strongly increased in the Sμ and Sα regions, but not the Sγ1 region in AER cells compared with AER cells treated with control siRNA (Fig. 2C). Similarly, the Top1 knockdown by the Tet-inducible microRNA in AER cells also enhanced cleavage in the Sμ but not the Sγ1 region (Fig. 2D). Thus, we concluded that Top1 reduction facilitates CSR by enhancing the S region cleavage. Importantly, the DSBs that were augmented by the Top1 knockdown were also inhibited by 25 and 50 nM CPT, indicating that DNA cleavage augmented by Top1 reduction is also catalyzed by Top1 (Fig. 2E).
Top1 Knockdown Induces Structural Changes in the S Region.
To explore the mechanism by which AID-mediated Top1 reduction augments S region cleavage and CSR, we speculated that aberrant supercoiling by Top1 protein reduction may induce non-B DNA structure in the S region so that Top1 cannot religate, resulting in irreversible cleavage by Top1. DNA structural alterations can be estimated by the DNA's sensitivity to bisulfite modification from dC to dU, which occurs in the single-stranded regions (6, 7). The presence of consecutive C bases is suitable for determining the single-strandedness by the bisulfite assay (7). We found that the three major C clusters (Fig. 6A and B I, II, and III) within the 150-bp region 3′ to the core Sμ region in CH12F3–2 cells showed a higher sensitivity to the bisulfite treatment after Top1 knockdown. A similar increase in bisulfite sensitivity was observed in the same clusters after CIT stimulation. Because the Sμ locus is transcriptionally active in CH12F3–2 cells, we observed about a 16% basal reactivity estimated by the total conversion frequency (Fig. 6C), which was increased to 27% upon Top1 knockdown. These results suggest that Top1 reduction indeed affected the DNA structure of the Sμ region. Although Top1 normally cleaves and religates one strand of DNA, when Top1 levels are reduced, structural alterations in the S region may form non-B structures that block the rotation and religation step, resulting in irreversible cleavage by Top1.
Fig. 6.
Top1 knockdown alters the single-stranded area of the Sμ region. (A) Schematic representation of the PCR amplified sequence flanking the Sμ core region. Gray and black arrowheads indicate forward (bisulfite specific) and reverse primers, respectively. The C-rich (59 Cs) area (150-bp) was located at the 3′ end of the amplified fragment (721-bp) and indicated by dotted line. (B) The bisulfite reactivity of the 59 C bases, with or without Top1 knockdown in CH12F-3 cells. Each C base with its position number from the left end of the 721-bp amplified segment is indicated on horizontal axis and the percent converted at each position out of the total molecules analyzed is plotted on vertical axis. The three sensitive areas are indicated by horizontal bars labeled I, II, and III. (C) Summary of the total C to T conversion frequency.
Discussion
In the present study, we showed that CPT an intercalating inhibitor of the Top1 catalytic activity (15, 16) blocked both CSR and S region cleavage induced by AID activation in B cells. In addition, AID expression reduced the Top1 protein amount by suppression of Top1 mRNA translation. This activity of AID depends on its deamination activity but not on the C-terminal domain that is not required for DNA cleavage (20). Furthermore, Top1 reduction mediated by RNA knockdown enhanced AID-induced CSR and S region DNA cleavage. From these observations, we conclude that the activation of AID reduces the Top1 protein and this decrease in the Top1 protein enhances DNA cleavage in the S region. CPT also inhibited DSBs augmented by Top1 knockdown. It is therefore likely that Top1 is the enzyme that introduces the DSBs by staggered nick cleavage in the S region after AID activation in B lymphocytes.
The mechanism by which Top1 introduces DSBs requires inhibition of the religation step of Top1. Transcription elongation by RNA polymerase II introduces uneven DNA supercoiling leading to a positive supercoil in the front of the migrating transcriptional machinery and negative supercoil at the rear. Top1 reduction is likely to retard relaxation of negative supercoil. Excessive negative supercoil of DNA facilitates local structural alterations including R-loop, stem-loop, cruciform, triplex, and other non-B DNA structures in DNA rich in palindromes or G/C stretches like the S region, which probably inhibits the religation step of Top1 after cleavage (3, 8–10) (Figs. S7 and S8). In addition, Top1 phosphorylates and activates the alternative splicing factor ASF/SF2 (31). Thus, it is possible that Top1 protein reduction may disturb splicing of S region transcripts and prolong the half life of S region transcripts, leading to the formation of frequent R-loop and consequent non-B DNA structures in the S region (3, 32). In support of this hypothesis, the reduction of either Top1 or ASF induces genomic instability (17, 32). In fact, we observed that the single stranded fraction of the Sμ region was augmented by Top1 knockdown. Interestingly, a similar bisulfite reactivity pattern was observed at the chromosomal instability sites identified in the bcl-2 and c-myc loci (8, 9).
Furthermore, Top1 promotes the formation of G-quartet structures and binds to non-B forms of DNA, such as cruciform and G-quartet DNA, which can occur in G-rich single-stranded regions (33, 34). Non-B form structures may result in irreversible cleavage on either strand of the S region by Top1 because the DNA rotation around the helix may be disturbed after nicking, thus inhibiting the Top1's religation activity (Fig. S8). In other words, the Top1 reduction may induce the structural changes of the S regions that favor irreversible cleavage by Top1.
Given the half-life of 3.7 h and reduction rate of translation to 50%, the Top1 protein level at a given time point (t in hour) after initiation of translation reduction is equal to 0.5 A0 (1+e−0.187t) where A0 is the initial amount of Top1 protein. This equation predicts that the Top1 protein amount converges to a half approximately 24 h after initiation of translation reduction. The experimental results are in general agreement with this prediction. How can Top1 mRNA translation be modulated by AID? Because the Top1 mRNA is not directly edited, we speculate that AID may edit small molecular weight RNA, such as microRNAs. The edited microRNAs may change the target specificity to regulate Top1 mRNA translation (Fig. S9). In summary, we have shown that Top1 is involved in the DNA cleavage step of CSR although the details of the mechanism remain to be explored.
Materials and Methods
All data shown are representative of at least three experiments.
siRNA System.
TopI siRNA #33 or #35, or a low GC control RNA (all by Invitrogen) were electroporated into CH12F3–2, AER, or splenic B cells and incubated for 24 h before stimulation. miR system: miR-Top1 was designed by BLOCK-iT RNAi Designer (Invitrogen), and cloned into pcDNA™6.2-GW/EmGFP-miR (Invitrogen). pcDNA 6.2-GW/EmGFP/miR-neg control (miR-neg) plasmid was used as the negative control. The sequences of miR and siRNA are in Table S2. Tet-ON system: AER cells was transduced by pLenti6/TR (Invitrogen) and selected with 5 μg/mL of blasticidin to get the Tet repressor positive (TetR+) subclone. pCMV promoter of the miR vectors was substituted by Tet operator promoter from pcDNA 4/TO/LacZ (Invitrogen). The TetR+ cells were electroporated by the Tet-operative miR vector, sorted by flow cytometry and subcloned. In the experiments, the cells were exposed to Tet or dox for 24 h before AID activation.
DNA Break Assay by Biotinylated Linker-Ligation.
Biotinylated linker-ligation method is based on biotinylated dUTP method (20). DNA break site is blunted and ligated with biotinylated linker. Sonicated labeled DNA were trapped with streptavidin magnetic beads, followed by second linker ligation. DNA was further globally amplified with linker-specific primers, and purified with WIZARD SV Gel and PCR Reaction Clean-up system (Promega). Gene-locus specific quantitative PCR was performed with ABI 7900HT. Four percent of the total sample was taken as the input DNA, just before trapping by magnetic beads. Sμ, Sα, or Sγ1 locus signal, calculated by beads-bound/input, is further standardized by the signal (beads-bound/input) of β2-microglobulin locus or β-actin locus to compensate the recovery of beads and DNA. All of the sequences of linkers and oligos were described in Table S2.
Metabolic Labeling of Newly Synthesized Proteins.
Newly synthesized protein was labeled with Click-iT AHA for Nascent Protein Synthesis kit and Click-iT Biotin Protein Analysis Detection Kit (Invitrogen) according to manufacturer's instructions. One million AER cells were labeled with 500 μM of AHA for indicated times and lysed in the lysis buffer [50 mM Tris·HCl pH8.0, 1% SDS, and 1× Complete (Roche), 2.5kU/mL Benzonase (Novagen)]. AHA incorporated protein was biotinylated. After precipitation and dissolving, biotinylated protein was collected with Dynabeads M-280 (Invitrogen). Protein was eluted into SDS sample buffer by boiling and quantified by Western blot with αTopI antibody. Total protein biotinylation efficiency was measured by dot blot onto the nitrocellulose membrane. After blocking, the membrane was incubated with HRP-conjugated, streptavidin-labeled antibody, washed, and colored by chemi-luminescence.
Bisulfite Modification of Genomic DNA.
CH12F3–2 cells were either stimulated with CIT or left untreated 24 h after siRNA (GC-control and Top1) introduction, incubated for another 24 h and harvested for surface IgA expression assay and genomic DNA isolation. Genomic DNA bisulfite modification of the Sμ region was carried out as described with some modifications (7). High molecular weight genomic DNA was isolated using Qiagen genomic DNA isolation system and processing of bisulfite modified DNA was performed following the instruction of Bisulfite modification system (Human Genetic Signatures). Amplification of the sequence 3′ to the Sμ core region with FTH111 and FTH94 primers.
Materials and other conventional experimental procedures are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Ms. Y. Shiraki and T. Kanda for the preparation of the manuscript. This work was supported by a Grant-in-Aid for Specially Promoted Research (No. 17002015) and also by the Global Centers of Excellence program of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911879106/DCSupplemental.
References
- 1.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 2.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Yancopoulos GD, et al. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stavnezer-Nordgren J, Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang FT, et al. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai AG, et al. Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J Biol Chem. 2009;284:7157–7164. doi: 10.1074/jbc.M806866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghavan SC, et al. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 10.Collier DA, Griffin JA, Wells RD. Non-B right-handed DNA conformations of homopurine. homopyrimidine sequences in the murine immunoglobulin C alpha switch region. J Biol Chem. 1988;263:7397–7405. [PubMed] [Google Scholar]
- 11.Chen X, Kinoshita K, Honjo T. Variable deletion and duplication at recombination junction ends: Implication for staggered double-strand cleavage in class-switch recombination. Proc Natl Acad Sci USA. 2001;98:13860–13865. doi: 10.1073/pnas.241524898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rada C, et al. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 13.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 14.Sabouri Z, et al. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol. 2009;21:947–955. doi: 10.1093/intimm/dxp061. [DOI] [PubMed] [Google Scholar]
- 15.Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 16.Kerrigan JE, Pilch DS. A structural model for the ternary cleavable complex formed between human topoisomerase I, DNA, and camptothecin. Biochemistry. 2001;40:9792–9798. doi: 10.1021/bi010913l. [DOI] [PubMed] [Google Scholar]
- 17.Miao ZH, et al. Nonclassic functions of human topoisomerase I: Genome-wide and pharmacologic analyses. Cancer Res. 2007;67:8752–8761. doi: 10.1158/0008-5472.CAN-06-4554. [DOI] [PubMed] [Google Scholar]
- 18.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 19.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 20.Doi T, et al. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2009;106:2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okazaki IM, et al. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 23.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. A hallmark of active class switch recombination: Transcripts directed by I promoters on looped-out circular DNAs. Proc Natl Acad Sci USA. 2001;98:12620–12623. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doi T, Kinoshita K, Ikegawa M, Muramatsu M, Honjo T. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc Natl Acad Sci USA. 2003;100:2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ChuCC, Paul WE, Max EE. Quantitation of immunoglobulin mu-gamma 1 heavy chain switch region recombination by a digestion-circularization polymerase chain reaction method. Proc Natl Acad Sci USA. 1992;89:6978–6982. doi: 10.1073/pnas.89.15.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scaldaferro S, Tinelli S, Borgnetto ME, Azzini A, Capranico G. Directed evolution to increase camptothecin sensitivity of human DNA topoisomerase I. Chem Biol. 2001;8:871–881. doi: 10.1016/s1074-5521(01)00059-x. [DOI] [PubMed] [Google Scholar]
- 29.Christensen MO, et al. Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J Biol Chem. 2004;279:21873–21882. doi: 10.1074/jbc.M400498200. [DOI] [PubMed] [Google Scholar]
- 30.Muller MT, Pfund WP, Mehta VB, Trask DK. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenreich A, et al. Cdc2-like kinases and DNA topoisomerase I regulate alternative splicing of tissue factor in human endothelial cells. Circ Res. 2009;104:589–599. doi: 10.1161/CIRCRESAHA.108.183905. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Arimondo PB, et al. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000;28:4832–4838. doi: 10.1093/nar/28.24.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiyagarajan MM, Waldman SA, Noe M, Kmiec EB. Binding characteristics of Ustilago maydis topoisomerase I to DNA containing secondary structures. Eur J Biochem. 1998;255:347–355. doi: 10.1046/j.1432-1327.1998.2550347.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.