Abstract
In a genetic screen in a drnl-2 background, we isolated a loss-of-function allele in miR319a (miR319a129). Previously, miR319a has been postulated to play a role in leaf development based on the dramatic curled-leaf phenotype of plants that ectopically express miR319a (jaw-D). miR319a129 mutants exhibit defects in petal and stamen development; petals are narrow and short, and stamens exhibit defects in anther development. The miR319a129 loss-of-function allele contains a single-base change in the middle of the encoded miRNA, which reduces the ability of miR319a to recognize targets. Analysis of the expression patterns of the three members of the miR319 gene family (miR319a, miR319b, and miR319c) indicates that these genes have largely non-overlapping expression patterns suggesting that these genes have distinct developmental functions. miR319a functions by regulating the TCP transcription factors TCP2, TCP3, TCP4, TCP10, and TCP24; the level of RNA expression of these TCP genes is down-regulated in jaw-D and elevated in miR319a129. Several lines of evidence demonstrate that TCP4 is a key target of miR319a. First, the tcp4soj6 mutant, which contains a mutation in the TCP4 miRNA-binding site complementary to the miR319a129 mutation, suppresses the flower phenotype of miR319a129. Second, expression of wild-type TCP4 in petals and stamens (i.e., AP3:TCP4) has no effect on flower development; by contrast, a miRNA-resistant version of TCP4, when expressed in petals and stamens (i.e., pAP3:mTCP4) causes these organs not to develop. Surprisingly, when AP3:TCP4 is present in a miR319a129 background, petal and stamen development is severely disrupted, suggesting that proper regulation by miR319a of TCP4 is critical in these floral organs.
Keywords: DRNL, flower development, forward genetics, microRNA, stamen
MicroRNAs (miRNAs) are 21–24 nucleotide regulatory RNAs that function in diverse aspects of plant biology (1) such as biotic and abiotic stress responses (2), metabolism (3), hormone signaling (4), transcription (5), development (6), and the regulation of miRNA machinery itself (7, 8). Most studies of plant miRNAs focus on alleles that ectopically express the miRNA. In miRNA overexpression lines, the RNA levels of target genes are down-regulated resulting in a phenotype that mimics the loss-of-function phenotypes of miRNA target mutants (9, 10). Loss-of-function alleles in miRNA genes are rare because most often miRNAs are redundantly encoded in plant genomes. Thus, mutation of a single miRNA gene usually does not result in a mutant phenotype. In the few miRNA loss-of-function mutants that are available (5, 6, 11, 12), the RNA levels of target genes are elevated, which is expected if the miRNA can no longer function to degrade target mRNAs.
miR319a was initially characterized based on the dramatic leaf phenotype that results from overexpression of miR319a, as in the jaw-D allele that results in a jagged and wavy (jaw) leaf-phenotype (13). The best-characterized targets of miR319 are a subset of TCP transcription factors (14). The TCP genes are grouped into two subclasses: class I and class II. Class I TCP genes, such as TCP20, function as positive regulators of cell growth (15) while class II genes like the Antirrhinum genes CINCINNATA (CIN) (16), DICHOTOMA, and CYCLOIDEA (17, 18) function as negative regulators of cell growth. A subset of class II TCP genes contains a miR319 binding site including TCP2, TCP3, TCP4, TCP10, and TCP24. In jaw-D, due to ectopic expression of miR319a, mRNA levels for the target genes TCP2, TCP3, TCP4, TCP10, and TCP24 are 2- to 34-fold down-regulated in seedlings (13). A role for TCP genes in leaf development also comes from analysis of the cincinnata (cin) mutant in Antirrhinum majus. cin loss-of-function mutants result in uncontrolled cell growth in the leaf margins leading to buckling of the leaf margin (13, 16, 19). In addition, cin mutants also exhibit small, slightly curled and pale petals (16). Nath et al. (19) postulate that CIN affects leaf development by making cells more sensitive to a growth-arrest signal at the leaf margin.
In Arabidopsis, single loss-of-function tcp mutants have only very subtle developmental phenotypes. Higher order tcp mutants exhibit a phenotype that resembles jaw-D suggesting a role for TCP genes in leaf development in Arabidopsis (20). When an miRNA-sensitive version of TCP4 is expressed under the broadly expressed cauliflower mosaic virus 35S promoter (i.e., 35S:TCP4) the plants fail to exhibit a dramatic leaf or flower phenotype likely due to the presence of miR319, which is sufficient to regulate the ectopically expressed TCP4. But if the miRNA-binding site of TCP4 is mutated (i.e., 35S:mTCP4) resulting in a form of TCP4 that is resistant to miR319, then ectopic expression results in severe developmental defects including seedling lethality (21). These results underlie the importance of regulating the levels of active TCP4 in Arabidopsis development.
Several lines of evidence also suggest a role for miR319 in flower development. jaw-D mutants exhibit short stamens and reduced male fertility (21). This effect on fertility is postulated to be due to cross-regulation by miR319 of the MYB genes MYB33 and MYB65. Because MYB33 and MYB65 have a poor match to the consensus miR319-binding sequence, MYB33 and MYB65 are normally secondary targets of miR319. Instead, MYB33 and MYB65 are regulated more efficiently by miR159 (21), a miRNA family closely related in sequence to the miR319 family. While miR319 has the ability to regulate MYB genes, the mostly non-overlapping expression domains and the low expression level of miR319 makes miR319 regulation of the MYB genes biologically insignificant (21).
Based on the overexpression phenotype, miR319a was postulated to play a key role in leaf development (13). However, analysis of the expression pattern of miR319a indicates that miR319a is not expressed in leaves (22). This suggests that miR319a might play no role in leaf development, a surprising conclusion in the light of the dramatic leaf phenotype in jaw-D. However, it is possible that the other members of the gene family such as miR319b and miR319c function in leaf development, but whether these two genes are expressed in leaves is not known.
Here, we describe the isolation of a loss-of-function allele of miR319a, miR319a129, which exhibits defects in floral organ morphogenesis. A forward genetic screen was performed in a dornröschen-like-2 (drnl-2) mutant background to identify modifiers of the flower development phenotype; in drnl-2 mutants filamentous organs develop in whorl 3 in place of stamens and petals are irregularly shaped (23). In miR319a129 drnl-2 double mutants, in addition to stamens developing as filamentous structures, second whorl petals also develop as filamentous organs. miR319a129 single mutants exhibit strongest defects in petal and stamen development. Here, we demonstrate that miR319a, miR319b and miR319c exhibit distinct expression patterns in seedlings and inflorescences, and that miR319a is expressed most broadly in developing petals and stamens. We also demonstrate that TCP4 is a key target of miR319a in flowers.
Results
Isolation and Characterization of the miR319a129 Mutant.
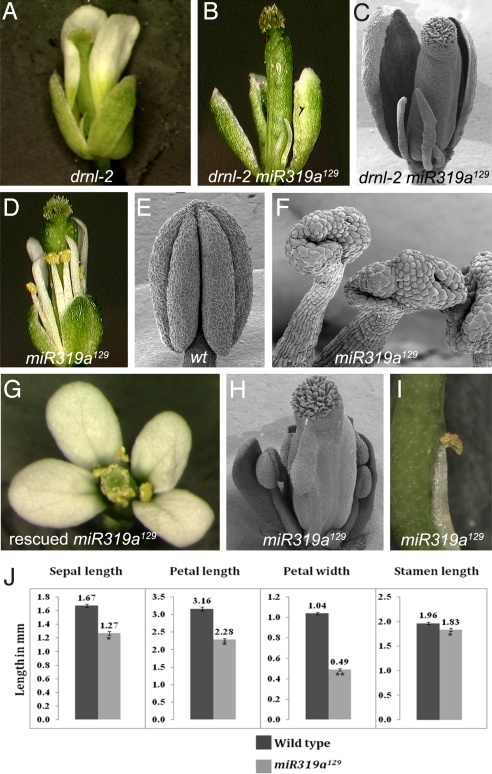
The AP2 domain transcription factor DRNL is critical for proper stamen growth in Arabidopsis (23). To isolate additional components that function together with DRNL in floral organ growth, we performed an enhancer screen in a drnl-2 background. We identified a strong enhancer (mutant 129) that exhibits dramatic defects in the second whorl of the flower. drnl-2 single mutants exhibit only minor defects in petal development characterized by irregular length of petals (Fig. 1A); by contrast drnl-2 129 double mutants (Fig. 1 B and C), exhibit either radialized filaments or dramatically reduced abaxialized petals in whorl 2. In 129 single mutants, developmental defects are present in all four whorls, but the most dramatic defects are in petals and stamens (Fig. 1D). Sepals of 129 single mutants are 18% shorter and often are not tall enough to fully enclose the developing flower (Fig. 1 D and J). On an average, mutant petals are 28% shorter and 50% narrower than wild-type petals at anthesis (Fig. 1 D and J). The stamens in 129 mutants are shorter (Fig. 1 D and J) and anthers are misshapen and lack an organized four-lobed structure (compare Fig. 1 E and F). Additionally, stamens are sometimes fused congenitally to the valve of the carpel (0.14 stamen-valve fusions per flower) (Fig. 1 H and I). Surprisingly, at least some of these fused stamens are able to produce viable pollen (Fig. 1I).
Fig. 1.
miR319a129 mutants exhibit dramatic defects in flower development. (A) drnl-2 mutant flower. Whorl 3 organs are either missing or develop as small filaments; whorl 2 petals are irregularly shaped. One petal has been removed. (B) drnl-2 miR319a129 flower. Both second whorl petals and third whorl stamens are converted to filamentous organs. One sepal was manually removed. (C) SEM of a drnl-2 miR319a129 flower. Two sepals have been removed. (D) miR319a129 flower. Sepals, petals and stamens are stunted; petals are narrow. (E) Wild-type anther exhibiting characteristic four-lobed structure. (F) Anthers from miR319a129 flowers are misshapen and lack an organized four-lobed structure. (G) Transformation of a T-DNA that contains a 3.7-kb genomic DNA containing the miR319a gene rescues the miR319a129 flower mutant phenotype. (H) SEM of a miR319a129 flower showing a stamen congenitally fused to the valve of the carpel. Some perianth organs were manually removed. (I) miR319a129 flower showing a stamen congenitally fused to the valve of the carpel. The fused anther produces pollen. (J) Floral organ size comparison between wild-type and miR319a129. Two-sample unequal variance directional t test was used to test significance of the difference. All of the numbers were significantly different between wild-type and mutant (*, P < 0.05; **, P < 0.005). P values were 0.013, 0.006, 0.000, and 0.020 for sepal length, petal length, petal width and stamen length respectively. Error bars, standard error of mean (SEM). n = 51–121.
We cloned the 129 gene by map-based approaches and found that the 129 mutant contains a G to A change at position 12 of the mature miR319a (At4g23713) (Figs. 2A and S1). The allele of miR319a we isolated (renamed miR319a129) is the only described loss-of-function allele. All other miR319a alleles described to date are overexpression alleles; the best characterized is jaw-D (13). The finding that line 129 contains a mutation in miR319a is surprising because loss-of-function miRNA mutants are rarely recovered in forward genetic screens due both to the small size of the target and to the redundancy of miRNA gene families. We are aware of only four other loss-of-function miRNA mutants in plants isolated based on mutant phenotype: miR164c/early extra petals in Arabidopsis (6), blind in Petunia, fistulata in Antirrhinum (5), and tasselseed4 in maize (12). Transformation of the wild-type miR319a gene into miR319a129 rescues the mutant phenotype providing formal proof that the mutation in miR319a is responsible for the miR319a129 phenotype (Fig. 1G).
Computational predictions (24) suggest that this mutation strongly reduces, and likely eliminates, the ability of miR319a to recognize its targets. Evidence that miR319a129 is non-functional comes from experiments where miR319a129 is ectopically expressed under the 35S promoter; while ectopic expression of wild-type miR319a results in a dramatic leaf phenotype (Fig. S2C) (like jaw-D), plants that ectopically express miR319a129 are phenotypically normal (Fig. S2 D and E).
miR319a, miR319b, and miR319c Exhibit Largely Non-Overlapping Expression Patterns.
The miR319a129 mutant exhibits flower-specific phenotypes despite the presence of highly similar and potentially redundant miR319 family members (Fig. 2A). Because the three miRNA products produced by the three miR319 genes are identical (or nearly identical), it is not possible to analyze the expression pattern of individual gene products using standard techniques such as northern or in situ hybridization. To ascertain if the miR319a129 phenotype could be attributed to the difference in expression of miR319 family members we analyzed expression of miR319a, miR319b, and miR319c using promoter-GUS fusions. Of the three miR319 genes, miR319b exhibits the most restricted expression pattern; GUS activity in miR319b:GUS lines is detected only in the sepal and stamen abscission zones of inflorescences late in flower development (Fig. 2J). By contrast, miR319a and miR319c are expressed in both seedlings and inflorescences (Fig. 2 B–I). In miR319a:GUS seedlings, GUS activity is detected at high levels in the stipules, but not in leaf primordia (Fig. 2 D and E). In miR319c:GUS seedlings, GUS activity is detected at highest levels at the base of leaf primordia and young leaves, but not in the stipules (Fig. 2 B and C). The seedling expression of miR319a and miR319c is completely non-overlapping and, miR319c, but not miR319a, is expressed in leaves, suggesting that miR319a might not play an important role in leaf development.
Fig. 2.
TCP and miR319 expression. (A) Mature miRNA sequence of three miR319 genes in Arabidopsis: miR319a, miR319b, and miR319c. miR319a and miR319b encode the identical miRNA. The mutation (G to A) in miR319a129 is indicated by the arrow above the miR319a sequence. The miR319 genes target a subset of TCP transcription factors such as TCP4. The tcp4soj6 mutation (C to T) is indicated by the arrow below the TCP4 sequence. The tcp4soj6 and miR319a129 mutations are complementary. (B) Whole-mount miR319c:GUS seedling. The base of young leaf primordia is strongly stained. (C) Section through a miR319c:GUS seedling. The base of young leaf priordia is strongly stained. (D) Whole-mount miR319a:GUS seedling. Stipules are strongly stained. (E) Section through miR319a:GUS seedling. Stipules are strongly stained. (F) Section through two miR319c:GUS flowers. In the stage 5 flower (Right), the pedicel and base of organ primordia are stained. In the stage 8 flower (Left), the petals, stamen filaments, and the base of the carpels and sepals are stained. Flowers staged according to Smyth et al. (25). (G) Section through a stage 10 miR319a:GUS flower. The proximal region of the petals is stained. (H) Section through a stage 8 miR319a:GUS flower. The petals are strongly stained. (I) Section through three miR319a:GUS flowers. In the stage 5 flower (Center) GUS activity is detectable at the base of the sepals and throughout developing second, third, and fourth whorl organs. In the stage 6 (Left) and stage 7 (Right) flowers, GUS activity is detectable at the base of the sepals and carpels, and at high levels throughout developing petals. In stamens, higher levels of GUS activity are detected in the stamen filament primordia compared to anther primordia. (J) Whole-mount miR319b:GUS inflorescence. The sepal and stamen abscission zones of late stage flowers are stained. (K) qRT-PCR demonstrates that TCP2, TCP3, TCP4, TCP10, and TCP24 RNA levels increase between 1.4- and 2.5-fold in inflorescences from miR319129 mutants as compared to wild-type. TCP genes are down-regulated in jaw-D inflorescences. Error bars, SEM. All values are significantly different from wild-type. P values for TCP2, TCP3, TCP4, TCP10, and TCP24 are 0.000, 0.028, 0.006, 0.029, and 0.029 respectively for miR319a129 and 0.026, 0.003, 0.000, 0.001, and 0.003 respectively for jaw-D. *, P < 0.05, ‡, P < 0.005.
In miR319a:GUS inflorescences, GUS activity is detectable from floral stages 4–11 (25), with highest levels and most persistent expression in developing petals (Fig. 2 G–I). In miR319c:GUS inflorescences, GUS activity is detectable from floral stages 1–12, with highest levels at the base of all four floral organs and in the pedicel (Fig. 2F). Thus, in the inflorescence, some spatiotemporal overlap of miR319a and miR319c expression is observed; but only miR319a is expressed persistently throughout developing petals. In summary, the expression patterns for the three miR319 genes in the inflorescence are distinct suggesting that these three genes may have largely unique developmental functions.
TCP Genes Are Up-Regulated in miR319a129.
The best-characterized targets of miR319 are five TCP genes that have the miR319 binding sequence: TCP2, TCP3, TCP4, TCP10, and TCP24 (13, 14, 21). In jaw-D, RNA levels of these five TCP genes are reduced 2–34-fold in seedlings (13) and 2- to 5-fold in inflorescences (Fig. 2K). Consistent with the prediction that loss-of-function of miR319a129 would lead to increased expression of the TCP genes, we observe that RNA levels of these five TCP genes are up-regulated between 1.4- and 2.5-fold in miR319a129 inflorescences (Fig. 2K).
Excess TCP4 Activity in the Flower Results in Dramatic Floral Defects.
To ascertain whether miRNA regulation of TCP4 is important for flower development, we expressed a miR319-resistant form of TCP4 (mTCP4) (13) under the control of the petal- and stamen-specific AP3 promoter (26, 27). Expression of a wild-type version of TCP4 under the AP3 promoter (AP3:TCP4) has very limited effects on flower development (Fig. 3G, inset) presumably because miR319 can still regulate TCP4. By contrast, strong AP3:mTCP4 lines exhibit a complete absence of petals and stamens (Fig. 3 A–C); the flowers are extremely small (Fig. 3C) and the sepals are abnormally fused both to themselves and to the pedicel (Fig. 3 A and C). Surprisingly, when AP3:TCP4 is present in a miR319a129 background (Fig. 3G) petals and stamens fail to develop resulting in a flower phenotype identical to AP3:mTCP4.
Fig. 3.
miR319a targets TCP4. (A) AP3:mTCP4 flowers consists of only sepals and carpels; petals and stamens are missing. (B) Scanning electron micrograph (SEM) of a AP3:mTCP4 flower. One sepal has been manually removed to allow visualization of the inner whorls. (C) AP3:mTCP4 flower (Right) is dramatically smaller than wild-type flower (Left). (D) 35S:TCP4 seedlings are normal. (E) 35S:mTCP4 seedlings are seedling lethal. (F) 35S:TCP4 miR319a129 seedlings are seedling lethal and resemble 35S:mTCP4 seedlings. (G) AP3:TCP4 miR319a129 flowers (Upper Left) and inflorescence (Right). AP3:TCP4 miR319a129 flowers resemble AP3:mTCP4 flowers (A–C) and are much smaller than wild-type (Center) or AP3:TCP4 flowers (Inset).
miR319a regulation of TCP4 is also important during seedling development. While 35S:TCP4 seedlings develop normally (Fig. 3D), ectopic expression of a miR319-resistant form of TCP4 (35S:mTCP4) results in seedling lethality (Fig. 3E) (13, 28, 29). Interestingly, similar to 35S:mTCP4, 35S:TCP4 leads to seedling lethality in the miR319a129 background (Fig. 3F). These lines of evidence strongly suggest that miR319a is critical for proper regulation of TCP4 during seedling and flower development.
Suppression of miR319a129 Phenotype by Compensatory Mutation in TCP4.
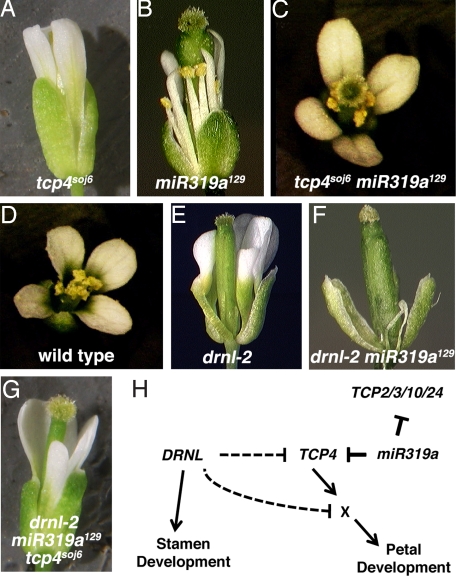
To investigate which of the five TCP genes is most critical for the miR319a129 phenotype, we made use of the tcp4soj6 allele which was isolated as a genetic suppressor of the curled leaf phenotype of jaw-D (21). tcp4soj6 contains a mutation in the miR319 binding site of TCP4, at a position complementary to miR319a129 mutation (Fig. 2A). tcp4soj6 single mutants are phenotypically normal (Fig. 4A). By contrast, miR319a129 tcp4soj6 double mutants exhibit a partial suppression of the second whorl narrow petal phenotype observed in miR319a129 (Fig. 4C and Fig. S3). The ability of tcp4soj6 to suppress the miR319a129 phenotype presumably is due to the restoration of TCP4 targeting by miR319a in the miR319a129 tcp4soj6 double mutant. The fact that the petal phenotype of miR319a129 mutants is not completely suppressed in miR319a129 tcp4soj6 double mutants suggests that miR319a regulates other genes in petals in addition to TCP4, likely TCP2, TCP3, TCP10, and TCP24. In contrast to miR319a129 single mutants, which have a strong flower phenotype, tcp4soj6 single mutants do not have a flower phenotype, suggesting that regulation of other TCP genes is critical for proper flower development. Despite this caveat, our results suggest that TCP4 is a key target of miR319a in petals. Another line of evidence that TCP4 is a key regulator in petal development comes from the analysis of drnl-2 miR319a129 tcp4soj6 triple mutants (Fig. 4G) in which the filamentous petal defects present in whorl 2 of drnl-2 miR319a129 double mutants are rescued (compare Fig. 4 F and G). Analysis of expression of the TCP genes via qRT-PCR in drnl-2 mutant inflorescences indicates that TCP4 RNA levels are not elevated (Fig. S4). Taken together, these results suggest that although miR319a acts directly on TCP4, DRNL likely functions on the petal development pathway downstream of TCP4 (Fig. 4H).
Fig. 4.
tcp4soj6 suppresses the phenotype of both miR319a129 and miR319a129 drnl-2. (A) tcp4soj6 flowers have a wild-type phenotype. (B) miR319a129 flowers have several defects including narrow petals and short stamens. (C) The petals of tcp4soj6 miR319a129 double mutants are taller and more lobed than miR319a129 single mutants. (D) Wild-type flower. (E) drnl-2 flowers exhibit strongest phenotypic effects in stamens while petals are only mildly affected. (F) drnl-2 miR319a129 flowers have dramatically reduced and filamentous petals and stamens. (G) Compared to the drnl-2 miR319a129 double mutant (F), the drnl-2 miR319a129 tcp4soj6 triple mutant develops petals in whorl 2. (H) A model for petal development mediated by DRNL, miR319a, and TCP4. drnl-2 single mutants exhibit only minor defects in petal development, whereas miR319a129 mutants have narrow petals that are half as wide as wild-type petals. miR319a129 drnl-2 double mutants exhibit a very dramatic defect in petal development characterized by filamentous petal formation. The petal defect in the double mutant is rescued by tcp4soj6 suggesting that the phenotypic enhancement in drnl-2 is mediated through TCP4. Thus, TCP4 (or a TCP4 target) is a critical convergence point in the petal development pathway controlled by both DRNL and miR319a. Although proper regulation of TCP4 is essential for petal development in a drnl-2 background, tcp4soj6 cannot rescue the filamentous stamen phenotype in whorl 3 of drnl-2 miR319a129 double mutants indicating the phenotypic defects in whorl 3 of drnl-2 are not mediated by TCP4.
Discussion
Previous studies that focused on overexpression alleles of miR319a such as jaw-D, suggested that miR319a played a key role in leaf development (13). Here, we demonstrate that a loss-of-function allele of miR319a results in phenotypic defects in flowers, but not leaves, demonstrating that miR319a is critical for proper flower development. In miR319a129 mutants, the most dramatic defects are in petals and stamens. The isolation of the loss-of-function miR319a129 allele in a forward genetic screen is surprising since miR319 is redundantly encoded in the Arabidopsis genome; thus, loss of a single miR319 gene would not be expected to result in a mutant phenotype because of this redundancy. However, we show that the three miR319 genes exhibit largely non-overlapping spatial expression patterns in seedlings and inflorescences, and that miR319a is the only one of the three miR319 genes that is broadly and persistently expressed in developing petals, likely explaining why we see the strongest phenotypic defects in these organs in miR319a129 mutants. Thus, the three members of the miR319 family exhibit distinct expression patterns suggesting that they possess unique developmental functions. The precise developmental roles of miR319b and miR319c will be clarified when loss-of-function alleles become available.
It is well-established that the key targets of miR319 are a subset of TCP transcription factors, specifically TCP2, TCP3, TCP4, TCP10, and TCP24 (13, 21). In jaw-D, these five TCP genes are dramatically down-regulated in seedlings as a result of targeting of TCP transcripts for degradation by miR319a. A similar, although less dramatic, reduction in TCP expression is observed in jaw-D inflorescences (Fig. 2K). In the loss-of-function miR319a129 mutant, we see an increase in TCP RNA levels in inflorescences (Fig. 2K). The increase (1.4- to 2.5-fold) that we observe is not as dramatic as the decrease observed in jaw-D; however, this is not surprising since miR319a is expressed in only a subset of cells in inflorescences (Fig. 2 G–I), so in essence we are observing an increase in TCP targets in a small number of cells of the inflorescence that express miR319a.
miR319 and Leaf Development.
The results of this study raise the question as to whether miR319a has a function in normal leaf development. Our analysis of the expression pattern of miR319a indicates that the miR319a promoter is not active in developing leaves (Fig. 2 D and E) (22). By contrast, the miR319c promoter is active in the proximal region of developing leaves (Fig. 2 B and C). This suggests that miR319c, but not miR319a might function in leaf development. Since miR319a and miR319c produce very similar miRNA products, the overexpression leaf phenotype of jaw-D might result from miR319a regulating normal miR319c targets in the leaf.
Although the TCP2, TCP3, TCP4, and TCP24 promoters are active throughout leaves (30), TCP4 RNA is detected only in the distal end of the leaf (13). miR319c is expressed in the proximal region of the leaf and likely targets TCP RNAs for degradation. Similar expression patterns are observed in tomato leaves, where there is an opposing and overlapping gradient of LANCEOLATE (LA) RNA (a TCP3/TCP4 ortholog) and a miR319 family member (28). The LA transcript accumulates at the distal end of the leaf whereas tomato miR319 is expressed at high levels in the proximal leaf region. It is hypothesized that the relative levels of miR319 and LA in different parts of the tomato leaf regulate the developmental fate of the leaf (28). In the proximal region of the leaf where the miR319 level is high and the LA level is low, organ initiation is promoted, while in the distal region of the leaf, where the miR319 level is low and LA levels are high, organ differentiation is favored (28). Since there is no overlap in expression of miR319 family members and TCP4 in the distal part of the leaf and miR319c is expressed strongly at the base of the leaves (13, 28), it is tempting to speculate that a similar proximal-distal specification circuit occurs in Arabidopsis as well.
Does TCP4 Function Non-Cell-Autonomously?
miRNA-resistant forms of TCP4 (mTCP4) have dramatic effects on plant development. Expression of mTCP4 under the control of the broadly expressed CaMV 35S promoter results in seedling lethality (Fig. 3E) (13). Previous studies suggest that TCP4 is a negative regulator of cell proliferation (16, 19). Thus, when constitutively active (i.e., miRNA-resistant) forms of TCP4 are expressed in plants, organ development and differentiation are suppressed. This activity of TCP4 explains, in part, the phenotype of plants that express mTCP4 under the control of the petal and stamen-specific AP3 promoter (Fig. 3 A–C). In pAP3:mTCP4 plants, petals and stamens do not develop (Fig. 3B), likely as a result of constitutively active forms of TCP4 being expressed during very early stages of flower development in petal and stamen primordia. Surprisingly, the phenotype of AP3:mTCP4 flowers is not confined to petals and stamens. AP3:mTCP4 flowers are much smaller than wild-type and the sepals are abnormally fused both to each other and to the pedicel (Fig. 3 A–C). These phenotypes outside of the petals and stamens suggest that mTCP4 may function non-cell-autonomously; that is, that expression of mTCP4 in the petals and stamens is altering the development of surrounding cells and tissues. It is unlikely that these effects are due to leakiness of the AP3 promoter, and subsequent low-level expression of mTCP4 in cells outside the petals and stamens. The best evidence for this comes from analysis of lines that ectopically express diptheria toxin (DT-A) under the control of the AP3 promoter (31). In the AP3:DT-A transgenic lines, petals and stamens fail to develop, but other floral organs (sepals and carpels) develop normally and the flowers are of normal size. Thus, the phenotype of AP3:mTCP4 likely is due to the fact that mTCP4 functions non-cell-autonomously in the flower, similar to what has been observed with other TCP genes such as TCP14 (32). However, the data that we have assembled at present falls short of formal proof that TCP4 functions non-cell-autonomously. It is also possible that one or more downstream targets of TCP4 functions non-cell-autonomously or that expression of excess TCP4 in whorl 2 and whorl 3 during very early stages of flower development grossly alters cell growth and division leading to a dramatic change in surrounding floral structures.
Controlling the Level of Active TCP4 Is Critical for Proper Petal and Stamen Development.
In miR319a129, the transcript levels for all five TCP targets are up-regulated. To identify which of the TCP targets mediate the defects observed in miR319a129, we used two different approaches. First we expressed a miRNA non-regulatable version of TCP4 in petals and stamens. This resulted in either severe reduction of petal and stamen formation or a complete failure of petal and stamen development (Fig. 3 A and B). Interestingly, expression of wild-type TCP4 using the petal- and stamen-specific AP3 promoter, in the miR319a129 background also led to a failure of petal and stamen development (Fig. 3G). This indicates that a failure to down-regulate TCP4 in petals and stamens leads to severe developmental defects. Additionally, a similar experiment using the 35S promoter suggests that excess TCP4 can only be tolerated in presence of functional miR319a during early seedling development (Fig. 3F).
A more convincing line of evidence that TCP4 is a key target of miR319a comes from the analysis of double mutants between miR319a and tcp4soj6. The mutation in tcp4soj6 is complimentary to the mutation in miR319a129 (Fig. 2A). In the tcp4soj6 miR319a129 double mutant, the prediction is that the mutant miR319a should be able to target the mutant TCP4 mRNA for degradation. We observe a partial rescue of the flower phenotype in the tcp4soj6 miR319a129 double mutant (Fig. 4C and Fig. S3) indicating that TCP4 mediates many aspects of the phenotype observed in miR319129. But the fact that a wild-type petal phenotype is not restored in miR319a129 tcp4soj6 double mutants suggests that miR319a regulates other genes in petals in addition to TCP4; likely some combination of TCP2, TCP3, TCP10, and TCP24.
A Model for Petal Development Mediated by DRNL, miR319a, and TCP4.
In this study, we have uncovered a role for miR319a in flower development, specifically in petal and stamen development (Fig. 4H). In miR319a129 single mutants, the petals are narrow and the stamens have defects in filament length and anther morphology. The miR319a129 petal phenotype is dramatically enhanced in the drnl-2 miR319a129 double mutant resulting in second whorl organs that develop as small filamentous organs. In both drnl-2 and miR319a129 single mutants, organs clearly recognizable as petals develop, but these organs are not completely wild-type. Thus, DRNL and miR319a function redundantly with regard to petal growth. The fact that tcp4soj6 can rescue the petal defects of drnl-2 miR319a129 double mutants (i.e., the drnl-2 miR319a129 double mutant lacks petals, but petals are restored in the drnl-2 miR319a129 tcp4soj6 triple mutants) suggests that the proper levels of active TCP4 are critical for proper petal development. miR319a directly targets TCP4 mRNA for degradation, but DRNL does not regulate TCP4 at the level of RNA accumulation (Fig. S4). Most likely, DRNL functions downstream of TCP4, although we have not ruled out the possibility that DRNL functions post-transcriptionally, for example, by altering the level or activity of the TCP4 protein.
Materials and Methods
Plant Growth Conditions.
Plants were grown under continuous light conditions in a 2:1:1 mixture of Promix:perlite:vermiculite at 23 °C. Transgenic plants were selected on Murashige and Skoog medium containing 50 μg/mL kanamycin sulfate or 50 μg/mL BASTA.
drnl-2 Modifier Screen.
drnl-2 homozygous seeds were mutagenized with 0.25% ethyl-methane-sulfonate (EMS). Because of the male fertility defects in drnl-2, it is difficult to generate a large number of homozygous seeds. Thus, we mutagenized a limited number of drnl-2 seeds and isolated seeds from 55 M1 plants, and 750 M2 plants were screened. After initial isolation, the 129 mutant was backcrossed to wild-type L-er, and in the F2 of the backcross, plants with the following phenotypes segregated: 179 wild-type: 63 drnl-2: 66 miR319a129 : 22 drnl-2 miR319a129, a ratio consistent with segregation of two unlinked nuclear genes in a 9:3:3:1 ratio.
Map-Based Cloning of miR319a129.
drnl-2 miR319a129 mutants were crossed to Columbia and DNA samples were prepared from individual mutant F2 plants (33). Two independent pools of 17 and 21 samples were subjected to bulk-segregant analysis (34). 411 independent miR319a129 homozygous mutants were used to perform fine mapping using SSLP, CAPS, and dCAPS markers (Table S1) designed using information available on TAIR (http://www.arabidopsis.org/) or reported by CEREON (34). Gene annotations from TAIR were used to identify candidate genes.
Scanning Electron Microscopy.
Samples were fixed and visualized as described in ref. 23.
Floral Organ Size Measurement.
Floral organs from anthesis stage flowers were used for scoring. The floral organs were flattened on sticky side of a cellophane tape and photographed. A photograph of a ruler was taken using same magnification was used as a calibration tool. SPOT-Advanced software (Diagnostic Instruments, Inc.) was used to calculate the length of floral organs.
Real-Time qRT-PCR Experiments.
One to two micrograms total RNA extracted using the RNeasy mini kit (Qiagen) was used for random-hexamer primed cDNA synthesis with SuperScript III 1st Strand Synthesis SuperMix (Invitrogen catalog no. 18080-400). The resulting cDNA was subjected to relative quantitative PCR in presence of iQ SYBR Green Supermix (Bio-Rad, catalog no. 170-8882) in a Bio-Rad Icycler or SYBR Premix Ex Taq II (Perfect Real Time) (TaKaRa, cat. #RR081A) in an ABI 7500 Real-Time PCR System. For each reported result at least three independent biological samples were subjected to minimum of three technical replicates. The results were normalized to β-tubulin (TUB2/TUB3). The primers used are reported in Table S2 (13).
Plasmid Constructs and Transgenic Lines.
The constructs used are listed in Table S3. The two complementation constructs containing 1.2- or 1.6-kb promoter sequence were both able to complement miR319a129 phenotype. For the promoter:GUS constructs we used 1.2-, 2.9-, and 2.6-kb regions 5′ to the predicted transcription start site for miR319a, miR319b, and miR319c respectively. Forty-five of 46 AP3:mTCP4 lines generated exhibit small flowers with reduced petals and stamens. All 21 independent AP3:TCP4 transgenic lines in miR319a129 background exhibit phenotypes similar to those described for AP3:mTCP4. The expression pattern we report here was observed in 12/14, 9/12, and 6/10 independent lines generated for the miR319a:GUS, miR319b:GUS, and miR319c:GUS constructs respectively.
GUS Staining.
Promoter:GUS fusion lines were stained using a standard protocol for 24–48 h (35).
Supplementary Material
Acknowledgments.
We thank D. Weigel and H. Wollman for generously providing tcp4soj6 seeds, TCP4 and mTCP4 constructs; C. Daghlian for his help with SEM; and the Arabidopsis Biological Resource Center for jaw-1D and jaw-3D seeds. This work was supported by the United States National Science Foundation Grants IBN-0516736 and IOS-0926347.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908718106/DCSupplemental.
References
- 1.Garcia D. A miRacle in plant development: Role of microRNAs in cell differentiation and patterning. Semin Cell Dev Biol. 2008;19:586–595. doi: 10.1016/j.semcdb.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Shukla LI, Chinnusamy V, Sunkar R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta. 2008;1779:743–748. doi: 10.1016/j.bbagrm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu PP, et al. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 5.Cartolano M, et al. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet. 2007;39:901–905. doi: 10.1038/ng2056. [DOI] [PubMed] [Google Scholar]
- 6.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulationg petal number in Arabidopsis. Curr Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 8.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 10.Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33, and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen RS, et al. Genetic analysis reveals functional redundancy and the major target genes of Arabidopsis miR159 family. Proc Natl Acad Sci USA. 2007;104:16371–16376. doi: 10.1073/pnas.0707653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 13.Palatnik JF, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 14.Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford BC, Nath U, Carpenter R, Coen E. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 2004;135:244–253. doi: 10.1104/pp.103.036368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 18.Luo D, et al. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/s0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- 19.Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 20.Schommer C, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palatnik JF, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Warthmann N, Das S, Lanz C, Weigel D. Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol. 2008;25:892–902. doi: 10.1093/molbev/msn029. [DOI] [PubMed] [Google Scholar]
- 23.Nag A, Yang Y, Jack T. DORNRÖSCHEN-LIKE, and AP2 gene necessary for stamen emergence in Arabidopsis. Plant Mol Biol. 2007;65:219–232. doi: 10.1007/s11103-007-9210-7. [DOI] [PubMed] [Google Scholar]
- 24.Zucker M. AMfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill TA, Day CD, Zondlo SC, Thackeray A, Irish VF. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development. 1998;125:1711–1721. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- 27.Tilly J, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- 28.Ori N, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39:787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 29.Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell. 2008;20:2293–2306. doi: 10.1105/tpc.107.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day CD, Galgoci BF, Irish VF. Genetic ablation of petal and stamen primordia to elucidate cell interactions during floral development. Development. 1995;121:2887–2895. doi: 10.1242/dev.121.9.2887. [DOI] [PubMed] [Google Scholar]
- 32.Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008;53:42–52. doi: 10.1111/j.1365-313X.2007.03308.x. [DOI] [PubMed] [Google Scholar]
- 33.Dellaporta SL, Wood VP, Hicks JB. A plant DNA mini-preparation: Version II. Plant Mol Biol Reporter. 1983;4:19–21. [Google Scholar]
- 34.Jander G, et al. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–550. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campisi L, et al. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. doi: 10.1046/j.1365-313x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.