Abstract
We have applied an optical super-resolution technique based on single-molecule localization to examine the peripheral distribution of a cardiac signaling protein, the ryanodine receptor (RyR), in rat ventricular myocytes. RyRs form clusters with a mean size of approximately 14 RyRs per cluster, which is almost an order of magnitude smaller than previously estimated. Clusters were typically not circular (as previously assumed) but elongated with an average aspect ratio of 1.9. Edge-to-edge distances between adjacent RyR clusters were often <50 nm, suggesting that peripheral RyR clusters may exhibit strong intercluster signaling. The wide variation of cluster size, which follows a near-exponential distribution, is compatible with a stochastic cluster assembly process. We suggest that calcium sparks may be the result of the concerted activation of several RyR clusters forming a functional “supercluster” whose gating is controlled by both cytosolic and sarcoplasmic reticulum luminal calcium levels.
Keywords: excitation-contraction coupling, fluorescence, heart, single molecule, super-resolution
Ryanodine receptors (RyRs) are widely expressed intracellular calcium channels that provide Ca2+ release in muscle, neurons, and many other tissues (1, 2). In cardiac myocytes, forceful contraction depends on the near-simultaneous cell-wide release of Ca2+ through RyRs located in junctions between the intracellular Ca2+ store (the sarcoplasmic reticulum or SR), and the cell surface and transverse tubular membranes (3). From electron microscopy, RyRs are thought to cluster within these junctions where they form regular “quasi-crystalline” arrays with a unit cell size of approximately 30 nm (1, 4). The arrangement of RyR clusters facing the narrow junctional gap between SR and sarcolemmal membranes (≈10–15 nm) is thought to critically determine the properties of microscopic release events called Ca2+ sparks that form elementary events in cardiac Ca2+ signaling (5, 6). Understanding the Ca2+-dependent interactions between RyRs within a cluster and the interaction between adjacent clusters (which are coupled via diffusion through the cytosolic space) requires knowledge of the morphology and distribution of RyR clusters. For example, it is unknown if abnormalities in RyR signaling (7) are associated with changes in cluster size or distribution.
Peripheral RyR clusters, which occur in junctions between terminal SR and the surface membrane, are functionally active and thought to represent the typical behavior of RyR clusters during excitation-contraction (EC) coupling (8, 9). They therefore provide a well defined and functionally characterized source of RyR clusters for structural studies. The development of imaging techniques based on the time-multiplexed observation of single fluorescent molecules allows high-contrast fluorescence imaging with a resolution limited only by signal to noise (10–12). Using standard fluorochromes (13–15) we have used this approach to investigate the distribution of peripheral RyRs with a resolution of approximately 30 nm. Quantitative analysis of our data shows that the size and morphology of RyR clusters in peripheral couplings is quite different from that deduced by previous studies. We also show that RyR clusters are often found in groups containing several closely spaced clusters and speculate that these groups may act synergistically as single calcium release units (CRUs).
Results
RyR Distribution in Peripheral Clusters.
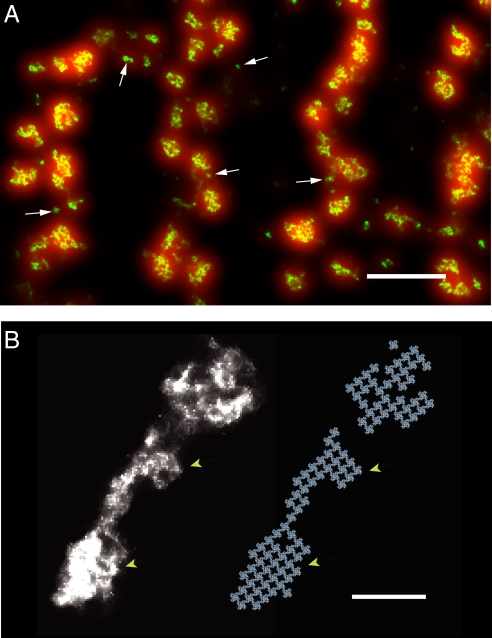
Rat ventricular myocytes, labeled with an antibody against the cardiac RyR2 and a secondary antibody carrying Alexa Fluor 647 (see Materials and Methods) were imaged using a custom-built localization microscope (13). Fig. 1A shows the typical peripheral RyR labeling pattern that was seen. Comparison between the diffraction-limited image (red) and the corresponding localization image (green) demonstrates the clear enhancement in resolution provided by localization microscopy. At conventional, diffraction-limited resolution (≈270 nm), the labeling shows only irregular double rows of puncta aligned with z-lines as described in ref. 16. From their apparent dimensions, such puncta would contain approximately 80–140 RyRs if uniformly filled by RyRs, and similar peripheral cluster sizes have been estimated from thin sectioning (17). In contrast, the high-resolution localization image shows that the red puncta are incompletely filled with RyRs, often containing several smaller clusters. Additionally, the shape and size of the RyR clusters revealed by the localization data are quite variable.
Fig. 1.
Imaging of peripheral ryanodine receptor (RyR) clusters with single protein resolution. (A) Image of a small region on the surface of a rat cardiac myocyte showing four successive z-lines with double rows of RyR clusters at each z-line. The conventional (diffraction-limited) fluorescence image in which all clusters appear as shapeless blurs is shown in red. Overlaid on this image is the localization image (green) revealing high-resolution structure within these blurs. Note the wide variation in cluster size and morphology. Arrows identify several smaller clusters which could be identified in the localization image but not in the conventional image. (Scale bar, 1 μm.) (B) Magnified view of two clusters alongside a reconstruction with candidate positions for the underlying approximately 30 nm wide RyR channels (each shown as a quatrefoil structure adapted from ref. 18). Note the sharp, straight, cluster edges and the edge dislocations corresponding to the width of single RyRs. Based on the reconstructions the clusters contain 61 and 39 RyRs, respectively. (Scale bar, 200 nm.)
The RyR protein is a large homo-tetramer of approximately 560 kDa subunits, with a quatrefoil structure that occupies a volume of approximately 29 × 29 × 12 nm as measured in the skeletal RyR1 isoform (1). RyR2 has a similar shape and size (18), and both cardiac and skeletal isoforms are thought to form quasi-crystalline arrays in the SR membrane (1, 19). Formation of regular arrays appears to be an intrinsic property of RyRs as shown by their spontaneous formation in solution and lipid bilayers (4, 20) as well as their occurrence in preparations from skeletal muscle (21, 22). Within these arrays, the RyRs have a center-to-center spacing of approximately 30 nm. The precise two-dimensional arrangement of RyRs in mammalian cardiac muscle has not been shown to date, evidence from frog myocardium suggests a similar regular array structure (23).
With the assumption of a 30-nm unit cell, we assigned candidate RyR positions to produce a model for RyR location which matched the localization data (Fig. 1B). Note the presence of sharp cluster edges and dislocations in the data matching the model. While the achieved effective resolution of approximately 30 nm was not quite sufficient to directly observe the regular grid structure of RyRs, the presence of sharp edges and steps in the outline of the clusters is clearly compatible with a regular array that has an approximate 30-nm unit size. This is direct evidence for a two-dimensional array-like arrangement of RyRs in cardiac muscle.
RyR Clusters Are Small and Closely Spaced.
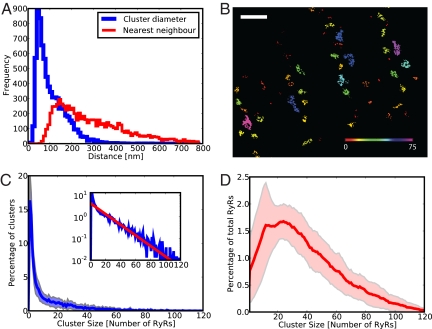
Because of the high contrast provided by fluorescent labeling, it was possible to automatically segment the localization data (see Materials and Methods) and obtain statistical data on cluster morphology. A total of 7,675 clusters were measured in 22 cells. Fig. 2A shows the resulting distribution of cluster diameters and nearest neighbor distances between cluster centroids, with mean values of 107 nm and 308 nm, respectively. While intercentroid distances convey the distribution pattern of clusters on the membrane, cross-signaling between RyR clusters (via calcium-induced calcium release, CICR) would depend strongly on the edge-to-edge distances between RyR arrays and about one third of clusters were within 50 nm of their nearest neighbors (see Figs. S1 and S2).
Fig. 2.
Quantitative analysis of RyR cluster properties. (A) Distribution of cluster diameters and nearest neighbor distances. Clusters are considerably smaller than previously thought and neighbouring clusters are often close enough to be inseparable at conventional imaging resolution. (B) A region of the surface sarcolemma of a cardiac myocyte in which segmented RyR clusters are shown color-coded according to the number of RyRs they contain. Note the presence of many small clusters (red corresponds to NRyRs ≤ 7). (Scale bar, 1 μm.) (C) The number of RyRs per cluster follows an approximately exponential distribution with a mean RyR number of 13.6. Shaded area represents ± 1 SD (n = 22 cells). Inset: the same data on a log scale with a maximum-likelihood fit to an exponential distribution (red). (D) Fraction of the total number of RyRs in clusters of a given size. This is interesting from a functional perspective as it tells us which sizes of clusters are likely to make the largest contribution to calcium signaling. The shaded area represents ± 1 SD.
The peripheral clusters were imaged in membrane areas in close contact with the coverslip and are therefore essentially flat and parallel to the image plane. Therefore, the apparent cluster area can be directly converted into the number of RyRs in the cluster (based on 1 RyR per 900 nm2). Fig. 2B shows a representative region in which segmented clusters have been color coded according to the number of RyRs they contain. The calculated cluster sizes followed an approximately exponential distribution with a mean of 13.6 RyRs (n = 22 from three animals) per cluster (see Fig. 2C). It is notable, that this result is much smaller than previous estimates of approximately 100 RyRs per cluster. There was modest cell to cell variation in cluster size (shaded region in Fig. 2C), and very little variation between animals (see Fig. S3). Although the large number of very small clusters shown in Fig. 2C is striking, Fig. 2D shows that the majority of RyRs are found in somewhat larger clusters of ≈25 RyRs. Some clusters were incompletely filled with RyRs, an observation consistent with a recent electron microscopic study which has shown interior junctions to be incompletely filled with RyRs (24).
Because some groups of clusters are in close proximity it is possible that several smaller clusters may act functionally as one calcium release unit or “supercluster” (see also Fig. S4). We therefore also calculated the total number of receptors in cluster groups that were within 100 nm of each other (edge-to-edge) and obtained a mean supercluster size of 21.6 RyRs (n = 10 cells from two animals). In agreement with a previous study (16), we find that virtually all RyR clusters occur within an approximately 700 nm wide band centered on z-lines with only few located between z-lines.
RyR Clusters Have Complex Geometries.
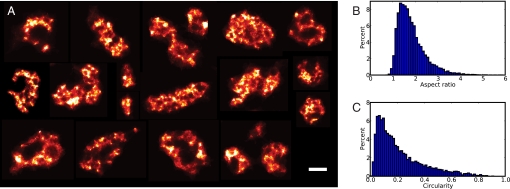
A random selection of larger clusters is shown in Fig. 3A, illustrating the diversity of peripheral RyR cluster shapes. One commonly occurring feature is a crescent or doughnut shaped cluster. In some cases such shapes would be consistent with the presence of a t-tubule opening (≈200 nm) and RyR clusters at the mouth of t-tubules have been seen previously in EM thin sections (25).
Fig. 3.
Morphology of RyR clusters. (A) A collage of randomly selected larger RyR clusters that illustrate the variable morphology. (Scale bar, 200 nm.) (B) The distribution of RyR cluster aspect ratios shows that clusters are typically elongated (mean ratio = 1.9). (C) The circularity, which quantifies how compact RyR clusters are, is much smaller than expected for a circle (circularity = 1) with a mean of 0.2 and reflects the complex outlines of these clusters.
To quantify these cluster geometries we measured RyR cluster aspect ratios and their “circularity” (a measure of compactness, with a perfect circle having a circularity of 1) as shown in Fig. 3 B and C. Most RyR clusters were moderately elongated (mean aspect ratio ≈1.9, see Materials and Methods). The mean circularity of RyR clusters was approximately 0.2, implying a very complex outline (note that a rectangle with the same average aspect ratio would have a circularity of 0.7).
Stochastic Self-Assembly Can Explain RyR Cluster Size.
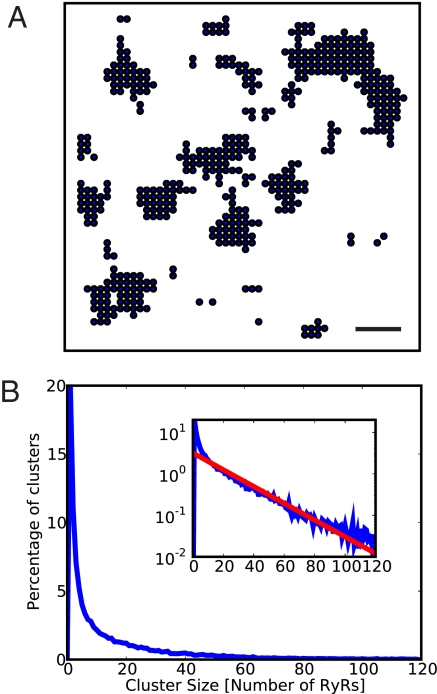
Given the highly ordered double-row structure seen in skeletal muscle (26), it is possible that RyR clusters are assembled onto a structural template. The approximately exponential size distribution seen here, however, argues against the idea that RyR cluster size is very tightly controlled. To test the idea that stochastic self-assembly could explain the observed RyR cluster size distribution we used a Monte-Carlo model of cluster growth. This model incorporated a nucleation process (to seed new clusters), accretion (that adds further RyRs), and turnover. Cluster shapes resulting from one Monte-Carlo simulation are shown in Fig. 4A, which are qualitatively similar to those observed. The size distribution obtained from 1,000 simulation runs (Fig. 4B) was a close match to the measured distribution and even exhibited a similar departure from exponential behavior at low cluster sizes (compare Fig. 2C). The histograms shown here were calculated with a simple model but more complex models that explicitly consider diffusion of receptors in the SR membrane and binding between RyRs yield similar results (see SI Text and Fig. S5).
Fig. 4.
Monte-Carlo simulation of stochastic self-assembly of RyR clusters using a simple growth model. The model yields cluster sizes and geometries which are very similar to those measured. (A) The simulation grid at the end of one simulation run (100 iterations) in which RyRs are shown schematically as circles. (Scale bar, 100 nm.) (B) The size distribution obtained from 1,000 runs. Inset: the same data on a log scale with a maximum-likelihood fit to an exponential distribution (red). The simulation parameters were (probabilities per pixel and iteration step): Pnucleation = 2.5 × 10−4, Pgrowth = 2.5 × 10−2 x Nneighbors, Pretention = 0.937.
Discussion
Using localization microscopy we have produced high-resolution immunocytochemical images of RyR distribution in the periphery of cardiac myocytes. These data show that the RyR clusters have widely varying sizes and shapes, although at a lower resolution, the data are compatible with previous immuno-fluorescence labeling.
Our data reveal that RyR cluster sizes vary widely from single RyRs to >100 RyRs, which seems at variance with the substantially larger estimates for the size of “couplons” within myocytes (17, 27), although the variability in junction size and feet (i.e., RyR) content has been noted in ref. 28. While this could reflect a genuine difference between peripheral and deeper RyR cluster size, it is possible that assumptions necessary for the estimation of cluster sizes from previous data led to an overestimate. For example, in both thin sectioning (17) and quantitative fluorescence imaging (27), the assumption of circularity and complete filling of the junction by close-packed RyRs has been used. Our data suggests that these key assumptions may be incorrect, and this conclusion is supported by some recent EM tomography (24, 29). An additional factor is the comparatively poor contrast in densely stained sections where cluster detection relies on the observation of a regular array of electron dense “feet” and which could make small clusters with only a few RyRs more difficult to identify. Increased confidence in our measurements is provided by comparing our data to stereological measurements from thin sections which suggest that ≈7% of sarcolemmal membrane area is associated with dyads (30). Our RyR clusters occupy approximately 6.4% of membrane area when small holes within the clusters are filled (and junctions would be at least as large as the enclosed RyR array). The similarity of these figures suggests that we have correctly identified the RyR clusters that form junctions within the cell.
The RyR clusters that we observed were frequently incompletely filled with RyRs in agreement with a recent study using electron microscopy which found that interior junctions were often incompletely occupied with RyRs (24). We calculated by image processing that filling the observed clusters completely with RyRs would only increase cluster areas by a factor of approximately 1.5 (increasing the mean cluster size to ≈21 RyRs). This would place a lower bound of 66% on the labeling efficiency (for 1 secondary per RyR). This efficiency is high enough to ensure that our segmentation and the resulting cluster size histograms are largely unaffected by (potentially) incomplete labeling, an idea supported by the only moderate increase in mean size when relatively widely spaced clusters (≤100 nm edge-to-edge) were regarded as one supercluster.
Analysis of peripheral spark amplitude has suggested that approximately six RyRs are involved in the genesis of a peripheral calcium spark (8). A higher figure (>18) was suggested from noise analysis of calcium sparks arising from deeper couplons (31). Our data (Fig. 2D) suggests that peripheral calcium sparks should most likely be triggered from clusters containing approximately 25 RyRs, a number which is in reasonable agreement with the latter estimate. This data can also be used to quantify the fraction of RyRs in the smallest clusters (32) (see Fig. 2D), but it is still unclear what the functional properties of such small RyR clusters in situ are. In connection with this point, the wide variation in cluster size (Fig. 2C) suggests that some form of autoregulation [such as local SR depletion (33) and/or luminal gating (34)] may be required to limit the site to site variability in calcium release. Were this not the case, the observed variation of mean cluster size would lead to a >5-fold variation in spark flux (8).
Our data shows that RyR clusters can be closely spaced with approximately 50% having edge-to-edge distances less than or equal to 100 nm (n = 10 cells). It is likely that clusters in such close proximity could trigger calcium release from each other. It has been estimated that a trigger level of ≈10 μM is needed to explain the approximately 104-fold increase in spark rates resulting from activated RyR clusters within the timescale of EC coupling (35). Computer simulations suggest that such calcium levels may exist within 100 nm of an activated cluster (32). With these figures in mind, we can envision a calcium spark incorporating more than one of the closely spaced RyR clusters. Such an effect may reconcile the quantal nature of peripheral calcium sparks (8) with our data without requiring a subpopulation of active RyRs (≤ 6) within a cluster. This reconciliation may require that the coupling between closely spaced clusters is reduced (perhaps by membrane distortion) to prevent them from always coactivating but allow stochastic variation in the number of activated clusters. It is also possible that several RyR clusters may be connected to the same SR terminal cistern which would result in tight coupling of calcium release between clusters via the cytosolic and luminal SR calcium concentrations. Our data raises the possibility of a new level of Ca2+ signaling via a “triggered saltatory” mechanism for calcium release wherein groups of neighboring clusters form a larger supercluster (see also Fig. S4), which acts in concert to a triggering event. This idea is consistent with data from Hayashi et al. who observed that interior couplings between SR and t-tubules generally consist of several closely spaced contact zones along t-tubules containing each only relatively few RyRs (24).
We have shown that our measured cluster size distribution could be explained by a stochastic process of cluster growth. A stochastic growth process could also explain observations in skeletal muscle derived BC3H1 cells, where smaller RyR clusters of variable orientation are packed into single larger clusters in peripheral junctions (36). Although our simulations assume spontaneous nucleation, it is likely that cluster nucleation in the cell may be secondary to mechanisms mediated by additional proteins [e.g., junctophilin (37)] to ensure the correct placement of junctions. Nevertheless our models show that with the placement of a few localizing proteins appropriately sized RyR clusters may form without requiring an explicit scaffold or other process that tightly controls cluster size. We envisage that future high resolution optical imaging of RyR and related junctional protein distributions (and their change in response to interventions and protein modifications) will yield further insight into the process of RyR cluster formation.
Finally, we have demonstrated that high-resolution immuno-cytochemical localization microscopy can provide information on a scale that fills the gap between FRET and related techniques and conventional fluorescence microscopy. Because this method should be generally applicable to other types of proteins and signaling pathways, we can expect it to provide insight for a variety of cell processes.
Materials and Methods
Sample Preparation.
Ventricular myocytes were enzymatically isolated from adult rats (6 weeks) as described in ref. 38 and fixed in 2% paraformaldehyde for 10 min. They were then labeled according to a standard immunofluorescence protocol (27) using a mouse-anti-RyR2 primary antibody (Ab, Affinity BioReagents, MA3–916) and an Alexa Fluor 647 conjugated goat-anti-mouse secondary Ab (Molecular Probes/Invitrogen).
To prepare a sample slide, approximately 5 μL of the labeled cell suspension was pipetted onto a clean coverslip, approximately 15 μL “switching buffer” (15) (0.5 mg/mL glucose oxidase, 40 μg/mL catalase, 10% wt/vol glucose, and 50 mM β-mercaptoethylamine in PBS, all obtained from Sigma–Aldrich) was added, a slide placed on top, and the edges sealed with nail varnish.
Microscope Setup.
Images were acquired on a commercial Nikon TE2000 inverted microscope with a Nikon 60×, 1.49NA oil-immersion TIRF objective (Nikon), and an Andor IXon DV887DCS-BV electron multiplying CCD camera (Andor Technology). A bandpass interference filter (HQ710/50m, Chroma) was used to select the emitted fluorescence. To the stock microscope we have added custom illumination optics allowing high intensity laser illumination at an adjustable angle, and a custom objective holder coupled directly to the microscope stage and designed to minimize thermal and mechanical drift. Focusing was performed using a piezo focusing unit (Physik Intrumente P-725). Laser illumination was provided by two solid state lasers (Viasho VA-I-LNS-532 and VA-I-N-671) providing focal plane intensities of approximately 107 W/m2 at 543 nm (green) and 109 W/m2 at 671 nm (red), respectively, over a 10-μm diameter field of view. The illuminator was adjusted to generate a highly inclined light sheet (39) at an angle just below the TIRF angle.
Image Acquisition.
Image acquisition was performed using a combination of the techniques described in refs. 12, 13, and 15. Cells were prebleached for approximately 5 s to push a large proportion of fluorophores into the dark state (13, 15) using only the red laser. Image sequences were then acquired, streamed to disk, and analyzed in real time using custom software. An electron multiplication gain of approximately 35 and an integration time of 27 ms were used. The acquisition was terminated when the total number of detected single molecule events had plateaued, typically at 105 to 2 × 105 events. In our experiments, molecules returned spontaneously from the dark state without needing to use an activation laser. Additional use of the green laser resulted in a 3–4-fold increase in the rate at which molecules returned from the dark state, reducing the number of frames required from ≈60,000 to 20,000. Total event numbers and the appearance of the reconstructed images were unaffected by the presence of additional 532-nm illumination.
Image Analysis and Reconstruction.
Single molecule signals were detected and fitted in individual frames as described in ref. 13. The mean estimated localization accuracy was approximately 13 nm, corresponding to an effective resolution of approximately 30 nm. This was consistent with the size of the smallest features present in the image.
Fluorophore density images were reconstructed from the single molecule positions using an approach based on triangulation whereby the point data are triangulated, and the resulting triangles rendered with an intensity proportional to their area (40). To improve the estimation of the underlying epitope distribution and to remove the faceted appearance of the visualization, 10 visualizations were averaged where the underlying points had been randomly jittered by half their mean neighbor distance, effectively blurring the image to its sampling limited resolution. This method reduces the extent to which structure arises purely from the stochastic nature of the distribution of single localization events.
Pseudo-wide-field images were obtained by rendering each localization event as a Gaussian with a half-width equal to that obtained from the fit. It should be noted that these images exhibit some optical sectioning akin to confocal images due to the rejection of out of focus single molecule events.
Cluster Analysis.
Connected clusters of labeling were detected by walking over the edges of a Delaunay triangularization of the fluorophore position data and grouping those points which were separated by edges shorter than our estimated resolution (30 nm). Clusters containing <10 points were discarded on the grounds of their poor signal to noise ratio and to avoid measuring objects generating fewer events than a typical single secondary antibody. In connection with this point we recorded several events per RyR (Fig. S6) consistent with the idea that there are approximately 1–4 Abs/RyR which carry several fluorophores each. When this algorithm was applied to images of no-primary Ab control cells, generally no clusters were detected, confirming very low levels of background staining.
The major and minor axes of a cluster were determined for the point cloud using principal component analysis, and cluster lengths and widths determined by taking the standard deviation along these axes and multiplying by 2.35 (corresponding to the ratio between standard deviation and full width at half maximum for a Gaussian distribution). Cluster aspect ratio was then obtained by taking the ratio of these two measures, and a cluster “diameter” was calculated as the average of major and minor axes.
To obtain a measure of size that was free from the assumption of an underlying distribution, we measured the cluster area by triangulating the fluorophore positions belonging to a cluster, discarding any triangles with edge lengths greater than our estimated resolution (to allow for concave shapes), and summing the area of the remaining triangles. A circumference was also extracted by summing the lengths of all of the exterior edges present in this triangulation, which was used to derive the circularity (4π×area/circumference2) of the clusters.
Cluster Growth Simulation.
Monte-Carlo simulations of a simplified cluster growth model were performed on a 50 × 50 unit rectangular grid (unit size 30 nm), where each grid cell could either contain an RyR, or be empty. Starting with an empty grid, the probability that a grid cell would contain an RyR in the next iteration was given as the sum of a small “nucleation” probability (2.5 × 10−4) which was independent of the RyR occupancy of neighboring cells, a “growth” probability (2.5 × 10−2 × Nneighbors) which was linearly dependent on the number of four-connected neighbors a newly placed RyR would have, and a “retention” probability (0.937 if there was already an RyR present) which allows for some RyR turnover (and was tuned to match the observed mean cluster size). One hundred MC steps were performed to produce each configuration used in the analysis. A more complex model was also implemented (see SI Text).
Supplementary Material
Acknowledgments.
We thank Dr. Patricia Cooper and Ms Cherrie Kong for assistance with cell isolation. This work was supported by grants from the Marsden Fund, the Health Research Council, and the University of Auckland Faculty Research Committee.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908971106/DCSupplemental.
References
- 1.Franzini-Armstrong C, Protasi F. Ryanodine receptors of striated muscles: A complex channel capable of multiple interactions. Physiol Rev. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 2.Sutko JL, Airey JA. Ryanodine receptor Ca2+ release channels: Does diversity in form equal diversity in function? Physiol Rev. 1996;76:1027–1071. doi: 10.1152/physrev.1996.76.4.1027. [DOI] [PubMed] [Google Scholar]
- 3.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 4.Yin CC, Lai FA. Intrinsic lattice formation by the ryanodine receptor calcium-release channel. Nat Cell Biol. 2000;2:669–671. doi: 10.1038/35023625. [DOI] [PubMed] [Google Scholar]
- 5.Cannell MB, Cheng H, Lederer WJ. The control of calcium release in heart muscle. Science. 1995;268:1045–1049. doi: 10.1126/science.7754384. [DOI] [PubMed] [Google Scholar]
- 6.Cannell MB, Soeller C. Numerical analysis of ryanodine receptor activation by L-type channel activity in the cardiac muscle diad. Biophys J. 1997;73:112–122. doi: 10.1016/S0006-3495(97)78052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehrens XH, Lehnart S, Marks A. Intracellular calcium release and cardiac disease. Ann Rev Physiol. 2005;67:69–98. doi: 10.1146/annurev.physiol.67.040403.114521. [DOI] [PubMed] [Google Scholar]
- 8.Shen JX, Wang S, Song LS, Han T, Cheng H. Polymorphism of Ca2+ sparks evoked from in-focus Ca2+ release units in cardiac myocytes. Biophys J. 2003;86:182–190. doi: 10.1016/S0006-3495(04)74095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SQ, Stern MD, Rios E, Cheng H. The quantal nature of Ca2+ sparks and in situ operation of the ryanodine receptor array in cardiac cells. Proc Natl Acad Sci USA. 2004;101:3979–3984. doi: 10.1073/pnas.0306157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 11.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baddeley D, Jayasinghe ID, Cremer C, Cannell MB, Soeller C. Light-induced dark states of organic fluochromes enable 30 nm resolution imaging in standard media. Biophys J. 2009;96:L22–24. doi: 10.1016/j.bpj.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fölling J, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 15.van de Linde S, Kasper R, Heilemann M, Sauer M. Photoswitching microscopy with standard fluorophores. Appl Phys B. 2008;93:725–731. [Google Scholar]
- 16.Chen-Izu Y, et al. Three-dimensional distribution of ryanodine receptor clusters in cardiac myocytes. Biophys J. 2006;91:1–13. doi: 10.1529/biophysj.105.077180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franzini-Armstrong C, Protasi F, Ramesh V. Shape, size, and distribution of Ca(2+) release units and couplons in skeletal and cardiac muscles. Biophys J. 1999;77:1528–1539. doi: 10.1016/S0006-3495(99)77000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X, et al. Three-dimensional localization of serine 2808, a phosphorylation site in cardiac ryanodine receptor. J Biol Chem. 2007;282:25929–25939. doi: 10.1074/jbc.M704474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dulhunty AF. Feet, bridges, and pillars in triad junctions of mammalian skeletal muscle: Their possible relationship to calcium buffers in terminal cisternae and T-tubules and to excitation-contraction coupling. J Membr Biol. 1989;109:73–83. doi: 10.1007/BF01870792. [DOI] [PubMed] [Google Scholar]
- 20.Yin CC, Han H, Wei R, Lai FA. Two-dimensional crystallization of the ryanodine receptor Ca2+ release channel on lipid membranes. J Struct Biol. 2005;149:219–224. doi: 10.1016/j.jsb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Paolini C, Protasi F, Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J Mol Biol. 2004;342:145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Wagenknecht T, Hsieh CE, Rath BK, Fleischer S, Marko M. Electron tomography of frozen-hydrated isolated triad junctions. Biophys J. 2002;83:2491–2501. doi: 10.1016/S0006-3495(02)75260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tijskens P, Meissner G, Franzini-Armstrong C. Location of ryanodine and dihydropyridine receptors in frog myocardium. Biophys J. 2003;84:1079–1092. doi: 10.1016/S0006-3495(03)74924-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi T, et al. Three-dimensional electron microscopy reveals new details of membrane systems for Ca2+ signaling in the heart. J Cell Sci. 2009;122:1005–1013. doi: 10.1242/jcs.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Maio A, Karko K, Snopko RM, Mejia-Alvarez R, Franzini-Armstrong C. T-tubule formation in cardiacmyocytes: Two possible mechanisms? J Muscle Res Cell Motil. 2006;28:231–241. doi: 10.1007/s10974-007-9121-x. [DOI] [PubMed] [Google Scholar]
- 26.Felder E, Franzini-Armstrong C. Type 3 ryanodine receptors of skeletal muscle are segregated in a parajunctional position. Proc Natl Acad Sci USA. 2002;99:1695–1700. doi: 10.1073/pnas.032657599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soeller C, Crossman D, Gilbert R, Cannell MB. Analysis of ryanodine receptor clusters in rat and human cardiac myocytes. Proc Natl Acad Sci USA. 2007;104:14958–14963. doi: 10.1073/pnas.0703016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tijskens P, Jones LR, Franzini-Armstrong C. Junctin and calsequestrin overexpression in cardiac muscle: The role of junctin and the synthetic and delivery pathways for the two proteins. J Mol Cell Cardiol. 2003;35:961–974. doi: 10.1016/s0022-2828(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 29.Asghari P, Schulson M, Scriven DR, Martens G, Moore ED. Axial tubules of rat ventricular myocytes form multiple junctions with the sarcoplasmic reticulum. Biophys J. 2009;96:4651–4660. doi: 10.1016/j.bpj.2009.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page E, Surdyk-Droske M. Distribution, surface density, and membrane area of diadic junctional contacts between plasma membrane and terminal cisterns in mammalian ventricle. Circ Res. 1979;45:260–267. doi: 10.1161/01.res.45.2.260. [DOI] [PubMed] [Google Scholar]
- 31.Bridge JHB, Ershler PR, Cannell MB. Properties of Ca2+ sparks evoked by action potentials in mouse ventricular myocytes. J Physiol. 1999;518:469–478. doi: 10.1111/j.1469-7793.1999.0469p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobie EA, et al. The Ca2+ leak paradox and rogue ryanodine receptors: SR Ca2+ efflux theory and practice. Prog Biophys Mol Biol. 2006;90:172–185. doi: 10.1016/j.pbiomolbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brochet DX, et al. Ca2+ blinks: Rapid nanoscopic store calcium signaling. Proc Natl Acad Sci USA. 2005;102:3099–3104. doi: 10.1073/pnas.0500059102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Györke S, Terentyev D. Modulation of ryanodine receptor by luminal calcium and accessory proteins in health and cardiac disease. Cardiovasc Res. 2008;77:245–255. doi: 10.1093/cvr/cvm038. [DOI] [PubMed] [Google Scholar]
- 35.Santana LF, Cheng H, Gomez AM, Cannell MB, Lederer WJ. Relation between the sarcolemmal Ca2+ current and Ca2+ sparks and local control theories for cardiac excitation-contraction coupling. Circ Res. 1996;78:166–171. doi: 10.1161/01.res.78.1.166. [DOI] [PubMed] [Google Scholar]
- 36.Protasi F, Franzini-Armstrong C, Flucher BE. Coordinated incorporation of skeletal muscle dihydropyridine receptors and ryanodine receptors in peripheral couplings of BC3H1 cells. J Cell Biol. 1997;137:859–870. doi: 10.1083/jcb.137.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: A novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 38.Evans AM, Cannell MB. The role of L-type Ca2+ current and Na+ current-stimulated Na/Ca exchange in triggering SR calcium release in guinea-pig cardiac ventricular myocytes. Cardiovasc Res. 1997;35:294–302. doi: 10.1016/s0008-6363(97)00117-x. [DOI] [PubMed] [Google Scholar]
- 39.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 40.Baddeley D, Cannell MB, Soeller C. Visualisation of localisation microscopy data. Microscopy & Microanalysis. 2010 doi: 10.1017/S143192760999122X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.