Abstract
Baculoviruses are ubiquitous insect viruses well known for their use as bioinsecticides, gene therapy vectors, and protein expression systems. Overexpression of recombinant proteins in insect cell culture utilizes the strong promoter of the polyhedrin gene. In infected larvae, the polyhedrin protein forms robust intracellular crystals called polyhedra, which protect encased virions for prolonged periods in the environment. Polyhedra are produced by two unrelated families of insect viruses, baculoviruses and cypoviruses. The atomic structure of cypovirus polyhedra revealed an intricate packing of trimers, which are interconnected by a projecting N-terminal helical arm of the polyhedrin molecule. Baculovirus and cypovirus polyhedra share nearly identical lattices, and the N-terminal region of the otherwise unrelated baculovirus polyhedrin protein sequence is also predicted to be α-helical. These results suggest homology between the proteins and a common structural basis for viral polyhedra. Here, we present the 2.2-Å structure of baculovirus polyhedra determined by x-ray crystallography from microcrystals produced in vivo. We show that the underlying molecular organization is, in fact, very different. Although both polyhedra have nearly identical unit cell dimensions and share I23 symmetry, the polyhedrin molecules are structurally unrelated and pack differently in the crystals. In particular, disulfide bonds and domain-swapped N-terminal domains stabilize the building blocks of baculovirus polyhedra and interlocking C-terminal arms join unit cells together. We show that the N-terminal projecting helical arms have different structural roles in baculovirus and cypovirus polyhedra and conclude that there is no structural evidence for a common evolutionary origin for both classes of polyhedra.
Keywords: in vivo crystallization, molecular arms, occlusion body, self-assembly, virus evolution
Polyhedrosis insect diseases were described in sericultural records in Europe as early as the 16th century and were first shown to be caused by viruses in 1913 (1). The diseases are characterized by the accumulation of large amounts of intracellular crystalline material. The crystalline material, which may constitute up to 17% of the protein content of infected larvae, consists of micrometer-sized protein crystals called viral polyhedra or occlusion bodies (2). Viral polyhedra contain numerous virus particles embedded within a crystalline protein lattice. Feeding larvae are infected by ingesting polyhedra, which dissolve in the alkaline midgut, releasing the virus particles. Viral polyhedra from diseased larvae are stable and can remain infectious in soil for several years. This unique mode of viral propagation within protein crystals is found only in insects and may have evolved to preserve the virus during diapause or periods when the seasonal larval population is too low to sustain continued virus replication (3).
Two unrelated virus families produce polyhedra. The polyhedra produced by cypovirus, a member of the Reoviridae family of double-stranded RNA viruses, typically contain thousands of embedded icosahedral virions (4). Members of the much larger and more varied baculovirus family of double-stranded DNA viruses produce polyhedra containing far fewer virus particles. These crystals are encased in an outer envelope of proteoglycans absent in cypovirus polyhedra (5). The smallest baculovirus polyhedra produced by granulovirus (β-baculovirus) are elongated crystals with maximum dimensions of ≈400 nm, and they each contain a single 60 × 280-nm rod-shaped membrane-bound virion. By contrast, polyhedra produced by the type species of α-baculoviruses, Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV), may contain hundreds of virus particles oriented randomly in small groups embedded in crystals up to 10 μm in diameter (3).
Despite the variety of shapes and sizes, and the very different types of virions they contain, the underlying crystalline symmetry of viral polyhedra is remarkably similar. All polyhedra characterized to date consist of crystallographic unit cells with ≈103-Å cell edges and body-centered cubic symmetry (6). Baculovirus polyhedrins are 243–249-aa proteins and are conserved (≈50% protein sequence identity between α- and β-baculoviruses). Cypovirus polyhedrins contain 247–252 amino acids and are less conserved (≈20% sequence identity among cypovirus clades). There is no significant sequence similarity between cypovirus and baculovirus polyhedrins. The common function, shared symmetry, and almost identical unit cell dimensions of baculovirus and cypovirus polyhedra suggest that despite the lack of sequence similarity, the underlying protein structures of polyhedra may be conserved (6).
Because polyhedra are small (typically 1–2 μm) and could not be recrystallized from purified protein, they have resisted structure determination for over 2 decades (2). The advent of novel microcrystallography techniques and synchrotron microfocus beamlines (7, 8) allowed the structure of silkworm cypovirus polyhedra to be solved recently, giving the first insights into the molecular organization of these infectious crystals. The structure revealed an intricate arrangement of polyhedrin trimers interlocked by extended N-terminal helices, explaining the remarkable stability of these polyhedra (9). Here, we use similar methods to determine the crystal structure of polyhedra produced by the baculoviruses AcMNPV and Wiseana nucleopolyhedrovirus (WNPV) (10). The polyhedrin amino acid sequences of these distantly related α-baculovirus species are 80% identical and unrelated to the amino acid sequence of cypovirus polyhedrin.
Results
Molecular Organization of Baculovirus Polyhedra.
To overcome the challenges of structure determination from native crystals smaller than 4 μm, we first analyzed polyhedra produced by a mutant of the AcMNPV polyhedrin (G25D) that forms larger 10–15-μm crystals in insect cell culture (11). The structure of these polyhedra was determined at a resolution of 2.3 Å by experimental phasing and required testing of over 200 crystals. These mutant polyhedrin crystals are not infectious and do not contain virus particles. We subsequently used this structure as a molecular replacement model to determine the 2.2-Å structure of infectious polyhedra directly purified from porina larvae infected with WNPV.
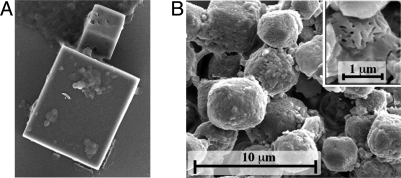
The mutant and WT crystals differ in both size and shape. G25D mutant polyhedra have a relatively smooth surface and sharp edges (Fig. 1A), whereas infectious WNPV polyhedra resemble truncated octahedra and have a pitted surface because of the presence of virus particles (Fig. 1B). They are also significantly smaller, with an upper size limit of 4 μm. Despite these differences, G25D AcMNPV and WNPV polyhedra both belong to the I23 space group with cell parameters of 103.2 Å and 102.4 Å, respectively. The crystalline matrix of these polyhedra consists of a dense arrangement of the polyhedrin molecules with density of 1.29 g·cm−3 and one of the lowest solvent contents known for protein crystals (22%). The crystals consist of essentially continuous protein except for small cavities at the origin and center of the unit cell.
Fig. 1.
Scanning electron microscopy of G25D AcMNPV polyhedra (A) and infectious WNPV polyhedra (B). (B, Inset) WNPV polyhedra in which imprints of virus particles are apparent. This is not observed for the G25D polyhedra, which do not contain virions and have a smooth surface.
The crystalline organization of polyhedra means that all polyhedrin molecules in the crystal are strictly equivalent and the whole assembly can be generated by applying the I23 crystallographic symmetries to a single polyhedrin protein. For clarity, we describe polyhedra here as an assembly around each lattice point of loose tetrahedral clusters, each of which is composed of four trimers of the polyhedrin. Two such tetrahedral clusters constitute the unit cell that is repeated 200 times in each direction for a typical 2-μm crystal. In the following sections, we initially describe the trimers and then the tetrahedral clusters of trimers (i.e., 12-mers) and the way the tetrahedral clusters interconnect to form polyhedra.
Polyhedrin Trimers.
The WNPV and G25D AcMNPV polyhedrin structures are very similar, with a rmsd of 0.441 Å over 167 residues, and the only notable difference is the absence of well-defined electron density for residues 29–48 in AcMNPV polyhedra. Thus, in the following description, the figures and residue numbers correspond to WNPV.
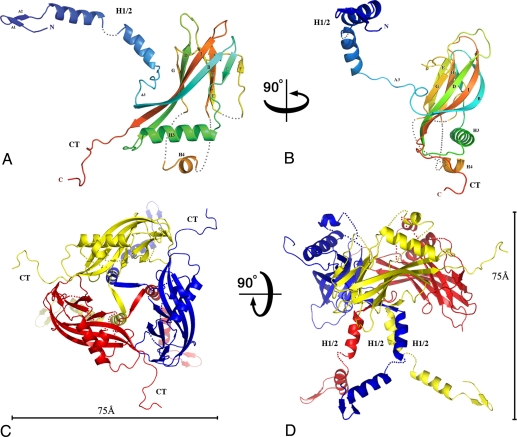
Individual baculovirus polyhedrin molecules fold into a central compact β-sandwich domain with a 52-aa extended projection at the N-terminus and a shorter 10-aa projection at the C-terminus. The central sandwich domain has the topology of a classic jelly-roll (BIDG-FCHE) (12) with strand F displaced. At one end of the long β-sheet, three of the strands (BID) extend and wrap around helix H3, giving a pronounced wedge shape to the central domain of the molecule (Fig. 2 A and B). The N-terminal residues 10–52 form a short β-hairpin followed by a long kinked helix (H1/2) that extends 60 Å away from the main body of the molecule perpendicular to the central sandwich (Fig. 2D). The C-terminal residues 233–243 also extend 20 Å from the central domain. Three regions exhibit various degrees of disorder in our structure, including the H1/2 helix for which only the main chain could be modeled with confidence (see details in Fig. 2 and Methods).
Fig. 2.
Structure of the baculovirus polyhedrin protein. Cartoon representations of the WNPV polyhedrin molecule (A and B) and its trimeric assembly (C and D) in orthogonal views. In A and B, the polypeptide chain is colored in a blue-to-red gradient from the N-terminus to the C-terminus (CT). The polyhedrin has a jelly-roll fold with two sheets, BIDG and FCHE. Helix H1/2 and the CT loop extend away from the main body of the trimer in orthogonal directions. Three regions are disordered, and dotted lines represent the missing loops. First, only weak electron density was visible for the H1/2 helix so that only the main chain was traced with a short break between residues 37 and 39. Second, the short loop comprising residues 142–145 had no interpretable electron density and was not modeled. Third, the region comprising residues 171–202 was disordered apart from a short helix (186–194). (C and D) Trimer is represented in orthogonal views, with scale bars representing 75 Å. The trimer has the shape of a chalice with a tripod base (H1/2) and prominent protrusions (CT).
The polyhedrin protein assembles into trimers located at crystallographic three-fold axes in polyhedra. The trimeric subunits have the shape of a chalice, with a central bowl supported by a tripod formed by three H1/2 helices (Fig. 2 C and D). The trimeric interface involves predominantly hydrophobic surfaces of the central jelly-roll domain corresponding to 1,470 Å2 or 10% of the surface of the polyhedrin protein. A short strand preceding the jelly-roll in the polyhedrin sequence plugs the center of the trimer.
Polyhedra Consist of Disulfide-Linked Tetrahedral Clusters of Four Trimers.
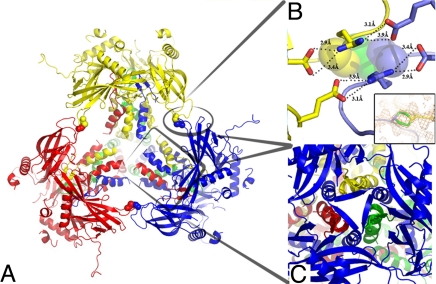
In polyhedra, four identical trimers are tightly associated into a stable cluster with tetrahedral geometry (Fig. 3A). At the periphery, this assembly is held together by two short loops extending from the central sandwich. The contact area is small (796 Å2) but involves strong bonds, including salt bridges and an intermolecular disulfide bond (Cys131-Cys131) formed across a crystallographic two-fold axis (Fig. 3B). Importantly, at the center of the tetrahedral cluster, N-terminal polyhedrin projections further strengthen the assembly by domain swapping, wherein each of the H1/2 helices from a trimer reaches out for one of the other three trimers in the cluster (Fig. 3 A and C). This results in the insertion of the N-terminal β-turn into cavities of the neighboring trimers so that the strands of the β-turn contribute two additional strands to the BIDG sheet in the jelly-roll domain. Overall, the extent and strength of these interactions suggest that the tetrahedral clusters represent the building blocks of polyhedra.
Fig. 3.
Tetrahedral clusters in polyhedra. (A) Cartoon representation of a tetrahedral cluster with different colors for each of the four WNPV polyhedrin trimers. (B) Close-up view of the contact area between trimers within the tetrahedral cluster highlighting salt bridges between residues Glu214 and Arg130 and a disulfide bond between two Cys131 residues (Inset). The side chain of the cysteine is in two alternate conformations, and conformation A in one molecule forms a disulfide bond with the symmetry-related cysteine in conformation B. (C) Close-up view of the interactions between the four trimers mediated by domain swapping of the N-terminal regions of three trimers (red, yellow, and green) with the central jelly-roll domains of the fourth trimer (blue).
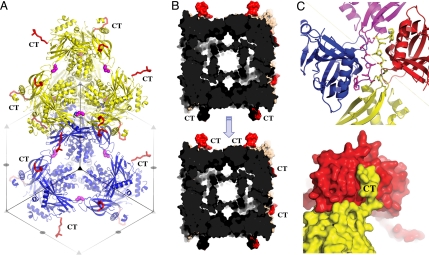
The tetrahedral clusters are tightly packed in polyhedra according to the crystallographic translations. The packing of polyhedrin molecules is intricate along the diagonal of the cubic unit cell (Fig. 4A). Interactions are also extensive and intimate along the faces of the unit cell, wherein the C-terminal extension of each polyhedrin molecule fits into a cavity in the face of a neighboring unit cell. This cavity is located between two clusters and accommodates two antiparallel C-terminal extensions from two other clusters. The C-terminal extensions mediate intimate interactions that interlock four different clusters (Fig. 4 B and C) but are not involved in the assembly of the trimer or tetrahedral cluster.
Fig. 4.
Architecture of baculovirus polyhedra. (A) Cartoon representation of two tetrahedral clusters (yellow and blue) showing their tight packing in polyhedra according to the I23 crystallographic translations. Symmetry elements are indicated as gray triangles and ellipses. The C-terminal (CT) loops project outward from the unit cell (red ribbon), and disulfide bonds stabilize the clusters (side chain represented as magenta spheres). (B) Surface representation of two unit cells with the CT loops highlighted in red. These cells pack as indicated by the arrow and interlock with extensive interactions mediated by the CT loop. This forms an extremely dense matrix with only a 30-Å wide cavity delimited by helices H1/2 and no continuous solvent channels. (C) Surface (Bottom) and ribbon (Top) representations of the anchoring interactions mediated by CT loops.
Sequence Conservation.
Baculoviruses are ubiquitous viruses that have been described in more than 600 species of insects, and more than 200 baculovirus polyhedrin sequences are available (National Center for Biotechnology Information Entrez search). To identify putative functional sites in baculovirus polyhedra, we constructed an alignment of 62 nonredundant full-length sequences and mapped the sequence conservation onto the surface of the polyhedrin structure.
Highly conserved surfaces are found at intertrimer interfaces, forming a rim of conserved residues at the periphery of the trimer. These include regions involved in stabilizing the tetrahedral cluster such as the loop containing the intermolecular disulfide bond and the N-terminal β-turn that is swapped between trimers (Fig. S1). Residues around the intermolecular disulfide bond (PNRC) are conserved in 59 of the 62 baculovirus polyhedrin sequences (Fig. S2). Regions involved in interactions among tetrahedral clusters such as the C-terminal extension are also conserved (Figs. S1 and S2).
In contrast, helix H1/2 appears to be an important functional part of the polyhedrin protein that exhibits considerable sequence variability. In our structure, this segment essentially forms a linker with constraints on the main chain fold and length but few interchain contacts. Indeed, this region is poorly ordered in both polyhedra we analyzed. Helix H1/2 may have several functions in the virus cycle. First, it contains a nuclear localization sequence (residues 31–34) that is essential to the correct localization of polyhedra in most baculoviruses (13). Second, mutations introduced in or near helix H1/2 affect the morphology and infectivity of polyhedra, suggesting a role for this helix in assembly and virus packaging. Mutations controlling the shape of polyhedra were previously identified at residues 24, 41, and 58 (mutations were reported as G25D (11), L43I (14), and P58L (15) in the original numbering). These mutations result in a change of the crystal morphology from a variety of polyhedral forms to cuboid crystals. In addition, the crystals are larger and contain fewer virus particles. Residues at position 24 and 58 are glycine and proline, respectively, which are known to alter the main chain flexibility. They have opposite effects, with G24 introducing flexibility at the base of helix H1/2 and P58 introducing rigidity at the C-terminal end of the helix. Both G24 and P58 are highly conserved in sequences of granulins and polyhedrins (Fig. S2). Position 41 is located within the most variable region of the polyhedrin close to the kink in helix H1/2, and both leucine and isoleucine residues are found in sequences of natural isolates of baculovirus. These differences represent the addition of a single methyl group to the residue, suggesting that even small rearrangements of helix H1/2 can lead to a change in the morphology of polyhedra and alter the efficiency of virus binding.
Structural Comparisons.
Structural comparison of the baculovirus polyhedrin with structures from the Protein Data Bank revealed weak similarity to other proteins containing a jelly-roll domain, especially viral capsid proteins. The structural similarities with viral capsids are restricted to the jelly-roll domain of the viral capsid proteins, and the extent of similarity is comparable to that for the jelly-roll domain in nonviral proteins (z scores of 5.6–6.3 in a Dali structural similarity search; Table S1). By contrast, z scores among homologous jelly-roll proteins from distantly related viruses are much greater (e.g., z = 10–20, refs. 16–19). Moreover, in contrast to these examples, the jelly-roll domains in viral capsids interact differently from polyhedrin jelly-roll domains in polyhedra, as described in Fig. S3. For these reasons, we do not infer structural evidence for homology between viral coat proteins and baculovirus polyhedra.
Interestingly, the search for structural homologues did not detect similarity between the baculovirus and cypovirus polyhedrin structures. This result was unexpected, because both proteins have similar function and the two structures are, at first glance, related: the polyhedrin proteins both contain a central jelly-roll domain with an extended N-terminal helical projection and form nearly identical crystal lattices.
Here, we compare the structures in detail and argue that despite the outward similarities, there is no evidence for common ancestry of baculovirus and cypovirus polyhedra. First, the folds of both polyhedrin proteins are distinct. Although they are both akin to jelly-rolls, cypovirus polyhedrin has a IBADGF-CHE topology that contrasts with the more classic BIDG-FCHE topology of the baculovirus polyhedrin (Fig. 5 A and D).
Fig. 5.
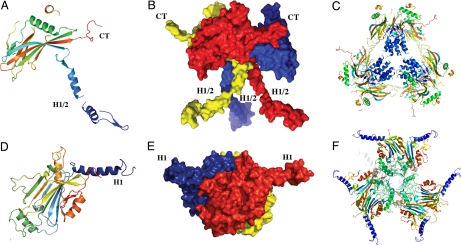
Comparison of baculovirus and cypovirus polyhedra. The distinct folds and trimeric organizations of nucleopolyhedrovirus and cypovirus polyhedrin proteins are shown in cartoon (A and D) and surface (B and E) representations. The cartoon representations of the tetrahedral clusters (C and F) highlight the different localizations and functions of the N-terminal helical regions of the two classes of polyhedrins. The central helices H1/2 stabilize the tetrahedral cluster in baculovirus polyhedra, whereas helices H1 extend away from the cluster in cypovirus polyhedra. Molecules are colored in a blue-to-red gradient from the N-terminus to the C-terminus (CT).
Second, the shapes of the molecules are fundamentally different. This is particularly clear in the trimers: On the one hand, trimers of the cypovirus polyhedrin are made of compact three-layer sandwiches and have a markedly pyramidal shape. On the other hand, baculovirus trimers present a deep depression at the center of the trimer and exhibit a chalice shape. Overall, this results in a concave trimer for the baculovirus polyhedrin departing from the convex trimer of cypovirus polyhedrin (Fig. 5 E and B).
Third, both polyhedrin proteins have a protrusion perpendicular to the trimer axis, but this extended molecular arm is located at opposite ends of the molecules and differs in its structure. Hence, the C-terminal extended sequence in baculovirus polyhedrin contrasts with its counterparts in the cypovirus polyhedrin consisting of an N-terminal helical extension. The N-terminal region of baculovirus polyhedrin is also α-helical, but it projects in a totally different direction to cypovirus polyhedron helix H1 along the trimer axis and plays a different role in polyhedra as described below (Fig. 5).
Beyond these differences in fold and shape, an analysis of the surface properties of both polyhedrin proteins reveals distinct charge distributions, with an overall negative electrostatic potential for the AcMNPV polyhedrin trimer and a weak positive electrostatic potential for the cypovirus polyhedrin trimer (Fig. S4).
Comparison Between Cypovirus and Baculovirus Polyhedra.
Polyhedra also differ in their molecular architectures: The proposed building blocks of cypovirus and baculovirus polyhedra are both clusters of 12 polyhedrin molecules with a tetrahedral geometry, but they adopt fundamentally different organizations (Fig. 5 C and F). In particular, cypovirus building blocks are compact and involve extensive buried surfaces with a high shape complementarity, taking advantage of the pyramidal shape of the cypovirus trimers. In contrast, baculovirus building blocks are only loosely connected at the center of the cluster and are linked by interactions at the periphery of the cluster. Contact areas are more restricted than in cypovirus polyhedra, but the bonds are strong and include salt bridges, a covalent disulfide bond, and domain swapping of the N-terminal helix. The resulting packing in baculovirus polyhedra creates a shell around loose helical bundles instead of the continuous matrix of cypovirus polyhedra (Fig. 4 A and B and Fig. 5C).
Discussion
We determined the structure of baculovirus polyhedra by x-ray microcrystallography from in vivo crystals purified from infected cells and larvae. Unexpectedly, polyhedra containing virus particles or not are isomorphous and diffract to similar resolution limits despite the presence of a large number of virions that interrupt the lattice in infectious crystals. This implies, first, that the ultrastructural differences observed between these polyhedra do not stem from a different lattice organization and, second, that incorporation of virus particles into polyhedra does not lead to long-range distortion of the crystals.
We found that polyhedra consist of tetrahedral clusters of four polyhedrin trimers. These clusters are covalently cross-linked by intermolecular disulfide bonds and are further stabilized by domain swapping from an N-terminal extension. The intimate and extensive interactions among the tetrahedral clusters confer a remarkable stability to polyhedra. In particular, a C-terminal extension forms a short molecular arm effectively interlocking four surrounding clusters.
The surfaces that stabilize tetrahedral clusters or are involved in the interactions among them are highly conserved. The surface conservation reflects the primary function of polyhedra to crystallize around virions in infected cells, such that crystal contacts represent functional sites of the polyhedrin protein. Interestingly, the bowl of the chalice is well conserved, although not involved in molecular contacts stabilizing polyhedra. Thus, this region is a possible candidate for the site of binding for virus particles or the polyhedron outer envelope. The proteoglycans of the envelope are proposed to be attached to the polyhedrin matrix via disulfide bonds (5), and the disordered loop located within the bowl area indeed contains a conserved cysteine residue that may be involved in envelope attachment. (Fig. S2).
As mentioned previously, covalent cross-links and domain swapping contribute to the remarkable stability of polyhedra. Although these features have not been previously described for polyhedra, they are frequently found in the icosahedral shell of viral capsids. Indeed, intermolecular disulfide bounds are present in a number of nonenveloped viruses, including the simian virus 40 (SV40), reovirus, and some picornaviruses. For these viruses, the disulfide bonds also form despite the normally reducing environment of the cellular cytoplasm. The contribution of these bonds to assembly, stability, and disassembly was studied in detail for SV40 (20) and reovirus (21). Intertwined N-terminal or C-terminal extensions are also common in icosahedral viruses and participate in viral morphogenesis, as shown in simple plant viruses (22). Similar extensions are also found in SV40, where they participate in the formation of more complex capsids from pentameric building blocks (23). In addition to orchestrating viral assembly, N-terminal extensions are important for the stability of a number of capsids, particularly by domain swapping, as observed in the virions of the rice yellow mottle virus (24).
Despite these analogies in the assembly strategy, we found no conclusive evidence of homology between the baculovirus polyhedrin protein and viral capsid proteins or, indeed, any viral or cellular protein. Admittedly, all the viral capsid proteins mentioned above have a jelly-roll domain like the baculovirus polyhedrin. However, the jelly-roll fold is ubiquitous in capsid proteins of unrelated viruses and is not necessarily indicative of evolutionary links (12). Importantly, beyond the presence of a jelly-roll domain, no significant similarity was found with other viral capsids in the specific use of the N-terminal and C-terminus or in the organization of the trimers and tetrahedral clusters.
The question of whether cypovirus and baculovirus polyhedra are homologous was recently raised by a study revealing the almost identical crystalline organization (6). We show here that these two classes of polyhedra are made of unrelated protein building blocks, achieving very different molecular architectures. Although the folds of both polyhedrins are akin to jelly-rolls, the two proteins differ in their topology, shape, and surface properties, resulting in trimers with contrasting features (Fig. 5 and Fig. S4). Importantly, the two classes of polyhedra also differ in their molecular organization. Although the pyramidal shape of cypovirus polyhedrin trimers allows a tight packing at the center of the tetrahedral cluster, the long tripod base of baculovirus polyhedrin trimers forms a loose bundle of helices in the equivalent location of baculovirus clusters.
This inside-out organization for baculovirus polyhedra compared with cypovirus polyhedra also means that the N-terminal helix H1 of cypovirus polyhedra projects away from the tetrahedral clusters (Fig. 5 C and F). Thus, in cypovirus polyhedra, helix H1 connects neighboring clusters but does not contribute to their stability. Conversely, the N-terminal helix H1/2 of baculovirus polyhedra stabilizes the tetrahedral cluster in its very center and is not involved in intercluster interactions.
In light of these fundamental differences in building blocks and architectures, we conclude that there is no structural evidence for a common ancestry of viral polyhedra.
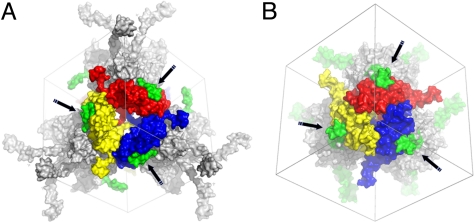
A striking illustration of a possible evolutionary convergence is the use in both classes of polyhedra of molecular arms to tie together neighboring clusters (Fig. 6). In both cases, these projections extend along the two-fold crystallographic axes of the crystal, reaching out to four neighboring tetrahedral clusters. These interactions involve surfaces of high complementarity that may anchor incoming blocks, ensuring efficient crystallization in the complex milieu of the cell. Accordingly, fusion of the cypovirus molecular arm helix H1 is sufficient for incorporation of foreign proteins into polyhedra (25), and alteration of the C-terminal molecular arms from the baculovirus polyhedrin protein abolishes the formation of polyhedra (26). Importantly, although functionally analogous, these arms are in opposite locations in the polyhedrin protein sequences and differ in their folds.
Fig. 6.
Molecular arms in baculovirus and cypovirus polyhedra. Surface representation of the interaction between a central trimer and a neighboring tetrahedral cluster in baculovirus (A) and cypovirus (B) polyhedra. The tetrahedral cluster is represented in gray except for the molecular arms clamping the central trimer, which are highlighted in green and indicated with black arrows. As described in the text, these arms are the C-terminus of baculovirus polyhedrin and the N-terminal helix of the cypovirus polyhedrin.
It will now be interesting to investigate further whether these similarities are a general feature of in vivo crystals produced by viruses. Such comparative studies may also shed light on the reason why the two classes of polyhedra converged to nearly identical lattices, an evolutionary conundrum reminiscent of the prevalence of the icosahedral symmetry in spherical virus particles.
Methods
Detailed methods are available in SI Text.
Production of Polyhedra.
The G25D AcMNPV polyhedrin was expressed in Sf21 cells by infection with a mutant baculovirus as described previously (11). Polyhedra were produced and purified according to established protocols. Crystals from selenomethionine polyhedrin (27) diffracted more consistently and to a higher resolution than the corresponding native polyhedra. Polyhedra were also purified from infected larvae of porina moths (Wiseana spp.) collected from pastures in the Otago region (New Zealand) (10). The concentration and quality of polyhedra were estimated by optical microscopy and scanning electron microscopy.
Structure Determination.
Diffraction experiments were carried out at the X06SA microfocus beamline (Swiss Light Source) using the MD2 microfocus diffractometer, with a beam of 15 × 5 μm focused on the detector. Data were collected at 100 K from polyhedra in 50% (vol/vol) ethylene glycol. Phases were determined from selenomethionine-substituted crystals and heavy-atom soaks. Sites were identified using SHELXc/d (28) and refined with SHARP (29) at a resolution of 2.8 Å (Table S2). Selenomethionine-substituted crystals had the strongest phasing power. The final model of the AcMNPV polyhedrin was refined to a resolution of 2.3 Å with BUSTER 2.8.0 (30), with individual temperature factors and tight geometrical restraints (see SI Methods). The structure of WNPV polyhedra was subsequently determined by isomorphous molecular replacement and refined at a resolution of 2.18 Å.
The WNPV and G25D AcMNPV polyhedrin models include residues 10 to 243 and 11 to 245, respectively, with three disordered regions (Fig. 2). The structures have very good geometry and crystallographic statistics, with 99% and 98% of residues in favored regions of the Ramachandran plot and R/Rfree of 0.16/0.19 and 0.16/0.21 for WNPV and AcMNPV polyhedrins, respectively (Table S3).
Supplementary Material
Acknowledgments.
We thank V. Young, C. Ferguson, C. Hobbis, and M. Middleditch for experimental assistance and R.E. Thorne for providing MicroMesh mounts (MiTeGen). This work was supported by the Royal Society of New Zealand Marsden Fund (P.M., V.W.), the University of Auckland Research Committee (P.M.), the Maurice Wilkins Centre for Molecular Biodiscovery (E.C.), and grants for Regional Consortium Research Development Work from the Kansai Bureau of Ministry of Economy, Trade and Industry (to K.I. and H.M.), CREST by Japan Science and Technology Agency (to H.M.), and a Japan Society for the Promotion of Science Invitation Fellowship for Research in Japan (to P.M.). E.C. is funded by a Tertiary Education Commission Bright Future Top Achiever Doctoral Scholarship and the University of Auckland Doctoral Scholarship, S.G. is funded by the Swiss National Centre of Competence in Research Structural Biology, and F.C. is funded by a National Health and Medical Research Council Career Development Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structures of G25D AcMNPV and WNPV polyhedra are deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3JW6 and 3JVB).
This article contains supporting information online at www.pnas.org/cgi/content/full/0910686106/DCSupplemental.
References
- 1.Glaser RW, Chapman JW. The nature of the polyhedral bodies found in insect cells. Biol Bull. 1916;30:367–390. [Google Scholar]
- 2.Rohrmann GF. Polyhedrin structure. J Gen Virol. 1986;67(8):1499–1513. doi: 10.1099/0022-1317-67-8-1499. [DOI] [PubMed] [Google Scholar]
- 3.Miller LK. The Baculoviruses. New York: Plenum; 1997. p. 447. [Google Scholar]
- 4.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitt MA, Manning JS. A phosphorylated 34-kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology. 1988;163:33–42. doi: 10.1016/0042-6822(88)90231-0. [DOI] [PubMed] [Google Scholar]
- 6.Anduleit K, et al. Crystal lattice as biological phenotype for insect viruses. Protein Sci. 2005;14:2741–2743. doi: 10.1110/ps.051516405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cusack S, et al. Small is beautiful: Protein micro-crystallography. Nat Struct Biol. 1998;5(Suppl):634–637. doi: 10.1038/1325. [DOI] [PubMed] [Google Scholar]
- 8.Riekel C, Burghammer M, Schertler G. Protein crystallography microdiffraction. Curr Opin Struct Biol. 2005;15:556–562. doi: 10.1016/j.sbi.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Coulibaly F, et al. The molecular organization of cypovirus polyhedra. Nature. 2007;446:97–101. doi: 10.1038/nature05628. [DOI] [PubMed] [Google Scholar]
- 10.Sadler T, Ward V, Glare T, Kalmakoff J. Examination of New Zealand's endemic Wiseana nucleopolyhedrovirus by analysis of the viral polyhedrin gene. Arch Virol. 1998;143:2273–2288. doi: 10.1007/s007050050460. [DOI] [PubMed] [Google Scholar]
- 11.Lin GY, Zhong J, Wang XZ. Abnormal formation of polyhedra resulting from a single mutation in the polyhedrin gene of Autographa californica multicapsid nucleopolyhedrovirus. J Invertebr Pathol. 2000;76:13–19. doi: 10.1006/jipa.2000.4934. [DOI] [PubMed] [Google Scholar]
- 12.Harrison SC. Principles of virus structure. In: Knipe DM, Howley PM, editors. Field's Virology. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 59–98. [Google Scholar]
- 13.Jarvis DL, Bohlmeyer DA, Garcia A., Jr Requirements for nuclear localization and supramolecular assembly of a baculovirus polyhedrin protein. Virology. 1991;185:795–810. doi: 10.1016/0042-6822(91)90551-l. [DOI] [PubMed] [Google Scholar]
- 14.Cheng XW, Carner GR, Fescemyer HW. Polyhedrin sequence determines the tetrahedral shape of occlusion bodies in Thysanoplusia orichalcea single-nucleocapsid nucleopolyhedrovirus. J Gen Virol. 1998;79:2549–2556. doi: 10.1099/0022-1317-79-10-2549. [DOI] [PubMed] [Google Scholar]
- 15.Carstens EB, Krebs A, Gallerneault CE. Identification of an amino acid essential to the normal assembly of Autographa californica nuclear polyhedrosis virus polyhedra. J Virol. 1986;58:684–688. doi: 10.1128/jvi.58.2.684-688.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulibaly F, et al. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 18.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li PP, Nakanishi A, Clark SW, Kasamatsu H. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc Natl Acad Sci USA. 2002;99:1353–1358. doi: 10.1073/pnas.032668699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odegard AL, Chandran K, Liemann S, Harrison SC, Nibert ML. Disulfide bonding among micro 1 trimers in mammalian reovirus outer capsid: A late and reversible step in virion morphogenesis. J Virol. 2003;77:5389–5400. doi: 10.1128/JVI.77.9.5389-5400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 A resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 23.Liddington RC, et al. Structure of simian virus 40 at 3.8 Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 24.Qu C, et al. 3D domain swapping modulates the stability of members of an icosahedral virus group. Structure. 2000;8:1095–1103. doi: 10.1016/s0969-2126(00)00508-6. [DOI] [PubMed] [Google Scholar]
- 25.Ijiri H, et al. Structure-based targeting of bioactive proteins into cypovirus polyhedra and application to immobilized cytokines for mammalian cell culture. Biomaterials. 2009;30:4297–4308. doi: 10.1016/j.biomaterials.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 26.Carstens EB, Williams GV, Faulkner P, Partington S. Analysis of polyhedra morphology mutants of Autographa californica nuclear polyhedrosis virus: Molecular and ultrastructural features. J Gen Virol. 1992;73:1471–1479. doi: 10.1099/0022-1317-73-6-1471. [DOI] [PubMed] [Google Scholar]
- 27.Bellizzi JJ, Widom J, Kemp CW, Clardy J. Producing selenomethionine-labeled proteins with a baculovirus expression vector system. Structure. 1999;7:R263–R267. doi: 10.1016/s0969-2126(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 28.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 29.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 30.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.