Abstract
Venomous animals immobilize prey using protein toxins that act on ion channels and other targets of biological importance. Broad use of toxins for biomedical research, diagnosis, and therapy has been limited by inadequate target discrimination, for example, among ion channel subtypes. Here, a synthetic toxin is produced by a new strategy to be specific for human Kv1.3 channels, critical regulators of immune T cells. A phage display library of 11,200 de novo proteins is designed using the α-KTx scaffold of 31 scorpion toxin sequences known or predicted to bind to potassium channels. Mokatoxin-1 (moka1) is isolated by affinity selection on purified target. Moka1 blocks Kv1.3 at nanomolar levels that do not inhibit Kv1.1, Kv1.2, or KCa1.1. As a result, moka1 suppresses CD3/28-induced cytokine secretion by T cells without cross-reactive gastrointestinal hyperactivity. The 3D structure of moka1 rationalizes its specificity and validates the engineering approach, revealing a unique interaction surface supported on an α-KTx scaffold. This scaffold-based/target-biased strategy overcomes many obstacles to production of selective toxins.
Keywords: mokatoxin, moka1, phage display, peptide toxin, animal venom
The voltage-gated potassium channel expressed on human T lymphocytes, Kv1.3, is a validated target for therapeutic modulation of the immune system (1–3). Thus, block of Kv1.3 on T cells by scorpion toxins counters the effects of anti-CD3/28 stimulation and suppresses effector cytokine secretion. This observation has motivated efforts to isolate native toxins specific for Kv1.3 from venoms and to design peptide and small molecule blockers (4–8). Regrettably, these natural and synthetic ligands have proven inadequate. For example, kaliotoxin-1 (KTX) (9) inhibits Kv1.3 to suppress T cell activity (10) but also blocks Kv1.1 and Kv1.2 (11) with sufficient potency to produce undesirable side effects such as diarrhea (12). Efforts to improve selectivity continue (2, 3, 13, 14).
The search for target-specific toxins is fueled by their proven utility and physical stability. Progress has been slow with current sources of new toxins—isolation from crude venom, shotgun venom gland sequencing, and site-directed mutation. This is because minute amounts of toxins are present in venoms, isolation is rarely coupled to known targets, effective strategies are lacking to link peptides predicted by sequencing or generated by combinatorial chemistry with targets of interest, and variation explored by point mutation is limited (15).
Here, we circumvent these obstacles by production of a scaffold-based/target-biased library and a high-throughput selection strategy. The library was constructed on the resilient scaffold found in scorpion α-KTx toxins (16, 17). This seemed prudent first because animal toxin scaffolds have evolved to tolerate extensive sequence diversity (18), and second because α-KTx toxins interact with potassium channels. Phage display and library sorting (19) were judged practical because proper folding of disulfide-rich proteins has been observed on phage (20), toxins remain active despite non-native residue variation (2, 6, 21, 22), and phage displaying random peptides have been sorted on ion channels (23). Design, isolation, and characterization of mokatoxin-1 (moka1), an avid and specific blocker of human Kv1.3 channels are described.
Results
Phage Display of a Neurotoxin.
Seeking a toxin specific for Kv1.3 we chose KTX, a scorpion venom peptide with an α-KTx scaffold that blocks Kv1.3 channels by a well-defined mechanism, as the lead for library design. α-KTx toxins bind directly in the potassium ion conduction pore to occlude the pathway (16) with affinities that are exquisitely sensitive to residues on the toxin and channel interaction surfaces (21, 24, 25). It follows that specific binding of phage to achieve library sorting demands that toxin variants (i) are synthesized and fold correctly (after proteolytic cleavage of the leader sequence that mediates surface expression), (ii) are accessible to target from the phage surface, and (iii) bind target in a stable manner despite their phage cargo.
To establish that phage could display KTX and bind to Kv1.3, nucleotides encoding the toxin were inserted upstream and in-frame with the gene for phage coat protein III (26). As a control, phage expressing a mutant KTX (DDD-KTX) that does not bind to Kv1.3 were also produced. DDD-KTX has three negatively charged Asp residues at sites where KTX has basic residues critical for binding: Arg24, Arg31, and Lys27—the last a conserved residue in α-KTx toxins with an ε-amino group that penetrates the ion conduction pore (16).
As a selection target, tetrameric channel complexes were synthesized bearing the pore-forming (P) domain from human Kv1.3 grafted into the homologous location of the bacterial potassium channel KcsA (to create KcsA-1.3), a strategy developed by others to study interaction of KTX and purified channel complexes (27). Wild-type KcsA channels were produced to test for nonspecific binding.
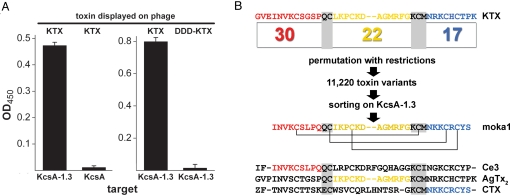
Phage expressing KTX were shown by ELISA to bind to immobilized KcsA-1.3 channels in a stable and specific manner because they were not recovered on wild-type KcsA (Fig. 1A). Binding was shown to require expression of wild-type toxin because neither KcsA-1.3 nor KcsA channels retained DDD-KTX phage. Specific binding of KTX phage to KcsA-1.3 channels argued that selective sorting of a toxin library was feasible.
Fig. 1.
KcsA-1.3 channels bind KTX phage and isolate moka1 phage from an α-KTx scaffold library. Phage preparation, library construction, sorting and ELISA protocols, and KcsA, KcsA-1.3, and toxin synthesis and purification are described in SI Materials and Methods. Single-letter codes for amino acids are standard with Z for pyroglutamate. (A) ELISA shows KTX phage bind to KcsA-1.3 but not wild-type KcsA channels (Left) and phage expressing KTX bind to KcsA-1.3 channels, whereas those expressing DDD-KTX do not (Right). Ninety-six well plates coated with KcsA-1.3 or KcsA were incubated with phage (108–1010/well). Data are mean ± SE. for three wells. (B) Library construction and sorting. Thirty-one scorpion toxins that share the α-KTx scaffold were aligned to define three domains (A, B, and C). In KTX, domains were from residues Gly1-Pro12 (A), Leu15-Gly26 (B), and Asn30-Lys38 (C). Domains were linked by sharing of nucleotide codes for amino acids QC (A and B) or KCM (B and C), thereby conserving the important KTX residue Lys27. This yields 30, 22, and 17 unique A, B, and C domains, respectively, and a calculated library diversity of 11,220. Moka1 (GQ153941), a unique toxin isolated from the library, is composed of residues present in the natural toxins Ce3 (red), AgTx2 (yellow), and CTX (blue). Móka is a Hungarian word that translates into English as fun. Isolation of two or more identical clones in 20 by sorting was the basis for further study because the probability of this in the absence of enrichment (e.g., randomly) is smaller than 10−8.
A Neurotoxin Library Yields Mokatoxin-1.
A combinatorial library was designed based on the sequences of 31 known or predicted α-KTx toxins (Table S1). To preserve scaffold architecture and favor folding of de novo toxins, the number and relative positions of disulfide bridges were preserved; further, two conserved regions corresponding to KTX residues Gln13Cys14 (QC) and Lys27Cys28Met29 (KCM) were maintained or introduced (Fig. 1B). Thus, toxin sequences were aligned at the six conserved Cys residues that form three disulfide bonds by allowance for insertions and deletions, and the QC and KCM regions were used to define three domains (A, B, and C) in all toxin sequences (Fig. 1B). The 31 parent toxins contain 30, 22, and 17 unique A, B, and C domains, respectively. Genes were produced by ligation of oligonucleotides for the domains using QC and KCM residues as shared linkers (Table S2). DNA sequencing showed incorporation of all domains in the library. Restricted permutation to form all toxins with the ABC pattern yields a calculated diversity of 11,200 unique toxins and reconstitution of 20 parental toxins.
Multiple independent phage expressing moka1 were isolated after two rounds of library selection using KcsA-1.3 as the target (Table S3). Moka1 was not observed in trials with wild-type KcsA as target. To confirm that isolation of moka1 was a specific process, phage expressing moka1 were diluted with those displaying DDD-KTX at a ratio of 1:15,000 to approximate their frequency in the library. After two rounds of selection on KcsA-1.3, 40% of phage carried moka1. Conversely, no enrichment was observed on wild-type KcsA where all recovered phage had DDD-KTX. Enrichment of moka1 phage in trials of this type was 75 to 350-fold per cycle (Tables S4–S6).
The A, B, and C domains in moka1 are derived from three scorpion species (Fig. 1B). Domain A is from toxin Ce3 of the Central American Centruroides elegans (5). Domain B is in agitoxin-2 (AgTx2) and agitoxin-3 (AgTx3) of the Middle Eastern Leiurus quinquestriatus (28) and kaliotoxin-3 (KTX3) of the North African Buthus occitanus (29). Domain C is in charybdotoxin (CTX) and Lq2 of the Middle Eastern L. quinquestriatus (30). Moka1 maintains the basic character of α-KTx toxins with four predicted net-positive charges at neutral pH.
Structure and Pharmacology of Mokatoxin-1.
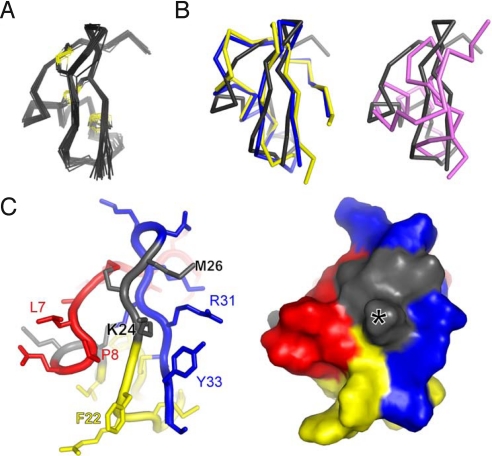
The solution structure of moka1 was determined by NMR spectroscopy (Fig. 2A and Table S7). The overall structure shows the peptide to preserve the α-KTx scaffold. Moka1 is more closely related to AgTx2 and CTX (Fig. 2B Left) than to KTX (Fig. 2B Right), although moka1 and KTX share a short and distorted helical region due to Pro at homologous sites (positions 14 and 17, respectively) (31). These structural features are consistent with the tripartite nature of moka1 (Fig. 2C).
Fig. 2.
Structure of moka1. Production of [U-13C, 15N]-moka1 and NMR spectroscopy are described in SI Materials and Methods. Statistics of the final structures are in Table S7 (A) Moka1 retains the α-KTx fold. NMR-derived solution structure of moka1 (PDB ID code 2kir). The Cα traces (gray) of 20 structures of moka1 are superimposed. The Cys residue side chains and the disulfide bonds are shown in yellow. (B Left) Superposition of the structure of moka1 (gray) with CTX (blue; PDB ID code 2crd) and AgTx2 (yellow, PDB ID code 1agt). (Right) Superposition of the structure of KTX (purple; PDB ID code 1ktx) and moka1 (gray). (C) The moka1 surface that is predicted to interact with the channel is shown en face in stick (Left) and surface representations (Right). Portions of moka1 originating from parental toxins are marked with different colors: Ce3 (red), AgTx2 (yellow), and CTX (blue). Residues common to all (QC and KCM) are shown in gray. Moka1 Lys24 equivalent to KTX Lys27 that is inserted in the channel pore is marked by an asterisk (*).
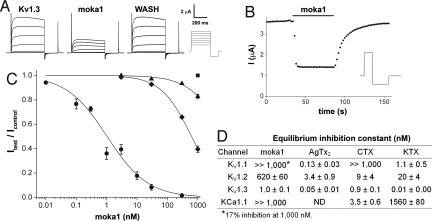
Moka1 was synthesized by solid-phase peptide synthesis, and the free product (without accompanying phage) found to block human Kv1.3 channels potently and selectively. Application of 1 nM moka1 suppressed half the current of channels expressed in Xenopus oocytes in reversible fashion (Fig. 3A), showing a rapid on-rate (39 × 106 ± 6 × 106/Ms) and slow off-rate (0.056 ± 0.006/s) as expected for a high-affinity interaction (Fig. 3B and Tables S8 and S9).
Fig. 3.
Moka1, a high-affinity and specific Kv1.3 channel blocker. Ion channel isoforms expressed in Xenopus laevis oocytes and studied by two electrode voltage clamp included rat Kv1.1, rat Kv1.2, human Kv1.3, and mouse KCa1.1. Bath solution was (in mM): 2 KCl, 96 NaCl, 1 MgCl2, 1.8 CaCl2, 5 Hepes (pH 7.5), and 0.1% bovine serum albumin (BSA). Equilibrium inhibition was determined by fitting dose-response curves for moka1 and calculated for other toxins from percent block. kon and koff were calculated. Methods are described in depth in SI Materials and Methods. (A) Representative current traces for human Kv1.3 channels before, in the presence of, and after washout of 3 nM moka1 at steady state. (Scale bars: 2 μA and 200 ms.) Voltage protocol: holding voltage −100 mV, 500 ms steps of 15 mV from −60 mV to 30 mV followed by a 200 ms step to −135 mV with a 30-s interpulse interval. (B) The time course for block and unblock of human Kv1.3 on acute application (bar) and washout of 3 nM moka1 during 100 ms steps to 0 mV from −100 mV followed by a 200-ms step to −135 mV every 2 s. (Inset) Voltage protocol. (C) Dose-response relationships for moka1 inhibition of human Kv1.3 (●), rat Kv1.1 (▴), rat Kv1.2 (◆), and mouse KCa1.1 (■), n = 3–11 cells. Kv1.1, Kv1.2, or Kv1.3 peak currents were recorded during a 500-ms step every 30 s to 0 mV from a holding voltage of −100 mV, followed by a 200-ms step to −135 mV. KCa1.1 currents were recorded with 50-ms steps every 3 s to 60 mV from −80 mV, followed by a 40-ms step to −100 mV. (D) Measured equilibrium inhibition (nM) for moka1, KTX, AgTx2, CTX on Kv1.3, Kv1.1, Kv1.2, and KCa1.1. Ce3 is reported to block human Kv1.3 with Ki of 366 nM (5). AgTx2 is reported to block KCa1.1 with Ki >1,000 nM (Table S8). Block of human Kv1.3 in human embryonic kidney (HEK293) cells showed inhibition with an affinity at equilibrium (Ki) of 4.4 ± 0.5 nM (Table S9).
Importantly, the pharmacological profile of moka1 was unlike any of its three parental toxins or KTX. Though moka1 blocked Kv1.3 in the nanomolar range, it was over 1,000-fold less potent on Kv1.1 (17% inhibition at 1 μM), 620-fold less potent on Kv1.2, and had no measurable effect on KCa1.1 at 1 μM (Fig. 3C). Conversely, nanomolar affinity was observed for AgTx2 with Kv1.1, Kv1.2, and Kv1.3, for CTX with Kv1.2, Kv1.3, and KCa1.1, and Ce3 is reported to be a low-affinity blocker of Kv1.3 with a Ki of 366 nM (5). While the library template toxin KTX binds Kv1.3 with high affinity and effectively differentiates Kv1.3 and Kv1.2, it is less able than moka1 to distinguish between Kv1.3 and Kv1.1 (Fig. 3D).
While the unique pharmacology of moka1 could not have been anticipated based on the traits of its three parent toxins, it can be rationalized by its unique 3D structure. Five residues that populate the majority of the channel-binding surface around the canonical pore-directed α-KTx Lys are from Ce3 (Leu7 and Pro8), AgTx2 (Phe22), and CTX (Arg31 and Tyr33) (Fig. 2C), an array of residues not seen in natural α-KTx toxins. Further, the N terminus in α-KTx toxins can modify toxin binding (21, 25, 32, 33), and the segment in moka1 is shorter and altered in position compared with AgTx2, CTX, and KTX (Fig. 2B).
Mokatoxin-1 Acts on T cells but Not the GI tract.
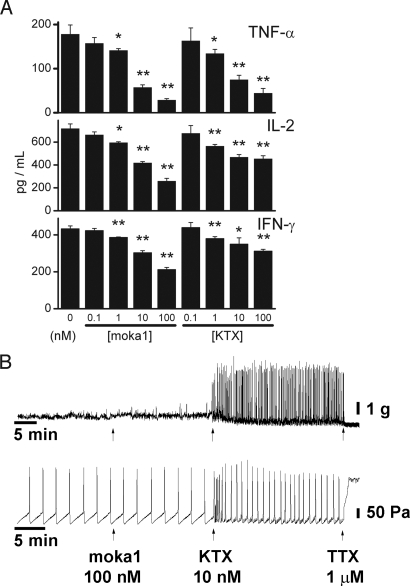
Human T cells were used to confirm that moka1 was active on Kv1.3 channels in their natural milieu. Consistent with expectations, moka1 inhibited secretion of TNF-α, IL-2, and IFN-γ at concentrations as low as 1 nM when CD3+ T cells were stimulated with anti-CD3/28 beads (Fig. 4A). Moreover, there was no reduction in cell viability with 1 μM moka1, indicating that suppression was not due to cytotoxicity and that toxin exposure was tolerated even at high concentrations. That moka1 inhibits T cells more potently than KTX (Fig. 4A) despite a lower affinity for human Kv1.3 channels expressed in oocytes (Fig. 3D) highlights the importance of native receptor milieu.
Fig. 4.
Moka1 inhibits secretion of effector cytokines by T cells and does not alter ileal motility and peristaltism. (A) Treatment with moka1 inhibits the secretion of effector cytokines by T cells. Human CD3+ T cells were purified from peripheral blood of healthy donors and 105 were treated with various doses of moka1 or KTX starting 1 h before stimulation for 16 h with anti-CD3/CD28 beads at a 1:1 ratio in triplicate. Supernatants were assessed for TNF-α, IL-2, and IFN-γ by ELISA. Data represent two independent experiments. Statistically significant differences between control (no toxin) and test conditions are indicated (*, P < 0.05; **,P < 0.01). Methods are described in-depth in SI Materials and Methods. (B) Effect of moka1 and KTX on isometric tension (Upper) and intraluminal pressure (Lower) of guinea pig ileum. Preparations were exposed to 100 nM moka1 which induced no discernable effect over 15–20 min. Subsequent addition of 10 nM KTX induced twitching and increased peristaltic activity—effects abolished by 1 μM tetrodotoxin (TTX). Ileal segments were from adult guinea pigs and studied in modified Krebs-Henseleit solution at 37 °C. For isometric tension assays, segments were mounted under tension and responses recorded using a force-displacement transducer coupled to a polygraph. Toxins were added after 60 min of equilibration (34). For assays of peristalsis, intraluminal perfusion from the oral end was continuous with pressure recording at the aboral end and the threshold for contraction used to quantify effects (12). Toxins were added after peristaltic activity was stable for >15 min. Methods are described further in SI Materials and Methods.
To confirm that moka1 did not cross-react with native gastrointestinal Kv channels, isolated ileal segments were studied (Fig. 4B). Whereas native α-KTx toxins have been shown to increase twitch frequency and lower the pressure threshold for eliciting peristaltic contractions (12), moka1 from 1 to 100 nM neither led to twitching nor altered the pressure threshold for contraction. In contrast, KTX at just 10 nM induced both effects significantly. These effects of KTX are caused by inhibition of Kv1.1 channels and abolished by tetrodotoxin, consistent with mediation via the enteric nervous system (34).
Discussion
Sushruta Samhita, a text in Vedic Sanskrit attributed to Sushruta (sixth-century BC) describes whole venoms as curatives. More recently, native, recombinant, and chemically modified peptide toxins and small molecule mimics have served as leads for drug development or found use in medical therapy, for example, as anticoagulants and in pain management (35). Here, a de novo toxin is made for the immunotherapeutic target Kv1.3 using a scorpion neurotoxin scaffold. Supporting its potential efficacy, moka1 exhibits a desirable profile that differs from other toxins and small molecules and inhibits human T cell cytokine release via Kv1.3 without increasing gastrointestinal motility due to cross-reaction with Kv1.1.
The attributes of moka1 validate scaffold-based/target-biased library design as a strategy to exploit the evolution-tested robustness of animal toxin scaffolds and their known associations with target classes. Combined with the power of phage display, the approach overcomes major obstacles to isolation of target-specific toxins. Thus, new toxins are isolated based on interaction with the target and are physically linked to their encoding gene, allowing toxin sequencing, overproduction, and characterization by standard methods. The strategy also provides a new source for biologically active neurotoxins. Though strategies such as depletion on secondary targets can be applied (36), isolation of moka1 is an example of achieving high target specificity by driving selection for high affinity (37).
Scaffold-based library design should prove applicable to other targets and selection criteria, for example, to isolate toxins that bind to receptors in defined functional states. Despite an apparent diversity of millions of toxins in the complete animal “venome,” nature employs a moderate number of molecular scaffolds. Moreover, toxins typically contain 20–100 amino acids, a size well handled by phage display. This suggests that other scaffolds will also prove amenable to designer toxin development for research, diagnosis, and therapy.
Materials and Methods
An extended version of Materials and Methods is available in SI Materials and Methods.
KTX Phage Particle and Library Construction.
Genes for KTX and DDD-KTX were constructed using Escherichia coli codon bias and cloned into pAS62 phagemid in frame with phage particle coat protein pIII. The combinatorial library was built based on 31 α-KTx toxin or toxin precursor sequences (Arachnida: Scorpionida) that exhibit a high sequence similarity to KTX, irrespective of the toxins' reported pharmacological property. The 31 sequences were aligned by the Cys residues forming disulfide bridges, allowing for insertions/deletions. This alignment defined three domains (Table S1) corresponding to Androctonus mauretanicus kaliotoxin-1 (KTX, α-3.1, GenBank accession no. P24662) residues Gly1-Pro12 (domain A), Leu15-Gly26 (domain B), and Asn30-Lys38 (domain C). In designing the library, N-terminal pyroGlu (Z) residues were substituted by Gly, and from eight toxins, 1–3 aa on either the N or C terminal were omitted (Table S1). This yielded 30, 22, and 17 unique sequences for domains A, B, and C, respectively. The sequences between the domains were modified to match KTX residues Gln13Cys14 (QC) and Lys27Cys28Met29 (KCM) to allow ligation of nucleotide duplexes and produce inserts with the ABC pattern. Complementary nucleotide pairs for each unique domain were synthesized, phosphorylated, and annealed individually. To achieve and monitor domain incorporation, 90 separate reactions were performed to ligate the ABC inserts into pAS62. Each of these reactions contained one domain A duplex, 7 or 8 domain B duplexes, and 17 domain C duplexes with the same total moles of A, B, and C. Ligation mixtures were transformed into E. coli XL1-Blue. This library resulted in de novo toxins ranging from 31 to 40 aa residues in length, and reconstitutes 20 original native toxins. Libraries by this method had 8–58% in-frame ABC toxin sequences, i.e., bona fide unique α-KTx scaffold toxins as confirmed by sequencing.
Phage Particle Binding, ELISA, and Library Screening.
For each binding determination, three wells in a NUNC-Immuno MaxiSorp 96-well plate (Nalge Nunc International/Thermo Fisher Scientific) were coated overnight at 4 °C with 1 μg of KcsA-1.3 or KcsA channels in 50 μL of 100 mM NaHCO3 per well, 1 mM n-dodecyl-β-d-maltopyranoside (DDM; pH 9.0). Thereafter at room temperature, wells were then washed once with 50 mM Tris-HCl, 150 mM NaCl (pH 7.5) containing 0.1% Tween 20, and 1 mM DDM (TBST), and blocked with 200 μL of 50 mM Tris-HCl, 150 mM NaCl (pH 7.5), and 1 mM DDM (TBS) containing 0.5% BSA before washing once with TBST.
For each well, an equal number of phage particles ranging between 108–1010 were added in 50 μL of TBS containing 0.5% BSA and incubated on a rocking shaker for 2 h. Following five washes with TBST, horseradish peroxidase conjugated mouse anti-M13 monoclonal antibody (GE Healthcare) diluted at ≈1:2,500 in 50 μL final volume of TBST with 0.5% BSA was added. Following incubation on a rocking shaker for 2 h, samples were washed five times with TBST and twice with TBS. Thereafter, 50 μL of 1-Step Turbo (3,3′,5,5′ tetramethylbenzidine) TMB-ELISA (Pierce Biotechnology) was added and the reaction stopped with 50 μL 2 M H2SO4. Absorbance was read at 450 nm. Data are mean ± SE for three wells.
In selections, wells were prepared, and phage particles added and incubated as above. Poorly adherent phage particles were removed by washing 3–20 times with 200 μL TBST. Bound phage particles were eluted with 100 μL of 0.1 M triethylamine by incubation for 10 min on a rocking shaker. The pH of the eluate was adjusted to between 7.0 and 8.5 with 1 M Tris-HCl (pH 8.0) before centrifugation at 12,000 × g for 5 min. The supernatant was used to infect E. coli XL1-Blue for reamplification of the library, determination of the recovery rate, and/or phage particle genotyping. The isolation of two or more identical clones in a sample of 20 was the basis for further study of a clone, as the probability of this occurring in the absence of enrichment (e.g., randomly) is less than 10−8. Furthermore, with a 50-fold enrichment per cycle, the probability of observing any one toxin sequence two or more times in a sample of 20 after two cycles is 0.954; conversely, the probability of observing that same toxin only once (and therefore our failure to study it further) is just 0.046. Phage particles were quantified by titering before and after selective library sorting and genotyped by DNA sequencing (Table S3–S6).
KcsA and KcsA-Kv1.3 Channel Synthesis.
The gene for KcsA-Kv1.3 was constructed as described by others (27), and the resultant ORF subcloned into pQE32. XL10 Gold cells (Stratagene) were transformed with cDNA for KcsA or KcsA-1.3 and grown overnight in LB media with 2% glucose and 200 μg/mL ampicillin. One milliliter of the overnight culture was inoculated into 1 L of LB media with 200 μg/mL ampicillin and 0.2% glucose. At OD 600 nm ≈1, protein expression was induced by addition of 0.5 mM IPTG and 10 mM BaCl2 at 30 °C. Cells were harvested the next day and lysed by passage through a cell disrupter in the presence of 50 mM Tris-Cl (pH 7.0), 150 mM KCl (buffer A), and proteases inhibitors. Centrifugation at 100,000 × g yielded a membrane preparation that was solubilized by addition of 10 mM DDM and incubation for 2 h at 4 °C. After clarification by centrifugation at 100,000 × g, the supernatant was loaded on a prepacked cobalt column (Clontech). The column was washed with 20 mM imidazole and protein eluted in a single step with buffer A containing 1 mM DDM and 500 mM imidazole. The oligomeric state of KcsA and KcsA-1.3 was corroborated by filtration over Superdex HR 200 followed by SDS/PAGE (38).
Toxins.
AgTx2 was purchased (Alomone Labs). KTX, DDD-KTX, CTX, and moka1 were synthesized using an optimized t-butoxycarbonyl(Boc) solid-phase protocol (39). Peptide folding reactions were quenched by acidification and products purified by reverse-phase HPLC. Fractions with the desired protein were identified by analytical LC-MS, lyophilized, and stored at −80 °C.
Human T Cell Studies.
CD3+ T cells were purified from peripheral blood of healthy donors. For IL-2, IFN-γ, and TNF-α assays, 105 freshly isolated cells were activated using anti-CD3/CD28 dynabeads in 96-well plates, in triplicate. Toxins were added 1 h prior to stimulation. After 16 h of activation, cells were counted and supernatants analyzed by ELISA.
Guinea Pig Ileum Studies.
Ileal segments from adult guinea pigs were studied in modified Krebs-Henseleit solution at 37 °C. For isometric tension assays, segments were mounted under tension and responses recorded using a force-displacement transducer coupled to a polygraph. Toxins were added after 60 min of equilibration (34). For assays of peristalsis, intraluminal perfusion from the oral end was continuous with pressure recording at the aboral end using the threshold for contraction to quantify effects (12). Toxins were added after peristaltic activity was stable for >15 min.
Electrophysiology.
For two-electrode voltage clamp, Xenopus oocytes were injected with cDNA encoding rat Kv1.1 (NM_173095), rat Kv1.2 (NM_012970), human Kv1.3 (NM_002232), and mouse KCa1.1 (NM_010610) subcloned into a dual-use laboratory vector (pMAX or pRAT) with 5′ and 3′ Xenopus laevis β-globin, a cytomegalovirus promoter for expression in mammalian cells, and a T7-promoter for in vitro transcription (40). Recording solution was (in mM): 2 KCl, 96 NaCl, 1 MgCl2, 1.8 CaCl2, 5 Hepes (pH 7.5), and 0.1% BSA. Equilibrium inhibition for moka1 was determined by fitting dose-response curves. Equilibrium inhibition constants for other toxins were calculated from the percentage of block achieved by 1–3 different toxin concentrations that inhibited between 20% and 80% of currents. kon and koff were calculated (21, 41).
NMR Spectroscopy.
[U-13C, 15N]-moka1 was produced in E. coli, purified, and dissolved in 10 mM sodium phosphate buffer (pH 6.0) in 93% H2O/7% D2O. NMR spectra were collected on Varian INOVA spectrometer at the 1H frequency of 600 or 500 MHz. Sequence-specific resonance assignments and structural restraints (distances, dihedral angles, and hydrogen bonds) were obtained from double- and triple-resonance spectra (42). NMR data were processed using the NMRPipe suite (43) and analyzed using NMRView (44). Backbone dihedral angle restraints were obtained from secondary chemical shift analysis using TALOS (45). Hydrogen bonding restraints were obtained using long-range HNCO (46). Structure calculation was performed using CYANA with automatic NOE assignment through the CANDID algorithm (47). Statistics of the final structures are in Table S7.
Supplementary Material
Acknowledgments.
We thank P. Lakner (New York University, New York) for advice on statistical analyses, and L. Plant, S. Kent, and D. Goldstein for advice during the course of the work. This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant T32HD007009 (to E.J.), National Institute of General Medical Sciences Grants GM72688 (to S.K.), and GM54237 (to S.A.N.G.). G.D. is a Fellow of the Leukemia and Lymphoma Society.
Footnotes
Conflict of interest statement: Based in part on this work, a patent application (“Identification of Toxin Ligands,” PCT/US2008/013385) has been filed.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910123106/DCSupplemental.
References
- 1.Price M, Lee SC, Deutsch C. Charybdotoxin inhibits proliferation and interleukin 2 production in human peripheral blood lymphocytes. Proc Natl Acad Sci. 1989;86:10171–10175. doi: 10.1073/pnas.86.24.10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beeton C, et al. Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc Natl Acad Sci. 2006;103:17414–17419. doi: 10.1073/pnas.0605136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard RJ, Garcia ML, Slaughter RS, Reuben JP. Selective blockers of voltage-gated K+ channels depolarize human T lymphocytes: Mechanism of the antiproliferative effect of charybdotoxin. Proc Natl Acad Sci USA. 1992;89:10094–10098. doi: 10.1073/pnas.89.21.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legros C, et al. Engineering-specific pharmacological binding sites for peptidyl inhibitors of potassium channels into KcsA. Biochemistry. 2002;41:15369–15375. doi: 10.1021/bi026264a. [DOI] [PubMed] [Google Scholar]
- 5.Olamendi-Portugal T, et al. Novel α-KTx peptides from the venom of the scorpion Centruroides elegans selectively blockade Kv1.3 over IKCa1 K+ channels of T cells. Toxicon. 2005;46:418–429. doi: 10.1016/j.toxicon.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Giangiacomo KM, et al. Revealing the molecular determinants of neurotoxin specificity for calcium-activated versus voltage-dependent potassium channels. Biochemistry. 2007;46:5358–5364. doi: 10.1021/bi700150t. [DOI] [PubMed] [Google Scholar]
- 7.Han S, et al. Structural basis of a potent peptide inhibitor designed for Kv1.3 channel, a therapeutic target of autoimmune disease. J Biol Chem. 2008;283:19058–19065. doi: 10.1074/jbc.M802054200. [DOI] [PubMed] [Google Scholar]
- 8.Pennington MW, et al. Engineering a stable and selective peptide blocker of the Kv1.3 channel in T lymphocytes. Mol Pharmacol. 2009;75:762–773. doi: 10.1124/mol.108.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crest M, et al. Kaliotoxin, a novel peptidyl inhibitor of neuronal BK-type Ca2+-activated K+ channels characterized from Androctonus mauretanicus mauretanicus venom. J Biol Chem. 1992;267:1640–1647. [PubMed] [Google Scholar]
- 10.Beeton C, et al. Selective blocking of voltage-gated K+ channels improves experimental autoimmune encephalomyelitis and inhibits T cell activation. J Immunol. 2001;166:936–944. doi: 10.4049/jimmunol.166.2.936. [DOI] [PubMed] [Google Scholar]
- 11.Grissmer S, et al. Pharmacological characterization of five cloned voltage-gated K+ channels, types Kv1.1, 1.2, 1.3, 1.5, and 3.1, stably expressed in mammalian cell lines. Mol Pharmacol. 1994;45:1227–1234. [PubMed] [Google Scholar]
- 12.Vianna-Jorge R, Oliveira CF, Garcia ML, Kaczorowski GJ, Suarez-Kurtz G. Shaker-type Kv1 channel blockers increase the peristaltic activity of guinea-pig ileum by stimulating acetylcholine and tachykinins release by the enteric nervous system. Br J Pharmacol. 2003;138:57–62. doi: 10.1038/sj.bjp.0705023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koo GC, et al. Blockade of the voltage-gated potassium channel Kv1.3 inhibits immune responses in vivo. J Immunol. 1997;158:5120–5128. [PubMed] [Google Scholar]
- 14.Abbas N, et al. A new Kaliotoxin selective towards Kv1.3 and Kv1.2 but not Kv1.1 channels expressed in oocytes. Biochem Biophys Res Commun. 2008;376:525–530. doi: 10.1016/j.bbrc.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 15.Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- 16.Miller C. The charybdotoxin family of K+ channel-blocking peptides. Neuron. 1995;15:5–10. doi: 10.1016/0896-6273(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez de la Vega RC, Possani LD. Current views on scorpion toxins specific for K+-channels. Toxicon. 2004;43:865–875. doi: 10.1016/j.toxicon.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Duda TF, Jr, Palumbi SR. Evolutionary diversification of multigene families: Allelic selection of toxins in predatory cone snails. Mol Biol Evol. 2000;17:1286–1293. doi: 10.1093/oxfordjournals.molbev.a026412. [DOI] [PubMed] [Google Scholar]
- 19.Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 20.Roberts BL, et al. Directed evolution of a protein: Selection of potent neutrophil elastase inhibitors displayed on M13 fusion phage. Proc Natl Acad Sci USA. 1992;89:2429–2433. doi: 10.1073/pnas.89.6.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein SA, Pheasant DJ, Miller C. The charybdotoxin receptor of a Shaker K+ channel: Peptide and channel residues mediating molecular recognition. Neuron. 1994;12:1377–1388. doi: 10.1016/0896-6273(94)90452-9. [DOI] [PubMed] [Google Scholar]
- 22.Ramu Y, Xu Y, Lu Z. Engineered specific and high-affinity inhibitor for a subtype of inward-rectifier K+ channels. Proc Natl Acad Sci USA. 2008;105:10774–10778. doi: 10.1073/pnas.0802850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M. Use of a modified bacteriophage to probe the interactions between peptides and ion channel receptors in mammalian cells. Nat Biotech. 1997;15:559–563. doi: 10.1038/nbt0697-559. [DOI] [PubMed] [Google Scholar]
- 24.Takacs Z, Wilhelmsen KC, Sorota S. Cobra (Naja spp.) nicotinic acetylcholine receptor exhibits resistance to Erabu sea snake (Laticauda semifasciata) short-chain alpha-neurotoxin. J Mol Evol. 2004;58:516–526. doi: 10.1007/s00239-003-2573-8. [DOI] [PubMed] [Google Scholar]
- 25.Ranganathan R, Lewis JH, MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 26.Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007;17:481–487. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Legros C, et al. Generating a high affinity scorpion toxin receptor in KcsA-Kv1.3 chimeric potassium channels. J Biol Chem. 2000;275:16918–16924. doi: 10.1074/jbc.275.22.16918. [DOI] [PubMed] [Google Scholar]
- 28.Garcia ML, Garcia CM, Hidalgo P, Lee A, MacKinnon R. Purification and characterization of three inhibitors of voltage-dependent K+ channels from Leiurus quinquestriatus var. hebraeus venom. Biochemistry. 1994;33:6834–6839. doi: 10.1021/bi00188a012. [DOI] [PubMed] [Google Scholar]
- 29.Meki A, et al. KTX3, the kaliotoxin from Buthus occitanus tunetanus scorpion venom: One of an extensive family of peptidyl ligands of potassium channels. Toxicon. 2000;38:105–111. doi: 10.1016/s0041-0101(99)00137-3. [DOI] [PubMed] [Google Scholar]
- 30.Miller C, Moczydlowski E, Latorre R, Phillips M. Charybdotoxin, a protein inhibitor of single Ca2+-activated K+ channels from mammalian skeletal muscle. Nature. 1985;313:316–318. doi: 10.1038/313316a0. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez I, et al. Kaliotoxin (1–37) shows structural differences with related potassium channel blockers. Biochemistry. 1994;33:14256–14263. doi: 10.1021/bi00251a038. [DOI] [PubMed] [Google Scholar]
- 32.Park CS, Hausdorff SF, Miller C. Design, synthesis, and functional expression of a gene for charybdotoxin, a peptide blocker of K+ channels. Proc Natl Acad Sci USA. 1991;88:1046–1050. doi: 10.1073/pnas.88.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stampe P, Kolmakova PL. Miller C Intimations of K+ channel structure from a complete functional map of the molecular surface of charybdotoxin. Biochemistry. 1994;33:43–50. doi: 10.1021/bi00168a008. [DOI] [PubMed] [Google Scholar]
- 34.Suarez-Kurtz G, Vianna-Jorge R, Pereira BF, Garcia ML, Kaczorowski GJ. Peptidyl inhibitors of Shaker-type Kv1 channels elicit twitches in guinea pig ileum by blocking Kv1.1 at enteric nervous system and enhancing acetylcholine release. J Pharm Exp Ther. 1999;289:1517–1522. [PubMed] [Google Scholar]
- 35.Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: A model for concerted pharmacological discovery. Mol Interv. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- 36.Dennis MS. Selection and screening strategies. In: Sidhu SS, editor. Phage Display in Biotechnology and Drug Discovery. Vol 3. Boca Raton, FL: CRC; 2005. pp. 143–164. [Google Scholar]
- 37.Li R, Hoess RH, Bennett JS, DeGrado WF. Use of phage display to probe the evolution of binding specificity and affinity in integrins. Protein Eng. 2003;16:65–72. doi: 10.1093/proeng/gzg002. [DOI] [PubMed] [Google Scholar]
- 38.Cortes DM, Perozo E. Structural dynamics of the Streptomyces lividans K+ channel (SKC1): Oligomeric stoichiometry and stability. Biochemistry. 1997;36:10343–10352. doi: 10.1021/bi971018y. [DOI] [PubMed] [Google Scholar]
- 39.Schnölzer M, Alewood P, Jones A, Alewood D, Kent SB. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int J Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 40.Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SAN. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell. 2005;121:37–47. doi: 10.1016/j.cell.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 41.Goldstein SA, Miller C. Mechanism of charybdotoxin block of a voltage-gated K+ channel. Biophys J. 1993;65:1613–1619. doi: 10.1016/S0006-3495(93)81200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 43.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 45.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 46.Cordier F, Grzesiek S. Direct observation of hydrogen bonds in proteins by interresidue 3hJNC′ scalar couplings. J Am Chem Soc. 1999;121:1601–1602. doi: 10.1021/ja038616m. [DOI] [PubMed] [Google Scholar]
- 47.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.