Abstract
Electrical signaling in biology depends upon a unique electromechanical transduction process mediated by the S4 segments of voltage-gated ion channels. These transmembrane segments are driven outward by the force of the electric field on positively charged amino acid residues termed “gating charges,” which are positioned at three-residue intervals in the S4 transmembrane segment, and this movement is coupled to opening of the pore. Here, we use the disulfide-locking method to demonstrate sequential ion pair formation between the fourth gating charge in the S4 segment (R4) and two acidic residues in the S2 segment during activation. R4 interacts first with E70 at the intracellular end of the S2 segment and then with D60 near the extracellular end. Analysis with the Rosetta Membrane method reveals the 3-D structures of the gating pore as these ion pairs are formed sequentially to catalyze the S4 transmembrane movement required for voltage-dependent activation. Our results directly demonstrate sequential ion pair formation that is an essential feature of the sliding helix model of voltage sensor function but is not compatible with the other widely discussed gating models.
Keywords: electrical excitability, gating
Electrical signaling in biology depends upon a unique electromechanical transduction process that couples small changes in the electrical potential across the cell membrane to conformational changes that open and close the pores of voltage-gated ion channels. The high sensitivity of voltage-gated ion channels to small changes in membrane potential depends on gating charges within their transmembrane structure, which are driven outward across the membrane by changes in the electric field and trigger a series of conformational changes that result in opening the pore (1–4). Approximately 12–16 positive charges move across the membrane during activation of the voltage sensors of voltage-gated sodium or potassium channels (5–10). Two major thermodynamic obstacles that the voltage-sensing process must overcome are stabilization of amino acid residues that serve as gating charges in the transmembrane environment of the protein and catalysis of their outward transmembrane movement.
Voltage-gated ion channels are composed of four subunits or domains that each contain six transmembrane segments (11). The S1–S4 segments serve as the voltage sensing module, and the S5 and S6 segment and their connecting P loop serve as the pore-forming module (2–4). The primary gating charges reside in the S4 segment in four to seven repeated motifs of a positively charged amino acid residue (usually arginine) followed by two hydrophobic residues (2–4). The crux of the problem of understanding the electromechanical coupling in voltage-gated ion channels is determining the structural and mechanistic basis for movement of these gating charges across the membrane and for coupling of this movement to pore opening. The sliding helix or helical screw models posit that sequential formation of ion pairs with negatively charged amino acid residues in the S2 and S3 segments serve to stabilize the S4 segments in the membrane and thereby catalyze their transmembrane movement (12, 13). Detailed structural versions of this gating model, developed using the Rosetta Membrane modeling method (14, 15) or a homology modeling strategy (16), require sequential formation of specific ion pairs during gating.
Previous studies of voltage-gated potassium channels using a modified intragenic-suppression strategy and charge-reversal mutations have suggested that the S4 gating charges form ion pair interactions with negatively charged amino acid residues in the S2 segment that are required for protein folding and function of the channel protein (17–19), and these interactions were used as constraints in structural modeling of the KV1.2 channel (14, 15). However, the sequential formation of multiple ion pairs on the millisecond time scale, a defining feature of the sliding helix model (12, 14), has not previously been tested.
The homotetrameric bacterial sodium channel NaChBac from Bacillus halodurans resembles individual domains of mammalian voltage-gated sodium and calcium channels, having six transmembrane segments with a voltage-sensing module consisting of S1–S4 segments and a pore-forming module of S5 and S6 segments (20). NaChBac activation is steeply voltage-dependent but slower than eukaryotic sodium channels (20). Its small size, homotetrameric structure, and lack of cysteine residues make NaChBac an ideal model for analysis of molecular mechanisms of voltage-dependent gating by disulfide locking.
Disulfide cross-linking strategies have proven to be a powerful tool to analyze the structures of the intermediates in protein folding reactions and of intermediate and fully activated states in enzymes (21–23). We previously demonstrated using a disulfide-locking method that the third gating charge (R3) in the S4 segment of NaChBac forms an ion pair with D60 near the extracellular end of the S2 segment (24). Here, we report disulfide locking of R4 in the NaChBac channel at two positions in the conformational pathway leading to the activated state using paired substituted-cysteine residues. Our results demonstrate rapid, sequential, state-dependent interactions of R4 with residues E70 and D60 in the S2 segment of NaChBac during voltage-dependent activation, which in turn determine the rate of inactivation. Based on these results, we developed high-resolution structural models of two intermediate states along the activation pathway using the Rosetta Membrane method. Sequential formation of ion pairs by the S4 gating charges, as demonstrated in this work, is an essential and unique feature of the sliding helix gating model and is not compatible with other models of voltage-dependent gating.
Results
Ion Pair Interactions in Voltage Sensors.
The positively charged arginine residues (R1–R4) positioned at three-residue intervals in the S4 segments of most sodium and potassium channels are conserved in NaChBac (Fig. S1A). In addition, the two negative charges in the S2 segment of NaChBac (D60 and E70) are in analogous positions to negatively charged aspartate or glutamate residues or hydrophilic asparagine residues that have a partially negatively charged carbonyl group in other voltage-gated sodium and potassium channels (Fig. S1A). These conserved residues contribute directly to gating charge transfer (6, 8, 9, 25, 26). According to the sliding helix gating mechanism, the conserved S4 gating charges and the negative charges in the neighboring S2 segment are predicted to form ion pairs sequentially during activation, which serves to stabilize and catalyze the transmembrane movement of the S4 segment (12, 13). The Rosetta Membrane structural version of the sliding helix model predicts sequential interaction of the third gating charge (R3) and the fourth gating charge (R4) in the S4 segment with the most highly conserved negatively charged residues on the S2 segment (E70 and D60) (Fig. S1B and movies S1–S3 in ref. 14) (14, 15).
Voltage-Dependent Disulfide Locking of R4 with E70.
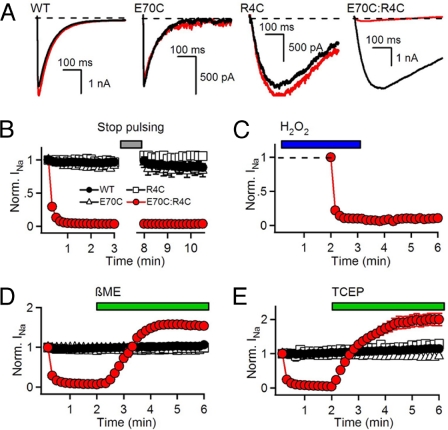
We expressed single and double cysteine mutants of R4, D60, and E70 in NaChBac and investigated the functional effects of disulfide bond formation. Repetitive depolarization of WT or the single cysteine mutants R4C and E70C from −120 to 0 mV for 500 ms results in inward sodium currents (INa) of constant size and shape (Fig. 1 A and B). In contrast, a single depolarization of E70C:R4C activated a large INa, but repetitive depolarizations caused nearly total loss of INa, which did not recover after 5 min at −120 mV (Fig. 1 A and B). Even after treatment with 10 mM hydrogen peroxide (H2O2) to enhance disulfide formation, INa remained stable without depolarizing pulses but was lost immediately during trains of repetitive depolarizations that activated the E70C:R4C channels (Fig. 1C). Consistent with disulfide bond formation, perfusion of the sulfhydryl reducing agent β-mercaptoethanol (βME) or the phosphine reducing agent Tris(2-carboxyethyl)phosphine (TCEP) reversed disulfide locking and restored INa (Fig. 1 D and E). The overshoot above the starting level of INa likely reflects reduction of disulfide bonds formed during biosynthesis. Together, these results demonstrate that activation of E70C:R4C immediately induces state-dependent disulfide bond formation, which locks the channel in a nonconducting state and can be reversed by sulfhydryl reducing reagents. Analysis of the decay of INa for WT and double-mutant channels upon repolarization to −120 mV revealed that Na current is lost because the disulfide-locked channels are unable to deactivate upon repolarization and enter the inactivated state (SI Results; Fig. S2), as we observed for the D60:R3 interaction (24).
Fig. 1.
Disulfide locking the E70C:R4C voltage-sensor. (A) INa from the first and last pulses in a 2-min 0.1 Hz train of 500-ms depolarizations to 0 mV from a holding potential of −120 mV. (B) Mean normalized peak currents during trains (n = 7). After 3 min, pulsing was stopped for 5 min to test for channel recovery. (C) Effect of 10 mM H2O2 (bar; n = 5). (D and E) Effects of 10 mM βME (D) or 1 mM TCEP (E) (Bars; holding potential = −140 mV; n = 6.)
Voltage-Dependent Disulfide Locking of R4 with D60.
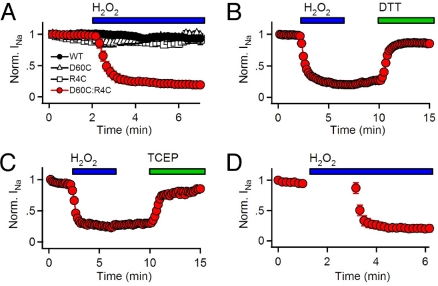
Continuing along the activation pathway, the S4 segment moves further outward where R4 is predicted to encounter D60, the outermost negative charge of the S2 segment. INa conducted by D60C:R4C channels was stable during repetitive stimulation under control condition (Fig. 2A). However, upon application of the extracellular oxidizing agents H2O2 or copper phenanthroline (CuP), INa from D60C:R4C diminished rapidly with repetitive pulsing and was restored by treatment with the reducing agents DTT or TCEP (Fig. 2 A–C and Fig. S3). Oxidization-induced disulfide locking required activation of the voltage sensor, because perfusion of H2O2 for 2 min at −120 mV had no effect by itself (Fig. 2D). These results indicate that R4 interacts with D60 during activation of NaChBac.
Fig. 2.
Disulfide locking the D60C:R4C voltage sensor. Mean normalized peak INa elicited by trains to 40 mV positive to the potential of half activation (V1/2 + 40 mV). Holding potential = −120 mV. (A) Effect of 10 mM H2O2 (n = 7). (B and C) Reversal of H2O2 effect with 10 mM DTT (B) or 1 mM TCEP (C) (V = +80 mV; n = 8). (D) Test for disulfide locking with H2O2 during 2 min (t = 1–3 min, bar) at −120 mV without pulsing.
Voltage-Dependent Reversal of Disulfide Locking.
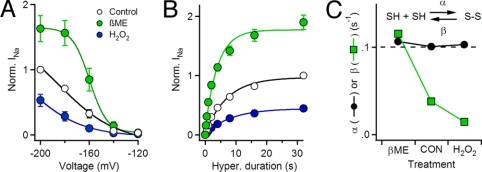
Because disulfide bonds have a free energy of 2–5 kcal/mol (27, 28), and the electrical free energy gained by movement of three positive charges through an electrical potential of −200 mV is 13.8 kcal/mol (SI Results), hyperpolarization can reverse disulfide locking (24). After inducing disulfide locking by repetitive depolarizations, hyperpolarization to potentials more negative than −150 mV caused progressive recovery of INa, which was enhanced by βME and hindered by H2O2. (Fig. 3 A and B). These results suggest that reducing conditions accelerate the reductive cleavage of the disulfide bond between E70C and R4C by increasing the concentration of reducing equivalents available for reaction, whereas oxidizing conditions slow the reductive cleavage reaction by reducing the availability of reducing equivalents. Consistent with this mechanism, the rate constant for reductive cleavage is increased by βME and diminished by H2O2, indicating that these effects are indeed caused by increasing or decreasing, respectively, the availability of reducing equivalents for the reductive cleavage reaction (Fig. 3C).
Fig. 3.
Reversal of disulfide locking of E70C:R4C by hyperpolarization. (A) Voltage dependence of reversal of disulfide locking. Before each trial, five 500-ms pulses to 0 mV were applied to disulfide-lock and fully inactivate all voltage sensors. Fully locked channels were hyperpolarized for 5 s to potentials from −200 to −120 mV followed by a 500-ms depolarization to 0 mV to assay the unlocked channels. Each cell was tested in control and in 10 mM βME or 10 mM H2O2, and currents were normalized to the largest current in control. Mean normalized peak currents were plotted against prepulse potential (n = 6; ±SEM). (B) Time-course of reversal of disulfide locking. Fully locked channels were hyperpolarized to −160 mV for the indicated times followed by a test pulse to 0 mV. Each cell was tested in control and then in the presence of 10 mM βME or 10 mM H2O2; peak test pulse currents were normalized to the peak current following the 32-s hyperpolarization in control. Mean normalized peak currents (±SEM) are plotted against prepulse duration and fit to a single exponential (n = 6). Error bars are smaller than data points where not visible. (C) Rate constants for disulfide locking. Assuming a bimolecular reaction, inset, forward α (black circles), and reverse β (green squares), rate constants were calculated from the rate and extent of reversal of unlocking at −160 mV from (B) as detailed in SI Methods.
Kinetics of Disulfide Locking of the R4 Gating Charge.
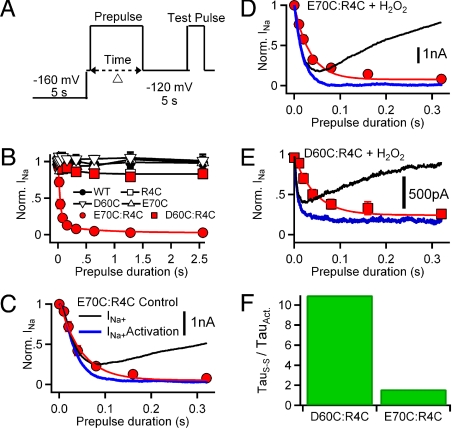
After a prepulse to −160 mV for 5 s to unlock the channels, we measured the rate of onset of disulfide locking of E70C:R4C at −40 mV (equivalent to V ≈ V1/2 + 40 mV), using irreversible loss of INa after repolarization to −120 mV as a criterion (Fig. 4A). For E70C:R4C channels, depolarization to −40 mV caused progressive disulfide locking and loss of INa, even after repolarization to −120 mV for 5 s, but depolarization had no effect on WT or single cysteine mutants (Fig. 4B). The rapid initial rate of loss of INa for E70C:R4C caused by disulfide locking was similar to the rate of activation of INa in both control and oxidizing conditions, when the effect of inactivation was compensated using an exponential function (Fig. 4 C and D). Depolarization of D60C:R4C channels to +80 mV (V ≈ V1/2 + 40 mV) in the presence of 2 mM H2O2 also caused progressive disulfide locking and approximately 80% loss of INa with a rate similar to activation (Fig. 4E). We normalized the time constants for disulfide locking of E70C:R4C and D60C:R4C to the time constants for pore opening of the two mutants to correct for their inherent difference in activation rate and thereby determine the sequence of interaction and disulfide locking of these interacting residues. The time constant ratio for E70C:R4C (1.65) was substantially shorter than for D60C:R4C (11.3), suggesting that the E70:R4 interaction occurs before the D60:R4 interaction during activation.
Fig. 4.
Time course of disulfide locking. (A) Voltage protocol. Channels were unlocked by a 5-s prepulse to −160 mV. Cells were then depolarized by prepulses to (V1/2 + 40 mV) of the indicated duration. Following 5 s at −120 mV, disulfide-locked channels were assayed with a 100-ms test pulse to V1/2 + 40 mV (WT, 0 mV; D60C, 20 mV; E70C, −30 mV; R4C, 60 mV; D60C:R4C, 80 mV; E70C:R4C, −40 mV). (B) Peak test pulse currents were normalized to the test pulse current in the absence of a prepulse. Mean values (±SEM) were plotted versus prepulse duration (n = 6). (C–E) Comparison of the rate of disulfide locking (red circles) and channel activation (blue line) for (C) E70C:R4C, control, (D) E70C:R4C, 2 mM H2O2, and (E) D60C:R4C, 2 mM H2O2. (F) Ratio of the time constant for disulfide locking (τS–S) to the time constant for activation (τAct) for E70C:R4C and D60C:R4C channels in 2 mM H2O2. For E70C:R4C, τS–S = 33 ± 2 ms, τact = 20 ± 2 ms (n = 6). For D60C:R4C, τS–S = 45 ms ± 2 ms (n = 7), τact = 4 ± 0.5 ms (n = 7).
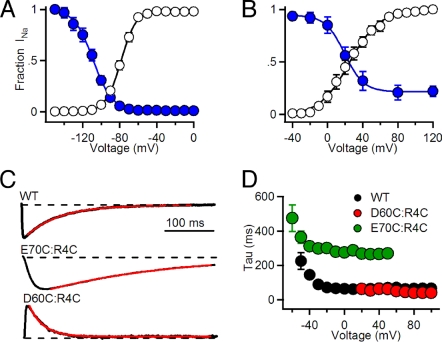
Voltage Dependence of Disulfide Locking of the R4 Gating Charge.
We induced disulfide locking of E70C:R4C during 500-ms prepulses to a range of voltages and recorded the extent of disulfide locking after repolarization to −120 mV for 5 s (Fig. 5A). The voltage dependence of disulfide locking [V1/2(S-S) = −108 ± 2 mV] for E70C:R4C was 22 mV more negative than the voltage dependence of activation (V1/2 = −86 ± 1 mV). By comparison, the voltage for half-maximal disulfide locking of D60C:R4C was 19 ± 2 mV, only 5 mV more negative than the voltage dependence of activation (V1/2 = 24 ± 2 mV; Fig. 5B). Evidently, the interaction of R4 with E70 occurs in an earlier step along the activation pathway of the S4 segment than its interaction with D60.
Fig. 5.
Voltage dependence of disulfide locking and rates of inactivation of disulfide-locked states. The voltage dependence of channel activation (white circles) and of disulfide locking (blue circles) measured in the presence of 2 mM H2O2 are compared for E70C:R4C (A) and D60C:R4C (B). The voltages for half-maximal channel activation and disulfide locking were determined from fits of a Boltzmann equation to the data. To measure the voltage dependence of disulfide locking of E70C:R4C (n = 9), channels were unlocked by a 5 s pulse to −160 mV, then depolarized with 500-ms prepulses to the indicated potentials. After 5 s at −120 mV, the number of channels locked during the prepulse was assessed with a 100-ms test pulse to +80 mV. Since H2O2-induced disulfide locking of D60C:R4C channels cannot be reversed by hyperpolarization, disulfide locking could only be tested at one potential for each cell (n = 5–6 for each potential). (C) Rates of inactivation of disulfide-locked channels. Sodium current traces from the indicated NaChBac channels elicited by 500-ms depolarizations to V = V1/2 + 40 mV from −120 mV (black traces). The time constant of inactivation was determined by an exponential fit to the decay of INa (red lines). INa is outward for D60C:R4C because its positive voltage dependence of activation requires test pulses to V = V1/2 + 40 mV, that are beyond the reversal potential. (D) Mean time constants of inactivation measured at the indicated potentials (WT, n = 10; D60C:R4C, n = 8; E70C:R4C, n = 10; ± SEM).
Kinetics of Inactivation of Disulfide-Locked Channels.
We compared the rate of inactivation of disulfide-locked E70C:R4C and D60C:R4C channels by depolarizing strongly to give rapid activation in the presence of H2O2 and fitting the decay phase of the current during 500-ms pulses to a single exponential (Fig. 5C). The E70C:R4C channel inactivated substantially slower than D60C:R4C channel or WT over a wide range of potentials (Fig. 5D). Evidently, partial outward movement of the S4 segment to the position allowing E70:R4C interaction is not sufficient to give the maximum rate of inactivation, suggesting that sequential outward movement of R4 into interactions with E70 and then D60 progressively increases the rate of inactivation.
Mutant Cycle Analysis of Gating Charge Interactions.
We used mutant cycle analysis to assess the energy of association of the ion-pair-forming residues during activation of NaChBac channels (29), as in our previous studies with R3 (24). The data in Fig. S4 and SI Results confirm that activation of double-cysteine mutants results in a substantial energy of interaction of R4C with E70C and D60C. The spontaneous formation of the disulfide bond in E70C:R4C adds approximately 6.5 kcal/mol of interaction energy that favors activation of this mutant (Fig. S4).
Specificity of Disulfide Locking and Energy Coupling.
To test the possibility that neighboring amino acid residues might form disulfide bonds nonspecifically, we studied double cysteine mutants of a positively charged S4–S5 linker residue (R132) in combination with D60 and E70 (D60C:R132C and E70C:R132C), plus the S4 hydrophobic residue adjacent to R1 (I114C) in combination with D60 (D60C:I114C). We found no evidence for state-dependent disulfide locking of any these pairs of substituted cysteine residues, supporting the specificity of disulfide locking of amino acid residues that form ion pairs during activation of the voltage sensor (SI Results; Fig. S5). Moreover, we found no energy coupling between these pairs of substituted cysteine residues in mutant cycle analysis (SI Results; Fig. S5). These results provide strong support for the specificity of interaction of R4 with E70 and D60.
Structures of Activated States of the Voltage Sensor.
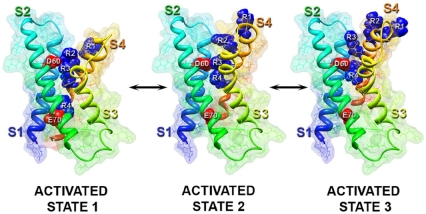
We developed structural models of the conformational changes that occur during activation of the voltage-sensing module of NaChBac, using the Rosetta Membrane method (14, 30) and the Kv1.2–Kv2.1 X-ray structure (31) as described in SI Methods. A homology/de novo model of the Kv1.2–Kv2.1 structure predicted that R3 in S4 interacts with D60 in S2 (Fig. 6, activated state 2), in agreement with our previous experimental data (24). To predict conformations of the voltage-sensing domain of NaChBac suggested by our experimental data, we constrained the side chains of R4 and E70 or R4 and D60 to be in close proximity to each other during simulations. Our model of R4 interaction with E70 (Fig. 6, activated state 1) shows ion pair interactions between R4 and E70 as well as R3 and D60. The R1–R3 residues are all exposed to the extracellular environment in this state (Fig. 6). This prediction of the model is in agreement with our experimental observation that the E70C:R4C sulfhydryls interact to form disulfide bonds only when the channel is activated. Comparison of our E70:R4 model (Fig. 6, activated state 1) to our D60:R3 model (Fig. 6, activated state 2) suggests that the S4 segment is positioned approximately 6 Å more inward in the interacting E70:R4 state compared to the D60:R3 interacting state, which is close to the magnitude of S4 vertical movement during gating predicted in models of the voltage-sensing domain of Shaker-type potassium channels (14, 32). Our model of R4 interaction with D60 (Fig. 6, activated state 3) shows ion-pair interactions between R4 and D60. R1-R4 are all exposed to the extracellular environment in the D60:R4 interacting state (Fig. 6, activated state 3), in agreement with our experimental observation that the D60C:R4C sulfhydryls interact only when the channel is activated. Comparison to our E70:R4 model (Fig. 6, activated state 1) shows that the S4 segment in the D60:R4 interacting state is positioned approximately 9 Å more outward than in the E70:R4 interacting state, setting a lower limit on the magnitude Cα movement in S4 between these states (14, 32). Together, these structural models reveal the probable conformational transitions of the voltage sensor in three sequential activated states in the voltage-dependent activation pathway.
Fig. 6.
Full atom and molecular surface representation of the voltage-sensing module of NaChBac in three sequential activated states. Segments S1 through S4 colored individually and labeled. Side chains of gating-charge-carrying arginines (labeled R1 through R4 and colored blue) in S4, and D60 and E70 (colored red) in S2 shown in space filling representation. A probe radius of 1.4 Å was used to scan the molecular surface of each structural model. Figure generated using Chimera.
Discussion
The Rosetta structural version of the sliding helix model proposes that the S4 gating charges interact with negatively charged amino acid residues in the S2 and/or S3 segments during voltage-dependent gating (12, 14, 15). These ion-pair interactions in the voltage sensor serve to stabilize the S4 segment in its transmembrane environment and catalyze its transmembrane movement by providing a low energy, hydrophilic pathway across the membrane. Our results provide strong support for this model of voltage-dependent gating by demonstrating state-dependent formation of disulfide bonds between cysteine residues substituted for the interacting partners in these ion pairs on the time scale of channel activation and by measuring the coupling energy for interaction of these residues during activation of WT and mutant channels by mutant cycle analysis. Disulfide-bond formation between substituted cysteine residues is a well-established method of analysis of the structures of intermediates in protein-folding pathways and of intermediate conformations in allosteric enzymes (21, 23, 33). Because the reactive sulfhydryls must approach within 2 Å at the time of formation of a disulfide bond, this is a high-resolution method of analysis of intra-protein interactions. The strict state dependence and the rapid rate of onset of disulfide locking exclude the possibility that an irrelevant conformation has been trapped by disulfide bond formation in our experiments, and studies of neighboring residues confirm that disulfide locking is specific. Therefore, these results provide strong support for formation of ion pairs between R4 in the S4 segment and E70 and D60 in the S2 segment during normal NaChBac gating.
Our results demonstrate that R4 forms an ion pair with E70 at more negative membrane potentials (relative to the voltage dependence of pore opening) and at a faster rate than with D60. This sequence of interaction is predicted in the sliding helix model of gating, in which the S4 gating charges move outward across the membrane by exchanging ion pair partners. Disulfide locking of E70C:R4C and D60C:R4C both trigger pore opening followed by inactivation and therefore must both stabilize one or more activated states of the voltage sensor. Activated state 1 stabilized by the E70:R4 interaction induces slower inactivation than the activated state 3 stabilized by the D60:R4 interaction, suggesting significant functional differences between these two conformational states in inducing inactivation. In our previous studies (24), the D60C:R3C disulfide bond was formed with a time constant ratio (compared to pore opening) of 1.65, comparable to that observed for E70C:R4C here; therefore, it is likely that these two states are identical or energetically very similar conformations along the activation pathway, as depicted in activated states 1 and 2 (Fig. 6).
How far does the S4 segment move in the plane of the membrane? The data presented here support sequential electrostatic interactions of R4 with E70 and then with D60 in the S2 segment. Structural modeling of the conformational changes of the voltage-sensing module of NaChBac using the Rosetta Membrane method revealed the probable structures of three distinct states of the S4 segment during gating. Transitions between activated state 1 and activated state 3 involve vertical S4 movement of approximately 9 Å, as measured by Cα–Cα distances.
Based on the Rosetta Membrane simulations, the sequential movement of R4 from interacting with E70 in S2 (activated state 1) to interacting with D60 in S2 (activated state 3) is only possible if the R3–R4 segment of S4 adopts a 310-helix conformation transiently. An α-helical secondary structure in R3–R4 region of S4 exposes R1 and R2 to the hydrophobic core of the membrane a highly unfavorable environment for arginine side chains (15). Thus, our models suggest that the 310-helix conformation of this segment of S4, which was also observed in the X-ray structure of Kv1.2–Kv2.1 chimera (31), is essential for sequential passage of R3 and R4 through the narrow gating pore in the voltage-sensing module. We propose that each turn of the S4 segment transiently adopts a 310-helix conformation as it moves through the narrow constriction in the gating pore, to allow productive energetic interactions of each S4 gating charge with the two negatively charged residues in S2 and thereby catalyze outward movement of S4.
S4 transition from activated state 1 to activated state 3 involves transfer of approximately one gating charge at the R4 position through the gating pore. However, the complete activation pathway from the initial resting state to the final activated state must involve transfer of three to four gating charges across the membrane in each voltage-sensing module (6–10). Consistent with this expectation, our models of the resting states of the KV voltage sensor have R1 in close proximity to the position of the first conserved negatively charged residue in S2 (equivalent to D60 in NaChBac) (14, 15), and our corresponding model of the resting state of NaChBac also reveals this interaction (34). Therefore, the complete series of conformational changes of the voltage-sensing module from the resting states to activated state 3, which transfer three to four gating charges, must involve outward movement of R4 by approximately 15 Å as well as the S4 segment rotation by approximately 180°, as suggested by the Rosetta Membrane version of the sliding helix model of the voltage-dependent gating (14).
Based on the sigmoid time course of sodium currents in the earliest voltage clamp studies by Hodgkin and Huxley (35), it has long been understood that voltage-gated ion channels must transit through multiple voltage-dependent states before opening. The 3-D structure of the KV1.2 channel and KV1.2– KV2.1 chimera show that the voltage sensor module and the pore-forming module are structurally independent, with relatively few interactions of amino acid residues between them (31, 36). There is strong evidence from kinetic analyses and structural studies that pore opening is a single concerted conformational change (37–40). Therefore, the multiple states in the activation pathway must represent a series of states of the voltage-sensing module. Our structural models developed from a combination of the structure of activated KV1.2 channel (36), the Rosetta Membrane method of structural prediction (30), and the structural constraints from results of disulfide locking and mutant cycle analyses (24) have so far yielded structural models of two resting states in which R1 interacts primarily with D60 or its equivalent (14, 15) and three activated states of the voltage sensor in which R3 and R4 interact with E70 and D60 (Fig. 6). We expect that there is at least one more conformational state in which R2 interacts primarily with D60, R1 is outside of the gating pore, and R3 remains lower than D60 in the gating pore. It is uncertain whether this state would favor pore closure or pore opening. Thus, our present studies favor a series of six states, where R1 indicates the resting state modeled in (15), R2 indicates the resting state modeled in (14, 34), Xn represents one or more intermediate states for which no structural model is available, and A1 through A3 represent the three activated states modeled here:
This way of thinking about voltage sensor function leads to the conclusion that there is no rigid coupling of voltage-sensor activation to pore opening. Instead, each of the four voltage sensors provides an increment of energy toward overcoming the barrier to pore opening. Kinetic models incorporating a series of conformational states of each voltage sensor followed by concerted opening have been developed, rigorously tested, and found to provide an accurate description of channel gating behavior (37–40). Our results provide the initial structural correlates for these gating models, based on the idea that voltage-dependent activation of all four voltage sensors contributes energy toward the pore-opening transition according to:
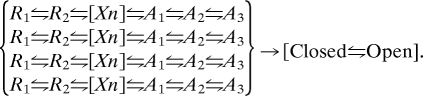 |
In this model, voltage sensors that have moved farther along the activation pathway make a larger contribution toward overcoming the energy barrier to activation, and when the sum of energetic contributions of all four voltage sensors is sufficient, the concerted transition to the open pore occurs.
Our results agree exactly with the predictions of the sliding helix mechanism of voltage-dependent gating but are not compatible with the paddle mechanism of gating. As originally proposed, the paddle gating mechanism envisions a sweeping paddle-like motion of the S3–S4 helical hairpin (the “gating paddle”) across the phospholipid bilayer (≈30 Å) from a position lying nearly horizontal along the inner surface of the membrane to a transmembrane position (41). Phospholipid head groups are proposed to provide stabilizing interactions for this movement of the gating charges (42). Based on the crystal structure of the KV1.2–KV2.1 chimera, the paddle model has been revised to accommodate ion pair interactions in the activated state of the potassium channel (31). However, the proposed rigid structure of the S3–S4 helical hairpin and its sweeping movement through the phospholipid bilayer of the membrane would not allow the multiple sequential ion-pair interactions that we have observed in our disulfide-locking experiments. The paddle mechanism is also incompatible with previous results on sodium channels showing that the S3–S4 linkers are located at the cell surface in the resting state and bind scorpion toxins there (43, 44); with measurements of gating-pore currents conducted by mutant gating pores in resting and activated states of sodium channel voltage sensors (45, 46); and with studies of fluorescent labeling and gating-pore currents in potassium channels (4, 32, 47). Disulfide-locking experiments showing motions of amino acid residues in the S4 segments relative to those in the S3 segments and the pore domain in potassium channels are also incompatible with a rigid gating-paddle structure (48, 49).
In contrast to the paddle model, elements of the transporter model of gating are compatible with our results and are incorporated into the Rosetta structural version of the sliding helix model (14). The transporter model posits that the transmembrane movement of the S4 segment is small (2–3 Å), whereas the surrounding S1, S2, and S3 segments rearrange to change the accessibility of the gating charges from internal to external (50). The Rosetta Membrane structural version of the sliding helix model incorporates this feature, in that the outward spiral movement of the S4 helical segment is accompanied by a counter-rotation of the S1 through S3 helical segments (14). The transporter model of gating does not include sequential ion pair interactions between the R4 gating charge and the two negatively charged residues in the S2 segment, and our results require a transmembrane movement of at least 9 Å of the S4 segment in the outward direction, much larger than proposed in the transporter model. Only the sliding helix model of gating is fully compatible with the results presented here and indeed requires the sequential ion-pair interactions that we have demonstrated.
Methods
Mutants of NaChBac were constructed and analyzed by whole-cell voltage clamp, and the structures of NaChBac in activated states were determined using the Rosetta ab initio modeling algorithm, as described in refs. 14 and 24 and in SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Yong Zhao for constructing some of the mutants used in this work. This research was supported by grants R01 NS15751 and U01 NS058039 from the National Institutes of Health (to W.A.C.) and by a National Research Service Award from training grant T32 GM07270 (to P.G.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912307106/DCSupplemental.
References
- 1.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 3.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 4.Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 6.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 7.Kuzmenkin A, Bezanilla F, Correa AM. Gating of the bacterial sodium channel, NaChBac: Voltage-dependent charge movement and gating currents. J Gen Physiol. 2004;124:349–356. doi: 10.1085/jgp.200409139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 9.Schoppa NE, McCormack K, Tanouye MA, Sigworth FJ. The size of gating charge in wild-type and mutant Shaker potassium channels. Science. 1992;255:1712–1715. doi: 10.1126/science.1553560. [DOI] [PubMed] [Google Scholar]
- 10.Hirschberg B, Rovner A, Lieberman M, Patlak J. Transfer of twelve charges is needed to open skeletal muscle Na+ channels. J Gen Physiol. 1995;106:1053–1068. doi: 10.1085/jgp.106.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Numa S, Noda M. Molecular structure of sodium channels. Ann N Y Acad Sci. 1986;479:338–355. doi: 10.1111/j.1749-6632.1986.tb15580.x. [DOI] [PubMed] [Google Scholar]
- 12.Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 13.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci USA. 1986;508:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K+ channels. Proc Natl Acad Sci USA. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathak MM, et al. Closing in on the resting state of the Shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Shafrir Y, Durell SR, Guy HR. Models of voltage-dependent conformational changes in NaChBac channels. Biophys J. 2008;95:3663–3676. doi: 10.1529/biophysj.108.135335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian DM. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari-Woodruff SK, Lin MA, Schulteis CT, Papazian DM. Voltage-dependent structural interactions in the Shaker K+ channel. J Gen Physiol. 2000;115:123–138. doi: 10.1085/jgp.115.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papazian DM, Shao XM, Seoh SA, Mock AF, Huang Y, Wainstock DH. Electrostatic interactions of S4 volatge sensor in Shaker K+ channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 20.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 21.Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: Increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 22.Siedler F, Rudolph-Bohner S, Doi M, Musiol HJ, Moroder L. Redox potentials of active-site bis(cysteinyl) fragments of thiol-protein oxidoreductases. Biochemistry. 1993;32:7488–7495. doi: 10.1021/bi00080a021. [DOI] [PubMed] [Google Scholar]
- 23.Wahlberg E, Hard T. Conformational stabilization of an engineered binding protein. J Am Chem Soc. 2006;128:7651–7660. doi: 10.1021/ja060933g. [DOI] [PubMed] [Google Scholar]
- 24.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci USA. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuhmer W, Parekh AB. The structure and function of Na+ channels. Curr Opin Neurobiol. 1992;2:243–246. doi: 10.1016/0959-4388(92)90110-7. [DOI] [PubMed] [Google Scholar]
- 26.Kontis KJ, Rounaghi A, Goldin AL. Sodium channel activation gating is affected by substitutions of voltage sensor positive charges in all four domains. J Gen Physiol. 1997;110:391–401. doi: 10.1085/jgp.110.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 28.Serrano L, Horovitz A, Avron B, Bycroft M, Fersht AR. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry. 1990;29:9343–9352. doi: 10.1021/bi00492a006. [DOI] [PubMed] [Google Scholar]
- 29.Serrano L, Neira J-L, Sancho J, Fersht AR. Effect of alanine versus glycine in α-helices on protein stability. Nature. 1992;356:453–455. doi: 10.1038/356453a0. [DOI] [PubMed] [Google Scholar]
- 30.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 32.Posson DJ, Selvin PR. Extent of voltage sensor movement during gating of Shaker K+ channels. Neuron. 2008;59:98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West JM, Tsuruta H, Kantrowitz ER. Stabilization of the R allosteric structure of Escherichia coli aspartate transcarbamoylase by disulfide bond formation. J Biol Chem. 2002;277:47300–47304. doi: 10.1074/jbc.M209913200. [DOI] [PubMed] [Google Scholar]
- 34.Yarov-Yarovoy V, DeCaen PG, Scheuer T, Catterall WA. Structural modeling of intermediate states of the gating pore of NaChBac. Biophys J. 2009:96. [Google Scholar]
- 35.Hodgkin AL, Huxley AF. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol. 1952;116:449–472. doi: 10.1113/jphysiol.1952.sp004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 37.Zagotta WN, Hoshi T, Aldrich RW. Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J Gen Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoppa NE, Sigworth FJ. Activation of Shaker potassium channels. III. An activation gating model for wild-type and V2 mutant channels. J Gen Physiol. 1998;111:313–342. doi: 10.1085/jgp.111.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ledwell JL, Aldrich RW. Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J Gen Physiol. 1999;113:389–414. doi: 10.1085/jgp.113.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horn R, Ding S, Gruber HJ. Immobilizing the moving parts of voltage-gated ion channels. J Gen Physiol. 2000;116:461–476. doi: 10.1085/jgp.116.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 43.Cestele S, et al. Structure and function of the voltage sensor of sodium channels probed by a beta-scorpion toxin. J Biol Chem. 2006;281:21332–21344. doi: 10.1074/jbc.M603814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA. Molecular determinants of high affinity binding of alpha-scorpion toxin and sea anemone toxin in the S3–S4 extracellular loop in domain IV of the Na+ channel alpha subunit. J Biol Chem. 1996;271:15950–15962. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 45.Sokolov S, Scheuer T, Catterall WA. Ion permeation through a voltage- sensitive gating pore in brain sodium channels having voltage sensor mutations. Neuron. 2005;47:183–189. doi: 10.1016/j.neuron.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Sokolov S, Scheuer T, Catterall WA. Gating pore current in an inherited ion channelopathy. Nature. 2007;446:76–78. doi: 10.1038/nature05598. [DOI] [PubMed] [Google Scholar]
- 47.Tombola F, Pathak MM, Gorostiza P, Isacoff EY. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 2007;445:546–549. doi: 10.1038/nature05396. [DOI] [PubMed] [Google Scholar]
- 48.Broomand A, Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008;59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 49.Broomand A, Mannikko R, Larsson HP, Elinder F. Molecular movement of the voltage sensor in a K channel. J Gen Physiol. 2003;122:741–748. doi: 10.1085/jgp.200308927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chanda B, Asamoah OK, Blunck R, Roux B, Bezanilla F. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.