Abstract
Bacteria use two-component signal transduction systems (TCS) extensively to sense and react to external stimuli. In these, a membrane-bound sensor histidine kinase (SK) autophosphorylates in response to an environmental stimulus and transfers the phosphoryl group to a transcription factor/response regulator (RR) that mediates the cellular response. The complex between these two proteins is ruled by transient interactions, which provides a challenge to experimental structure determination techniques. The functional and structural homolog of an SK/RR pair Spo0B/Spo0F, however, has been structurally resolved. Here, we describe a method capable of generating structural models of such transient protein complexes. By using existing structures of the individual proteins, our method combines bioinformatically derived contact residue information with molecular dynamics simulations. We find crystal resolution accuracy with existing crystallographic data when reconstituting the known system Spo0B/Spo0F. Using this approach, we introduce a complex structure of TM0853/TM0468 as an exemplary SK/RR TCS, consistent with all experimentally available data.
Keywords: direct coupling analysis, signal transduction, structure based simulations, transient protein complexes, two component system
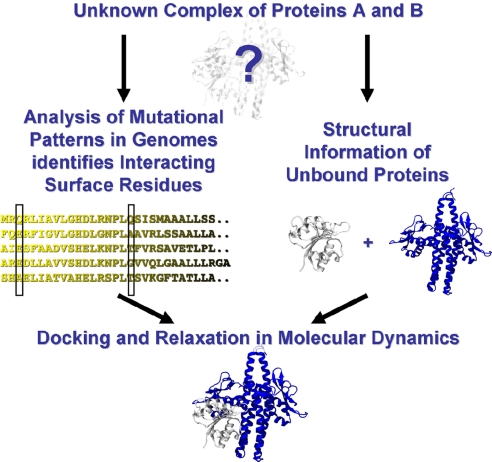
A protein's biological function is often dominated by transient interactions with other proteins with the resulting protein–protein interfaces being considered as targets for drug design (1). Experimental techniques like NMR or X-ray crystallography have proven tremendously successful in providing structural information but face problems when resolving transiently bound protein complexes. Structure prediction methods cannot readily close this gap, because database-driven methods like homology modeling (2) suffer from the lack of structural templates of protein complexes. Physics-based (3–5) approaches struggle with the accuracy of their force fields (6) and often have computationally prohibitive costs for these large complexes. Hybrid methods, which combine experimental data with structural information to generate predictions, have proven more successful (7). Going beyond approaches that rely on a single theoretical framework, the present study integrates information from genomic analysis into molecular dynamics simulation. A sequence-based genomic analysis investigates subtle statistical fluctuations accompanying the mutational patterns of coevolving proteins and suggests amino acids defining the interaction surface of the two proteins. This information is integrated into molecular dynamics simulations to predict the structure of the protein complex (Fig. 1).
Fig. 1.
Flow-chart of our approach. Given the target sequence of an unknown protein complex, direct-coupling analysis investigates the mutational pattern of sequential homologues in databases and suggests pair-wise contacts defining an interaction surface. Similarly, unbound structures for the given target sequences can be either directly extracted from a structural database or generated by structure-prediction methods like homology modeling. This information of the unbound structures and interaction surface contacts is sufficient information for docking simulations in computationally efficient structure-based models, providing both insight into the mechanism of docking and making a prediction of the protein complex. To improve the quality of the prediction, it can be additionally relaxed in physics-based empirical force fields.
Because the accuracy of genomic analysis increases with available sequence information, two-component signal transduction systems (TCS), which are ubiquitous and highly amplified within sequenced bacterial genomes, are ideally suited for statistical analysis. In TCS, a sensor histidine kinase (SK) and a response regulator/transcription factor protein (RR) connect an input signal to an appropriate response by forming a complex to facilitate a transphosphorylation reaction between the two proteins (8, 9). The signal modulates the autokinase activity of the SK and thereby controls the flux of phosphoryl groups through the system (10). Autokinase activity of the SK is mediated by two domains, a dimeric four-helix bundle domain termed HisKA, which contains the phosphorylatable histidine residue, and an ATP-binding ATPase domain. The phosphoryl group is transferred from the SK-histidine to an RR-aspartate residue. This results in activation of the transcription factor activity of the RR.
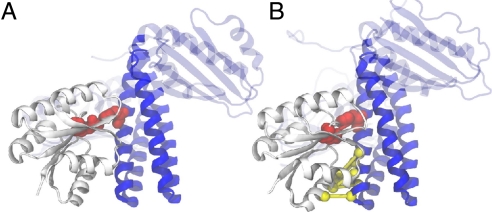
A well-known, albeit not ideal, structural representative of an SK/RR complex is Spo0B/Spo0F (11), part of the Bacillus subtilis sporulation phosphorelay. The phosphorelay comprises a tandem arrangement of a pair of TCS, where the phosphoryl group is transferred in a His-Asp-His-Asp cascade. The central kinase Spo0B has lost its autophosphorylation activity (11), and instead it serves as a phosphotransferase connecting the two RR proteins, Spo0F and Spo0A, in the pathway* (see SI Text and Figs. S1 and S2 for details). Consequently, although Spo0B retains the overall fold of the SK, it has mutated beyond sequence recognition, with only the helix α1 featuring the active-site histidine readily alignable against true SK proteins. In addition to the Spo0B/Spo0F complex (Fig. 2), the structures of the individual protein have been experimentally determined (12, 13). The complex structure has since been used extensively as a reference for interpretation of experimental as well as computational results, aimed at identifying surface residues as well as residues involved in interaction specificity of two-component system proteins (14–18).
Fig. 2.
Crystal structure and prediction of the Spo0B/0F complex. (A) The crystal structure of the complex [PDB ID code 1f51 (11)] of Spo0B (blue, mobile C-terminal region transparent) and Spo0F (white). His-30 and Asp-54, the phosphoryl transferring groups, are highlighted in red. (B) DCA predicts contacts between six amino acid pairs (yellow). Our docking simulations based on the unbound proteins [1ixm (13), 1pey (12)] and these additional contacts predict the bound complex with a backbone-RMSD of roughly 3 Å (floppy C/N termini and the mobile ATPase of Spo0B are disregarded) reaching crystallographic accuracy. Additional inclusion of the Spo0B-His Spo0F-Asp contact (red) improves the accuracy to 2.5 Å.
Here, we integrate two complementary computational techniques to simulate molecular docking of TCS proteins based on the structures of the individual proteins and predict the complexed structures for this important class of bacterial signaling system. Native structure-based simulations (SBS) were chosen as a molecular simulation technique, because their concise Hamiltonian allows their easy adaption to new scientific questions and makes very large proteins and protein complexes computationally accessible. Based on the energy-landscape theory (19), SBS coarse grain a protein's physical/chemical interactions into smoothened effective interactions, providing access to time scales sufficient to simulate (un)folding and large-scale conformational motions (20, 21). The required information about the intermolecular protein–protein surface, introduced as additional contacts within SBS, is provided by a purely sequence-based direct-coupling analysis (DCA) (16) of the interacting proteins in a large set of sequential homologues. DCA distinguishes coevolved residue pairs with statistically significantly linked substitutions arising from direct vs. indirect interactions. The directly interacting pairs predicted by DCA have been shown to lie reliably at the interface of coevolving proteins (16).
Applying the combined methods of DCA with SBS and structures of individual proteins, we demonstrate that the crystallographic complex between Spo0B and Spo0F can be reconstituted with crystal resolution accuracy. We introduce a model of a true SK/RR complex consistent with all available experimental data. Both, this validation and the prediction are important steps toward the ultimate aim of fully integrating genomic information into molecular dynamics, facilitating simulations of systems where sufficient structural information is unavailable or experimentally inaccessible.
Results and Discussion
Docking Simulations of the Spo0B/Spo0F Complex.
Bacillus subtilis Spo0B and Spo0F proteins crystal structures have been determined for both individual proteins as well as for the complex of the two proteins trapped in the act of phosphotransfer (11–13). For this reason, the system makes a perfect test case for the validation of our method. The DCA identifies six highly correlated residue pairings between helix α1 of Spo0B and helix α1 of Spo0F that are lined up almost linearly alongside each other in the complex structure (16) (see Fig. 2). The individual pairings are Spo0B/Spo0F residues: 37/15, 38/14, 41/18, 42/14, 42/18, and 45/22. To verify whether these sequence derived residue contacts are sufficient to generate predictive models of protein complexes, SBS-docking simulations (see Methods) based on the structures of the isolated proteins were performed (see Movie S1).
When performing these simulations, one has to choose a reasonable representation of the DCA residue pairs. To maintain a simplistic Hamiltonian for these simulations, we choose to implement them as regular contacts between the Cα atoms of the corresponding residues with a 5-fold increased contact strength and a contact distance of 7 Å.† Other values for the two parameters have been explored and revealed that the structure predictions from multiple implementations are highly similar and are robust toward the details of implementation. This robustness has been observed previously for geometric features of protein folding (22) and appears to be a property of protein energy landscapes.
One crucial limit for the application of SBS docking is that the individual proteins do not undergo substantial conformational changes during docking. SBS are based on the structures of the unbound individual proteins. During docking, however, the physical environment of the interaction surface changes, which should be reflected by dissimilar orientations of the local side chains. We therefore probe the conformational stability and optimize side-chain packing of the predicted structure by a 25-ns simulation in an empirical all-atom force field with explicit water representation (see Methods). This additional simulation aims at removing such artifacts resulting from the coarse-grained representation in SBS. After ≈10 ns, the structure seems sufficiently equilibrated and the last 15 ns only lead to relatively minor further changes. Considering the size of the entire system, which possesses several hundred amino acids, this indicates the general stability of the predicted complex. Especially the side-chain rotamers of the contact surfaces residues change to better reflect the new physical environment.
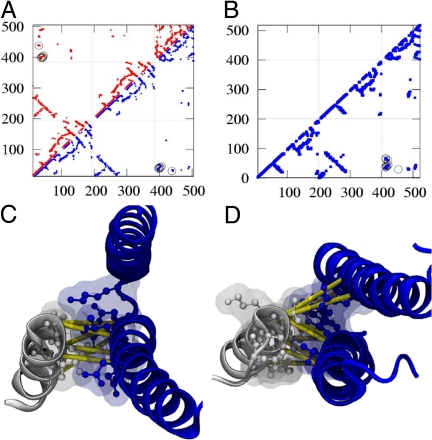
The resulting structural model is in excellent agreement with the crystallographic data with accuracy comparable to the experimental resolution [≈3 Å backbone-RMSD, crystallographic resolution 3 Å (11)] and high agreement of the contact maps (Fig. 3).‡
Fig. 3.
Contact maps and helical interfaces for Spo0B/Spo0F and TM0853/TM0468. (A) The axes denote the consecutively numbered amino acids of the Spo0B/Spo0F system (1–192, 193–384, 385+). The contact maps¶ derived from the complexed crystal structure (red) and the predicted structure (blue) agree well. The DCA-contacts are highlighted by circles (six contacts+His-Asp). (B) The prediction of the TM0853/TM0468 (1–200, 201–400, 401+) system shows a similar contact map (blue, nine contacts+His-Asp). (C) For the SpooB/Spo0F-system, only helix 1 of Spo0B has significant contacts with helix 1 of the Spo0F (coloring scheme as in Fig. 2, the interfacial amino acids are shown additionally as ball-and-stick). (D) For the predicted complex structure of TM0853/TM0468, both helices 1 and 2 of the HK are oriented tighter toward helix 1 of the RR. DCA accordingly predicts contacts between helix 1–helix 1 and helix 2–helix 1.
The included DCA-identified contacts are a sufficient subset of the interprotein contact map because the simulations are able to reconstitute both the complex structure and the entire contact map. This indicates that these statistically correlated contacts are not a random subset but the most crucial part of the interprotein contacts contributing to the binding affinity of the two proteins. The quality of the prediction is increased to 2.5 Å backbone-RMSD by additionally adding a contact for the crucial phosphoryl-group transferring His-Asp pair (Fig. 2), which because of its perfect conservation, cannot be detected by DCA. Because a phosphoryl group is ∼4 Å in diameter, this can be included as an additional contact of 11 Å. The strength of this contact is increased 2-fold compared with the other DCA contacts.
Docking Simulations of the TM0853/TM0468 Complex.
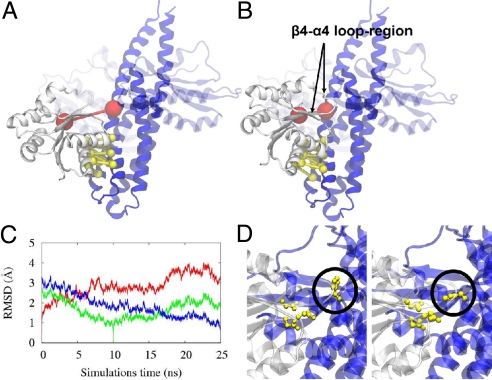
Having successfully validated the method, the approach is applied to docking simulations of a more representative SK/RR pair, the Thermotoga maritima signaling system of SK TM0853 and paired RR TM0468 (23), to generate a predictive complex. TM0853 represents the best available structural example of an SK (24). The structure of the RR TM0468 could be easily generated by homology modeling (2) thanks to a large available and structurally relatively invariable dataset of RR structures with high sequence similarity (we use PDB 1mvo with 45% sequence identity, the closest sequence homolog to TM0468). DCA identified nine potential contact pairs alongside helices α1 and α2 of TM0853 with helix α1 of TM0468 with additional contacts compared with Spo0B/Spo0F between helix α2 of TM0853 and helix α1 of TM0468 (TM0853/TM0468 contacts are: 268/13, 272/13, 294/13, 298/13, 267/14, 271/17, 272/17, 291/20, and 275/21 (Fig. 3). The crucial His-260-Asp-53 interaction, subject to phosphoryl group transfer, is included as an additional contact for docking simulations. The simulations converge into a docked complex, which is relaxed with an empirical force field (Fig. 4). During the relaxation, we observe fast equilibration and little change in the overall structure after 10 ns.
Fig. 4.
Prediction of the TM0853/TM0468 complex. The cytoplasmic domain of the sensor histidine-kinase protein TM0853 [blue, C-terminal ATP-binding domain transparent, PDB ID code 2c2a (24)] and the response regulator TM0468 [white, homologue model based on the structure of 1mvo (26), 45% sequence identity] transfers a phosphoryl group between the two residues. The crucial interaction between TM0853-His-260 and TM0468-Asp-53 (red) is responsible for this transfer and requires the two residues to be in direct contact. Both residues are perfectly conserved over a large set of homologues and are therefore impossible to detect as a contact by DCA. (A) Docking simulations based purely on the contacts predicted by DCA (yellow) converge to a structure with high similarity to the Spo0F/Spo0B complex, but with a large distance between the His-Asp pair (20 Å between Cα). (B) When additionally including the crucial His-Asp interaction as a contact, TM0468 docks at a slightly different angle, which brings His and Asp in direct contact (12 Å between Cα). (C and D) After an additional relaxation in an empirical force-field (C) and rotamer-correction (D), we introduce this structure as a docked complex (PDB file in SI Text), whereas structure (A) might represent an intermediate step during the RR SK binding event. (C) The graph shows a 25-ns relaxation in an empirical force field. The trajectories are compared by the Cα-RMSD (floppy C and N termini as well as the ATPase region are excluded) to the structure right after structure-based docking (red), the structure after 10 ns (green), and the final structure after 25 ns (blue). The relaxation aims at removing artifacts from the structure-based docking procedure and has only limited effect on the backbone because a 3 Å shift can be considered small given the size of the system. (D) The rotamers D9, D10, K105, and H260 (yellow, H260 highlighted by a black circle) of the predicted TM853/TM0468 complex need slight correction to facilitate the phophortransferase reaction: Right after docking TM0853-H260's rotamer is not in line with the TM0468-D53. This can be easily corrected leaving sufficient space for phosphoryl between the two residues.
As contained within the template of the unphosphorylated SK (24), His-260 stays in an unlikely rotamer conformation for the phosphoryl transfer, likely because of the absence of a phosphoryl group in the MD simulations. Furthermore, RR catalytic residues Asp-9 and 10 are not complexed with a catalytic ion as in the Spo0B/Spo0F complex, resulting in a different rotamer conformation for Asp-10. This is easily fixed by adjusting the rotamer conformations of these residues (Fig. 4D). The result is a phosphotransfer active site in excellent spatial agreement with the Spo0B/Spo0F cocrystal structure (PDB file as supplemental data in Dataset S1, His-Asp distance 5.7 and 5.6 Å, respectively), a first validation of the accuracy of the predicted structure. A key feature of the predicted structure for the SK/RR complex are contacts between SK (α1-α2) loop and helix α2 with RR helix α1, which are not observed in the Spo0B/Spo0F complex. The importance of these regions for functionality and interaction specificity is in full agreement with experimental observations (18, 25).
Evaluation of the Accuracy of the Predicted TM0853/TM0468 Complex.
The high accuracy of the Spo0B/Spo0F predictive complex when compared with the existing cocrystal structure (2.5 Å RMSD, Fig. 2) validates our method, and suggests that an equally high accuracy might be obtained for the predictive SK/RR complex of TM0853 with TM0468 (Fig. 4, the structure is provided as SI Text). This is also reflected in the high agreements of the contact maps for the crystal structure and the predicted structure (Fig. 3). We caution, however, that unlike the existing Spo0F structure, the TM0468 structure had to be homology modeled using the structure of the closest sequence homolog, B. subtilis PhoP [PDB ID code 1mvo (26)], which shares 45% sequence identity. Although this structural model is expected to be mostly accurate, slight discrepancies are to be expected. The most obvious feature of concern is the (β4-α4)-loop/helix α4-region, which is known to be very dynamic in RR proteins (14, 27, 28) (Fig. 4B). In the structural template, and hence in our TM0468 model structure, the (β4-α4)-loop is quite extensive and the helix α4 is quite short (two turns) in comparison with three to four turns in the average RR structure. This might not accurately reflect the TM0468 structure in this region, and contacts with TM0853, involving this region might be slightly perturbed.
The most important functional aspect of the complex is the formation of an active site for phosphoryl-group transfer. Active-site residues should be in similar distances as those observed for the Spo0B/Spo0F complex. Residues involved include His-260 (phosphoryl group donor) from the SK and RR residues D53 (phosphoryl group acceptor), D9, D10 (metal coordination), and K105 (D53 activation). The distance between His-260 and D53 is 5.7 Å, positioned perfectly for phosphoryl group transfer and similar to the 5.6 Å distance observed in the Spo0B/Spo0F cocrystal structure (29). D53 and D9 are in hydrogen bonding distance with K105, as generally observed in RR structures and important for active-site activity. The DI contacts between helix α1 of the SK and helix α1 of the RR used for the assembly of both, Spo0B/Spo0F and TM0853/TM0468 complexes are naturally very similar in the individual models.
Forthcoming contacts in the predictive model of possible functional importance are observed between residues E282 with K24 (a potential salt bridge), between S279 with K24 (a potential hydrogen bond), and between L283 and F20 (hydrophobic) with minimal distances during MD simulations of 2.7, 2.7, and 3.6 Å, respectively. These contacts are interesting in the light of published results of specificity swapping experiments aimed at rewiring an SK to phosphorylate noncognate RR (18). These authors found that in addition to our identified covarying residue positions (14, 15), most swapping experiments required replacement of the entire (α1-α2)-loop region, which includes the S279/E282/L283 residues. These results can now be explained through the additional contacts observed in the model structure. Because the (α1-α2)-loop region is variable in length and sequence in different SK, it is not surprising that no covariance signal could be observed for these contacts. Nevertheless, these and similar contacts surely contribute to interaction specificity between SK and RR proteins.
Three contacts between SK helix α2 and RR helix α1 identified by DCA were included in the docking simulations of the TM0853/TM0468 and are in contact in the final model structure. These contacts were omitted for the Spo0B/Spo0F docking simulations, since Spo0B helix α2 orients differently and cannot readily be aligned with SK helix α2 (SI Text). The importance of the helix α2 contacts in SK/RR interaction has been demonstrated by Skerker et al. (18), and these contacts could not have been forthcoming from the Spo0B/Spo0F structure, where they are not realized (Fig. 3 C and D).
Whereas the intramolecular orientation between helices α1 and α2 differs for SK TM0853 and Spo0B, the intermolecular orientation of helix α1 to α2′ within the dimer remains conserved (see SI Text). The most likely explanation for this structural conservation is that this surface has a conserved functional importance in the phosphotransfer reaction for both Spo0B/Spo0F as well as true SK/RR pairs. In the Spo0B/Spo0F cocrystal structure contacts are made with this interface that were implied in sealing of the phosphotransfer active site from solvent access (11, 29, 30). In particular, contacts between Spo0B residues K63 and K67 in helix α2 and Spo0F residue Y84 in the (β4-α4)-loop have been experimentally validated to be important (25). The observed structural conservation of this interface between Spo0B and SK TM0853 suggest that true SK proteins interact with their paired RR in a similar manner. If this were true, one would have expected that our DCA would find some highly correlated residues at this interface, which is not the case. DCA does not pick up conserved residues, but the residues of interest are variable. Another possibility is that the contacts in this region of the protein can be made in a number of different ways, i.e., the involved residue positions differ from SK/RR pair to another SK/RR pair. This would result in dilution of the correlation signal. Consistent with this notion, we do observe such contacts, most notably between TM0853 residues N300 and E303 with TM0468 residue K85. Because these contacts involve the dynamic RR (β4-α4)-loop/helix α4-region, we suggest caution with these results. As discussed above, it is possible that this region is perturbed in our TM0468 homology model, based on the PhoP template structure.
The accuracy of the predicted complex will ultimately be revealed when a cocrystal structure for this complex becomes available. This is likely to be imminent, given that a diffraction quality crystal for this complex has been described (23), which was one reason why we chose to complex this particular protein pair. In the meantime, the predicted complex is consistent with existing structural, computational, and experimental data. Structural analysis of Spo0B in comparison with SK TM0853 revealed that the Spo0B/Spo0F structure is, indeed, an adequate model structure for SK/RR interaction. The important differences are found in contacts made between helix α2 with the RR, which are not realized in Spo0B (see Fig. 3 C and D).
Summary
TCS are an important pathway enabling bacteria to sense and react to extracellular stimuli. The crucial transfer of a phosphoryl group between an SK and an RR requires the formation of a protein complex, which is ruled by transient interactions and has proven an elusive target for structure determination techniques. The present study exemplifies the feasibility of integrating sequence-based genomic analysis with molecular simulation to solve this challenge and predict the protein complex in the absence of a complex template. The predicted structural model is consistent with all existing experimental data on the SK/RR system. The validation of our approach on the Spo0B/Spo0F complex suggests that our prediction of the SK/RR TM0853/TM0468-system should possess high similarity to the crystallographic complex. Since the existence of diffraction quality crystals of this complex has been reported (23) a crystal structure will likely be forthcoming soon allowing for independent verification of the predicted structure. We are confident that further refinement of this approach will successfully introduce other short-lived complexed structures ruled by transient interactions and allow concurrent simulation of the conformational and functional motions of the complex, such as those during the autophosphorylation reaction or phosphoryl-transfer reaction.
Methods
DCA for the Identification of Residue Contacts in Protein–Protein Interaction.
DCA is a computational approach to identify residue contacts between interacting proteins based on sequence information (16). The basic idea is that, given two interacting proteins, a mutation in the interaction surface of one of the proteins will most likely induce a deleterious effect on the interaction affinity. It may, however, be possible to compensate for this effect by substituting also the contact amino acids in the other protein. Residue contacts should therefore show up via correlated amino acid occurrences in pairs of residue positions in large multiple-sequence alignments of homologous interacting protein pairs. Correlation itself is not sufficient to indicate residue contacts, since correlation can result both from a strong direct coupling of the positions, but also from indirect coupling effects via intermediate residue positions (16). DCA disentangles direct and indirect correlations, and identifies interprotein residue contacts connected by strong direct statistical coupling. The main steps of this analysis on SK/RR are as follows.
Data extraction.
As a first step, as many as possible bacterial genomes (31) were scanned with the Pfam Hidden Markov Models (32) for the protein domain families HisKA (PF00512, describing the most abundant class of bacterial SK that this work exclusively concentrates on) and RR (PF00072). Pairs of SK/RR domains appearing inside a single operon but in different proteins are known to interact and are here referred to as cognate SK/RR pairs. Seven hundred sixty nine included bacterial species lead to M = 8,998 cognate pairs. They are collected in a large multiple-sequence alignment (MSA) of M rows and L = LSK + LRR columns, with the aligned single-domain sequences having length LSK = 87 in the case of SK, and LRR = 117 for RR.
Covariance analysis.
As the next step, statistical correlations between the amino acid occupations of different MSA columns, i.e., different residue positions in the proteins, are identified. To this aim, single-column counts fi(Ai) of the relative number of occurrences of amino acid Ai in column i, and column-pair counts fij(Ai,Aj) for the cooccurrence of amino acids (Ai,Aj) in columns i and j are constructed. Correlation between columns is measured by the mutual information (MI),
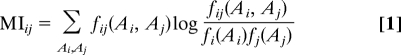 |
which equals zero if and only if the two MSA columns are completely uncorrelated, otherwise MI takes positive values. Interdomain column pairs (i,j) with high mutual information are candidates for residue contacts, but as shown in ref. 16, this information is not sufficient and leads to a substantial fraction of false positives.
DCA.
One major reason for such false positives is indirect statistical correlation introduced via intermediate residues. In ref. 16, a method was introduced to disentangle direct and indirect correlations, here referred to as DCA. Residue-position pairs are considered in their sequence context, and a global statistical model for concatenated sequences (A1,…,AL) of cognate SK/RR pairs has been formulated,
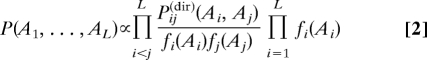 |
Model parameters are the column-pair distributions Pij(dir)(Ai,Aj) measuring the direct coupling of columns (i,j). These parameters have to be fitted to reach consistency of the global statistical model with the empirical frequency counts,
The knowledge of the direct coupling terms Pij(dir)(Ai,Aj) allows for determining the so-called direct information,
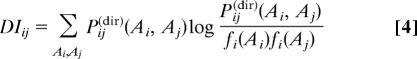 |
which eliminates all effects of indirect coupling. It was shown, that the highest DI column pairs are accurate predictors for actual interprotein residue contacts and are used for the structure-based modeling of the SK/RR cocrystal (16).
Molecular Dynamics.
Native structure-based methods.
Protein evolution shaped a smooth and funneled energy landscape by ensuring a dominance of interactions present in the native state during the entire folding process (19, 33, 34). This prevents entrapment in local minima and provides a degree of robustness permitting protein folding and function despite moderate environmental changes or mutations. Native structure-based models§ represent the ideal case of a perfectly funneled energy landscape where only interactions present in the native state are taken into account. Typically, each amino acid is condensed to a Cα-bead, but variants include multiwelled Gaussians for the contacts (35), Cα Cβ (22, 36), or all-atom representations (20, 37–39). Because the latter incorporate the details of packing best while maintaining computational tractability, we choose ref. 20 as a basis for our docking simulations. An additional weak center-of-mass force for all atoms (k = 0.25·10−6 kBT/Å2) enforces protein complex formation (21). The predicted direct interactions at the surface between the proteins are included as additional contacts between the corresponding Cα atoms with a 5-fold increased strength at a distance of 7 Å. We run the docking simulations at a temperature of 2/3 ϵ/kB, all well below folding temperature to ensure fast convergence (“kinetic simulations”).
The simulations use the GROMACS software package (40, 41). The temperature is kept constant by Langevin coupling with a coupling constant of 1. Each docking simulation runs 2.5 Mio stochastic dynamics time steps of 0.0025 by using the described center-of-mass force and subsequent 0.5 Mio time steps without the center-of-mass force, running for a total of ≈20 h on a typical CPU (Spo0F/Spo0B system, 4,164 atoms).
Amber F99 simulations for relaxation.
Subsequently, the docked complexes were additionally relaxed in an empirical all-atom force-field for refinement. We used AmberF99 (42) with explicit Tip3p solvent and counter ions (43), stochastic dynamics with a time step of 0.002 fs, and Particle Mesh Ewald electrostatics (44). We use a Langevin coupling (300 K, inverse friction constant 0.1 ps) and an additional Berendsen pressure coupling. This refinement aims at removing artifacts from different physical environments for the isolated and docked proteins.
Supplementary Material
Acknowledgments.
We thank A. Beath, B. Lunt, and J. A. Hoch for insightful discussions. This work was supported by the Center for Theoretical Biological Physics sponsored by National Science Foundation (NSF) Grant PHY-0822283 with additional support from NSF Grant MCB-0543906, MCB-0746581, and National Institutes of Health Grant R01GM077298. H.S. was funded by National Institute of General Medical Sciences Grant 019416 (to J. A. Hoch).
Note Added in Proof.
A seminal manuscript that reports the X-ray diffraction structure of the predicted SK/RR complex between TM0853 and TM0468 at 2.8 Å resolution appeared in print (45), while the current manuscript was in review. The experimental structure now allows the evaluation of our blind structural prediction. The individual structures of the two proteins within the complex overlay to an RMSD of 1.9 Å for the dimeric SK HisKA domain and 2.1 Å for the RR domain. The entire complexes overlay to an RMSD of 3.3 Å (Fig. S3A). Not surprisingly the SK/RR interfaces that feature the DCA contacts that were utilized for assembly are very similar for the experimental and predictive complexes. As expected, most discrepancies between the predicted and the experimental structures are found in the orientation of the dynamic (β4-α4)-loop/helix α4 region, and are largely due to the homology template, that was used to model the RR0468 structure. Repeating the analysis utilizing the isolated experimental RR0468 structure, which has been published along with the complex structure (45) is likely to result in significant improvements and a complex structure of comparable accuracy to that achieved for the Spo0B/Spo0F complex. Of note, the (α1-α2)-loop region in the SK undergoes some conformational changes upon RR binding. These conformational changes were perfectly predicted by our structural model (Fig. S3B) and occured during the final relaxation step in an empirical force-field. The contacts formed between this region and helix-α1 of the RR were described above and accurately captured what is observed in the experimental structure.
Footnotes
The authors declare no conflict of interest.
Note that Spo0B along with SK proteins are structurally distinct from Hpt-type phosphotransferases, commonly utilized as intermediary proteins in phosphorelays.
Typically, two interacting residues possess three to six contacts on the all-atom level, making a 5-fold increase in contact strength a reasonable number. The contact distance of 7 Å represents an average distance for interprotein contacts.
We want to emphasize that this prediction does not use any information from the protein complex crystal, which is used as comparative reference only.
We define a contact when any heavy atoms from two different amino acids are within 4.5 Å and the residues more than three residues in sequences apart.
Structure-based models are often referred to as Go-models.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912100106/DCSupplemental.
References
- 1.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Eswar N, Eramian D, Webb B, Shen M-Y, Sali A. Methods in Molecular Biology. Totowa, NJ: Humana; 2008. pp. 145–159. [DOI] [PubMed] [Google Scholar]
- 3.Fujitsuka Y, Takada S, Luthey-Schulten ZA, Wolynes PG. Optimizing physical energy functions for protein folding. Proteins. 2004;54:88–103. doi: 10.1002/prot.10429. [DOI] [PubMed] [Google Scholar]
- 4.Hansmann UHE, Okamoto Y. Prediction of peptide conformations by multicanonical algorithm—New approach to the multiple-minima problem. J Comput Chem. 1993;14:1333–1338. [Google Scholar]
- 5.Schug A, Herges T, Wenzel W. Reproducible protein folding with the stochastic tunneling method. Phys Rev Lett. 2003;91:158102. doi: 10.1103/PhysRevLett.91.158102. [DOI] [PubMed] [Google Scholar]
- 6.Best RB, Buchete NV, Hummer G. Are current molecular dynamics force fields too helical? Biophys J. 2008;95 doi: 10.1529/biophysj.108.132696. 07–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 8.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 9.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17:706–715. doi: 10.1016/j.sbi.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure (London) 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay D, Sen U, Zapf J, Varughese KI. Metals in the sporulation phosphorelay: Manganese binding by the response regulator Spo0F. Acta Crystallogr D. 2004;60:638–645. doi: 10.1107/S0907444904002148. [DOI] [PubMed] [Google Scholar]
- 13.Varughese KI, Madhusudan, Zhou XZ, Whiteley JM, Hoch JA. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol Cell. 1998;2:485–493. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- 14.Szurmant H, et al. Co-evolving motions at protein–protein interfaces of two-component signaling systems identified by covariance analysis. Biochemistry. 2008;47:7782–7784. doi: 10.1021/bi8009604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White RA, Szurmant H, Hoch JA, Hwa T. Features of protein–protein interactions in two-component signaling deduced from genomic libraries. Methods Enzymol. 2007;422:75–101. doi: 10.1016/S0076-6879(06)22004-4. [DOI] [PubMed] [Google Scholar]
- 16.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein–protein interaction by message passing. Proc Natl Acad Sci USA. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burger L, van Nimwegen E. Accurate prediction of protein–protein interactions from sequence alignments using a Bayesian method. Mol Syst Biol. 2008;4:165. doi: 10.1038/msb4100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Whitford PC, et al. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins Struct Funct Bioinf. 2009;75:430–441. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schug A, Whitford PC, Levy Y, Onuchic JN. Mutations as trapdoors to two competing native conformations of the Rop-dimer. Proc Natl Acad Sci USA. 2007;104:17674–17679. doi: 10.1073/pnas.0706077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira LC, Schug A, Onuchic JN. Geometrical features of the protein folding mechanism are a robust property of the energy landscape: A detailed investigation of several reduced models. J Phys Chem B. 2008;112:6131–6136. doi: 10.1021/jp0769835. [DOI] [PubMed] [Google Scholar]
- 23.Casino P, Fernandez-Alvarez A, Alfonso C, Rivas G, Marina A. Identification of a novel two component system in Thermotoga maritima. Complex stoichiometry and crystallization. Biochim Biophys Acta. 2007;1774:603–609. doi: 10.1016/j.bbapap.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzeng YL, Hoch JA. Molecular recognition in signal transduction: The interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 26.Birck C, Chen Y, Hulett FM, Samama JP. The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface. J Bacteriol. 2003;185:254–261. doi: 10.1128/JB.185.1.254-261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 28.McLaughlin PD, et al. Predominantly buried residues in the response regulator Spo0F influence specific sensor kinase recognition. FEBS Lett. 2007;581:1425–1429. doi: 10.1016/j.febslet.2007.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varughese KI, Tsigelny I, Zhao H. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J Bacteriol. 2006;188:4970–4977. doi: 10.1128/JB.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoch JA, Varughese KI. Keeping signals straight in phosphorelay signal transduction. J Bacteriol. 2001;183:4941–4949. doi: 10.1128/JB.183.17.4941-4949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding—A synthesis. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 34.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 35.Lammert H, Schug A, Onuchic JN. Robustness and generalization of structure-based models for protein folding and function. Proteins Struct Funct Bioinf. 2009;77:881–891. doi: 10.1002/prot.22511. [DOI] [PubMed] [Google Scholar]
- 36.Finke JM, Cheung MS, Onuchic JN. A structural model of polyglutamine determined from a host–guest method combining experiments and landscape theory. Biophys J. 2004;87:1900–1918. doi: 10.1529/biophysj.104.041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimada J, Kussell EL, Shakhnovich EI. The folding thermodynamics and kinetics of crambin using an all-atom Monte Carlo simulation. J Mol Biol. 2001;308:79–95. doi: 10.1006/jmbi.2001.4586. [DOI] [PubMed] [Google Scholar]
- 38.Linhananta A, Zhou YQ. The role of sidechain packing and native contact interactions in folding: Discontinuous molecular dynamics folding simulations of an all-atom G(o)over-bar model of fragment B of staphylococcal protein A. J Chem Phys. 2002;117:8983–8995. [Google Scholar]
- 39.Zhou Y, Zhang C, Stell G, Wang J. Temperature dependence of the distribution of the first passage time: results from discontinuous molecular dynamics simulations of an all-atom model of the second beta-hairpin fragment of protein G. J Am Chem Soc. 2003;125:6300–6305. doi: 10.1021/ja029855x. [DOI] [PubMed] [Google Scholar]
- 40.Kutzner C, et al. Speeding up parallel GROMACS on high-latency networks. J Comput Chem. 2007;28:2075–2084. doi: 10.1002/jcc.20703. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comp Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) perform in calculating conformational energies of organic and biological molecules? J Chem Phys. 2000;21:1049–1074. [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 44.Essmann U, et al. A smooth particle Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 45.Casino P, Rubio V, Marina A. Structural insights into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.