Abstract
Huntington disease (HD) is an inherited neurological disorder caused by a polyglutamine expansion in the protein huntingtin and is characterized by selective neurodegeneration that preferentially occurs in striatal medium spiny neurons. Because the medium spiny neurons are innervated abundantly by glutamatergic axons from cortical neurons, the preferential degeneration in the striatal neurons supports the glutamate excitotoxicity theory for HD pathogenesis. Thus, glutamate uptake by glia may be particularly important for preventing glutamate excitotoxicity in HD. Although mutant huntingtin is expressed ubiquitously in various types of cells, it accumulates and forms aggregates in fewer glial cells than in neuronal cells. It remains largely unknown whether and how mutant huntingtin in glia can contribute to the neurological symptoms of HD. We generated transgenic mice that express N-terminal mutant huntingtin in astrocytes, a major type of glial cell that remove extracellular glutamate in the brain. Although transgenic mutant huntingtin in astrocytes is expressed below the endogenous level, it can cause age-dependent neurological phenotypes in transgenic mice. Mice expressing mutant huntingtin show body weight loss, have motor function deficits, and die earlier than wild-type or control transgenic mice. We also found that mutant huntingtin in astrocytes decreases the expression of glutamate transporter by increasing its binding to Sp1 and reducing the association of Sp1 with the promoter of glutamate transporter. These results imply an important role for glial mutant huntingtin in HD pathology and suggest possibilities for treatment.
Keywords: excitotoxicity, glia, neurodegeneration, polyglutamine, glutamate
In Huntington disease (HD), selective neuronal loss occurs preferentially in medium spiny neurons of the striatum and then extends to other brain regions as the disease progresses (1). Because medium spiny neurons are innervated by glutamatergic axons from cortical neurons (2), they are particularly vulnerable to glutamate excitotoxicity, a possible pathogenic mechanism for the preferential neurodegeneration seen in the striatum of HD patients (3). In support of this theory, excitotoxicity of the NMDA receptor, an ionotropic receptor for glutamate, is now associated with HD in various animal models (4, 5).
The majority of cells in the brain are glia that support the survival of neuronal cells. Astrocytes are the major type of glia and express glutamate transporters that uptake extracellular glutamate to prevent glutamate neurotoxicity (6–8). Although mutant huntingtin (htt) is expressed in glial cells in the brains of HD mice and patients (9, 10), whether and how mutant htt in glia contributes to neuropathology in vivo remains unknown. Because glial cells can be therapeutic targets, establishing a transgenic mouse model expressing mutant htt specifically in glia can help develop treatment for HD.
Current HD mouse models have limitations for studying glial htt contribution, because transgenic htt in these HD mice is either overexpressed in neurons or widely expressed in neuronal and nonneuronal cells. While overexpression of polyQ proteins in the astrocytes of flies (11) or in mice (12) can induce neuropathology, it remains to be investigated whether mutant htt in glia at the endogenous level can induce neurological phenotypes. This is particularly important for validating the pathogenic role of mutant htt in glial cells, as less mutant htt accumulates and forms aggregates in glial cells than in neurons in HD mouse brains (10, 13). Another important issue is how mutant htt affects glial function if glial mutant htt does contribute to HD neuropathology. Although our early study has shown that mutant htt can affect glutamate uptake in cultured glial cells (10), the mechanism underlying this defect remains to be understood.
To examine the in vivo contribution of glial mutant htt to HD neuropathology and to investigate the mechanism by which mutant htt affects glial function, we generated transgenic mice that express mutant N-terminal htt fragments in astrocytes. Although transgenic mutant htt is expressed at a lower level than endogenous normal htt, it can induce age-dependent neurological phenotypes. Furthermore, we show that mutant htt in astrocytes binds strongly to the transcription factor Sp1 and reduces the association of Sp1 with the promoter of the glutamate transporter gene, leading to lower levels of glutamate transporters and reduced glutamate uptake. Our findings demonstrate that glial mutant htt can contribute to neurological phenotypes and suggest that improving glial function could be an effective route to therapies for HD.
Results
Expression of Mutant htt in Astrocytes of Transgenic Mice.
We previously found that mutant htt is expressed in glial cells in HD mouse brains and that fewer glial cells than neurons display nuclear htt aggregates (10, 14). To generate a mouse model that selectively expresses mutant htt in glial cells, we constructed a vector that expresses htt under the control of the human glial fibrillary acidic protein (GFAP) promoter (Fig. 1A), an astrocytic promoter that has been widely used to express a variety of genes in astrocytes (15, 16). We verified that this promoter can specifically drive the expression of transfected htt in cultured astrocytes, but not in non-glial HEK293 cells (Fig. S1). Furthermore, transfected mutant htt formed small aggregates in a cultured glioma cell line (Fig. S1), confirming the expression and accumulation of mutant htt in glial cells.
Fig. 1.
Selective expression of mutant htt in glial cells by the GFAP promoter. (A) DNA vector for expressing N-terminal htt (1–208) with either 23Q or 160Q under the control of the human GFAP (gfa2) promoter. An intron and polyadenylation signal are provided by a fragment of the mouse protamine-1 gene (MP-1). (B) EM48 Western blot analysis reveals the specific expression of transgenic htt (arrow) in different brain regions of GFAP-HD mice. Cereb, Cerebellum; B.S., brainstem; Stra, striatum; Ctx, cortex. Arrow indicates transgenic htt. (C) 1C2 Western blot analysis of cultured astrocytes from WT and GFAP-HD mice. Transfected htt in HEK293 cells served as a control. (D) RT-PCR analysis of the transcript levels of transgenic htt in mouse brain cortex. Primers for transgenic human htt were used for PCR. GAPDH was also amplified and served as an internal control in the same PCR to generate the ratios of transgenic htt to GAPDH. RT, reverse transcriptase. (E) Cultured astrocytes from wild-type (WT) and GFAP-HD mouse cortex tissues were analyzed using the primers crossing over the CAG repeat for distinguishing htt-160Q (arrowhead) and endogenous mouse (arrow) htt transcripts. (F) RT-PCR analysis of cultured astrocytes using the primers that are common to both mouse and human htt sequences. Note that the transcriptional levels of htt in two cultures of htt-160Q astrocytes are slightly higher than in wild-type astrocytes, suggesting that additional transgenic htt expression is at a low level.
We then began pronuclear injection of GFAP-htt vectors to generate transgenic mice. Based on the findings that N-terminal htt fragments are more toxic than full-length mutant htt (13, 17), we used cDNAs encoding the first 208 amino acids with 23 or 160 glutamines (Q) in the polyQ domain and generated three founders for htt-23Q and five founders for htt-160Q (GFAP-HD) mice. By using Western blot analysis or immunocytochemistry, we identified that three GFAP-HD mouse lines (line-1, -11, and -31) show detectable mutant htt in their brains. Western blot analysis revealed that htt-160Q is expressed in various brain regions including the cerebellum, brainstem, striatum, and cortex (Fig. 1B). Similarly, htt-23Q is also expressed in various brain regions (Fig. S2). To verify the expression of transgenic htt in astrocytes, we cultured astrocytes from GFAP-HD mouse brains. Western blots with 1C2 clearly detected the expression of transgenic mutant htt (Fig. 1C).
Immunostaining of GFAP-HD transgenic mice shows an obviously low density of mutant htt-positive cells in their brain sections (Fig. S3). Htt-160Q is expressed in glial cells in various brain regions, including the brainstem and striatum, but accumulates and forms more nuclear aggregates in glial cells in the brainstem and spinal cord. This is consistent with the finding that the GFAP promoter drives the expression of transgenes in astrocytes in the spinal cord of adult mice (18). The low density of mutant htt labeling, which was not seen in wild-type mouse brain under the same staining condition, reflects the restricted expression of mutant htt in glial cells.
Level of Transgenic htt-160Q Is Below That of Endogenous Mouse htt.
Using primers that specifically amplify transgenic human htt transcript, we found that the transcription level of htt-160Q is lower than in htt-23Q and N171-82Q mouse brains, as well as lower than in HD KI mouse brains, which express mutant htt at the endogenous level (Fig. 1D). To compare the expression levels of transgenic htt and endogenous mouse htt in the same astrocytes, we isolated astrocytes from wild-type and htt-160Q transgenic mice for RT-PCR. The cultured astrocytes allowed us to perform RT-PCR with primers that amplify the repeat region, such that the transgenic htt and endogenous mouse htt could be distinguished in the same cells under the same PCR conditions. The results also showed that the levels of transgenic htt are lower than endogenous mouse htt in cultured astrocytes (Fig. 1E). We further used different primers that can amplify mouse and human htt transcripts together of the same sequences and length. We observed that the level of htt in transgenic astrocytes is slightly higher than in wild-type astrocytes, also suggesting that the additional expression of transgenic mutant htt is at a low level (Fig. 1F).
The low expression level of transgenic mutant htt is also suggested by the limited number of brain cells showing mutant htt staining (Fig. S3). To confirm this, we compared htt staining in the same brain cortex regions from GFAP-HD mice and N171-82Q mice that express N-terminal mutant htt under the control of the neuronal prion promoter (19). It is clear that htt-160Q is expressed in few glial cells in the GFAP-HD mouse brain whereas N171-82Q htt is much more abundant in neuronal cells (Fig. 2A). The low density of mutant htt staining in GFAP-HD mice also suggests that the expression of transgenic htt is restricted to glial cells.
Fig. 2.
Expression of transgenic htt in HD mouse brains. (A) Comparison of mutant htt staining density between GFAP-HD and N171-82Q mouse brain cortex. The front cortex region was stained with 1C2 at a 1:3,000 dilution. Smaller nuclear size with htt labeling in glial cells and reactive astrocytes (arrow) are seen in GFAP-HD mouse brain, whereas N171-82Q mouse brain shows more abundant and larger neuronal nuclear staining as well as small neuropil aggregates. (Scale bar, 20 μm.) (B) High-magnification (×63) micrographs of brain sections. (Scale bar, 10 μm.) Brain sections were stained with 1C2. (C) Double immunostaining of GFAP-HD mouse brain with mouse antibody 1C2 for mutant htt (red) and rabbit antibody for GFAP (green). The merged image also shows nuclear staining by Hoechst dye (blue). (Scale bar, 10 μm.)
Increased Gliosis in Aged GFAP-HD Mouse Brain.
Examining the brain morphology of GFAP-HD mice at light or electron microscopic levels revealed no obvious morphological abnormalities or death of neurons or glial cells. However, in older GFAP-HD mice (>1.5 years), there is an increase in GFAP staining in the cortex, striatum, brainstem, and cerebellum (Fig. S4A). Increased GFAP levels were also seen in cultured astrocytes from GFAP-HD mice as compared to littermate wild-type controls (Fig. S4B). In the brain, increased GFAP staining reflects reactive astrocytes, an early event in neurodegeneration. High-magnification graphs show that mutant htt is distributed and forms small aggregates in both the cytoplasm and nucleus of large-sized glial cells (Fig. 2B), which are reactive astrocytes. Double immunostaining clearly demonstrates that GFAP-positive astrocytes express htt-160Q in the large astrocytes (Fig. 2C). Such double labeling was not seen in the glial cells that had a similar morphology in wild-type mouse brain, and NeuN-positive neurons do not display mutant htt staining. Although the GFAP promoter can also drive transgene expression in a subset of neurons during embryogenesis (20), we did not detect the expression of mutant htt in neurons or microglial cells in our GFAP-HD mice, which is consistent with the fact that the GFAP promoter primarily expresses transgenes in astrocytes (15). Electron microscopic examination of the brain of an htt-160Q-1 mouse at 26 months of age did not reveal any obvious evidence of cellular degeneration (Fig. S5). It is possible that expression of mutant htt in both neurons and glia is required for prominent cell degeneration. Because we found no obvious degeneration of neurons and glia in GFAP-HD mouse brain, we believe that the expression of mutant htt in glial cells is more likely to cause cellular dysfunction than degeneration. This possibility is also consistent with the fact that neuronal dysfunction rather than overt neuronal loss mediates severe neurological phenotypes and early death of some HD mouse models (19, 21) and that neuronal dysfunction can be induced by defective astrocytes in the absence of neurodegeneration (18).
Age-Dependent Neurological Phenotypes and Glutamate Transport Deficiency in GFAP-HD Mice.
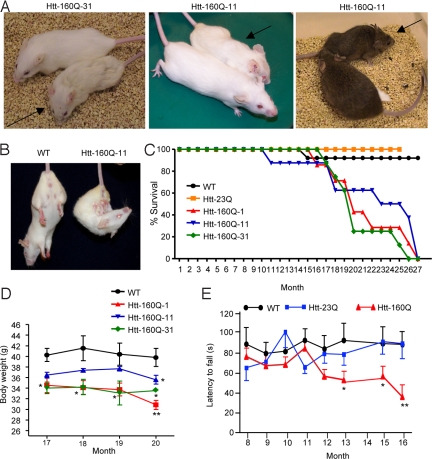
GFAP-HD mice developed late-onset neurological symptoms. Three lines of GFAP-HD mice all show similar neurological phenotypes, which are body weight loss, clasping, and hunchback appearance (Fig. 3 A and B and Movie S1). Such phenotypes also occurred in the mice that had been crossed with mice of the B6C3 genetic background and carried the same htt-160Q (right panel in Fig. 3A). Earlier death occurs in all three GFAP-HD mouse lines (Fig. 3C). Body weight loss is seen when mice are >16 months old, but motor deficit as measured by rotarod performance could be detected earlier, at the age of 1 year (Fig. 3 D and E). Once GFAP-HD mice show obvious motor deficit, their behavioral phenotypes progressively worsen, and they often die 1–2 weeks after the onset of severe symptoms. Thus, there is clearly an age-dependent neurological phenotype in GFAP-HD mice.
Fig. 3.
Progressive neurological phenotypes of GFAP-HD mice. (A) Photos of GFAP-HD mice. Htt-160Q-31 (20 months old) and htt-160Q-11 mice (18 months old) on FVB background and their littermate controls are shown in the left and middle panels. Htt-160Q-11 mouse and its littermate control (11 months old) that also carried the B6C3 genetic background are shown in the right panel. Arrows indicate mutant mice. (B) Old GFAP-HD mouse (18 months old) showing the clasping phenotype. (C) Surviving plots for wild-type (WT), htt-23Q, and three GFAP-HD mouse lines. (D) Body weight loss of male GFAP-HD mice is observed after 16 months. (E) Rotarod performance of wild-type, htt-23Q, and htt-160Q mice. *, P < 0.05; **, P < 0.01 compared with WT or htt-23Q mouse line.
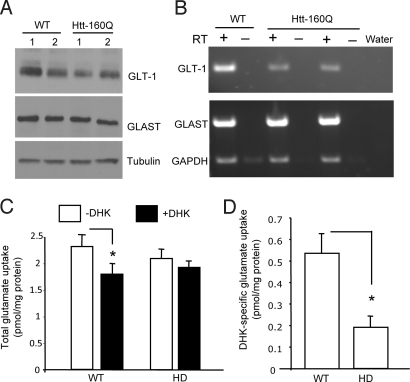
Because glutamate transporter (GLT-1 or EAAT2) transcripts are decreased in other HD mouse models (22, 32) and in cultured glial cells expressing mutant htt (10), we were interested in investigating whether GFAP-HD transgenic mice also show decreased glutamate transporters in their brain. Western blot analysis showed that GLT-1, but not glutamate/aspartate transporter (GLAST), is reduced in GFAP-HD mouse brains (Fig. 4A). RT-PCR analysis of cultured astrocytes also demonstrates that GLT-1, but not GLAST, is reduced specifically in GFAP-HD astrocytes (Fig. 4B). Furthermore, there is significantly reduced glutamate uptake in the brain cortex-striatum slices from GFAP-HD mouse brains (Fig. 4 C and D).
Fig. 4.
Decreased expression of GLT-1 in GFAP-HD mice. (A) Representative Western blot analysis of GLT-1 in the cortex of htt-160Q transgenic (GFAP-HD) mice. Two mice (1 and 2) for each group were shown. The blots were also probed with antibodies to tubulin or GLAST. (B) RT-PCR analysis of GLT-1 and GLAST transcripts in cultured astrocytes from two htt-160Q pups and one wild-type littermate control pup. RT: reverse transcriptase. (C) [3H]-glutamate uptake assays of cortico-striatal brain slices from 20-month-old htt-160Q or littermate wild-type mice (n = 4 each group). Glutamate uptake (pmol/mg protein/15 min) was measured in the absence (−DHK) and presence (+DHK) of the GLT-1 specific blocker DHK (1 mM). (D) The differences between DHK treated and nontreated samples were obtained and show that DHK-specific glutamate uptake is significantly reduced in htt-160Q brain slice. *, P < 0.05.
Mutant htt Reduces Transcriptional GLT-1 by Affecting the Association of its Promoter with Sp1.
Although the above findings and previous reports have found deficient GLT-1 expression in HD mice, we lack mechanistic insight into this phenomenon. Based on the facts that the promoter of the GLT-1 gene carries multiple Sp1 binding sites (23), we asked whether mutant htt in astrocytes binds Sp1 and affects the Sp1-dependent transcription of GLT-1. To better analyze the repeat-dependent effect in glial nuclei, we tagged transfected htt proteins with NLS to ensure that they had equal access to the nucleus. Immunostaining shows that mutant htt (150Q) forms nuclear aggregates, whereas the normal htt fragment is diffuse in the nucleus of cultured astrocytes (Fig. 5 A and B). We then performed Sp1 immunoprecipitation from the transfected astrocytes. Although there was less htt-150Q than htt-20Q in the input, more htt-150Q than htt-20Q was coprecipitated with Sp1 (Fig. 5C). Quantification of the ratios of immunoreactive 20Q or 150Q to tubulin verified that the input of 150Q was much lower than that of 20Q while similar amounts of 20Q and 150Q were precipitated (Fig. S6). Thus, more 150Q than 20Q was precipitated by anti-Sp1, which was verified by measuring the ratio of the precipitated htt to its input (Fig. S6).
Fig. 5.
Mutant htt decreases the association of Sp1 with the GLT-1 promoter. (A and B) EM48 immunostaining of cultured wild-type mouse astrocytes that were transfected with NLS-exon1 htt (green) containing 150Q or 20Q. The high-magnification graphs in (B) show that mutant htt (150Q) forms nuclear aggregates, whereas the normal htt fragment (20Q) is diffuse in the nucleus (blue). (C) Immunoprecipitation of Sp1 in transfected astrocytes shows that more mutant htt (150Q) than normal htt (20Q) is coprecipitated with Sp1. (D) Coexpression of the GLT-1 promoter reporter with N-terminal htt fragments containing different numbers of amino acids (67, 208, 508) plus either 20Q, 150Q, or 120Q. Note that mutant htt fragments (67–150Q and 208–120Q) significantly inhibit the reporter activity. ***, P < 0.001. (E) Chromatin immunoprecipitation (ChIP) assay showing decreased association of Sp1 with the GLT-1 promoter in the frontal brain tissue of htt-160Q transgenic mice compared with wild-type littermate controls. Rabbit anti-Sp1 and IgG, which served as a control, were used for immunoprecipitation. C, no template. (F) Quantification of the ratios of PCR products from immunoprecipitated (IP) to input. **, P < 0.01 compared to wild-type (WT) control (n = 4).
Next we assessed the influence of transfected htt on the promoter activity of the human GLT-1 gene in transfected astrocytes. Small N-terminal mutant htt (67–150Q and 208–120Q) produced more inhibitory effects on GLT-1 promoter activity than a larger N-terminal htt fragment (508–120Q) (Fig. 5D), which also supports the idea that protein context can modulate htt toxicity (24, 25). Although transfection of normal exon1 htt (N67–20Q) also elicited some inhibition of the GLT-1 promoter activity, mutant htt inhibited significantly more GLT-1 promoter activity, indicating a repeat-dependent inhibition (Fig. 5D).
To investigate whether mutant htt reduces Sp1 occupancy of the GLT-1 promoter, we performed a chromatin immunoprecipitation (ChIP) assay using brain tissues from GFAP-HD transgenic mice and littermate wild-type controls. This assay revealed that the presence of transgenic htt-160Q reduces the association of Sp1 with the GLT-1 promoter (Fig. 5E). Quantitative analysis of ChIP results further confirmed that this reduction was specific to the GLT-1 promoter, but not to the Sp1-regulated PCNA promoter (Fig. 5F). Taken together, these findings suggest that mutant htt binds more Sp1 and reduces Sp1-mediated GLT-1 expression in astrocytes, which can lead to defective glial glutamate uptake and increased neuronal excitotoxicity.
Discussion
Huntingtin is expressed in various types of cells, including neurons and non-neuronal cells. While our recent studies and others have demonstrated the presence of mutant htt in glial cells (9, 10, 13, 26, 27), the in vivo role of glial htt in the HD pathology of mice remains unknown. In this study, we show that mutant htt in astrocytes can mediate neurological symptoms in mice.
Our findings support the idea that cell–cell interaction plays an important role in HD pathogenesis (10, 28). In the brain, glia-neuron interactions are important for maintaining the normal function of neurons. In other neurological disorders that are also characterized by selective neurodegeneration, such as Alzheimer's disease and ALS, non–cell-autonomous effects of mutant proteins and glial dysfunction involvement are well documented (18, 29, 30). Importantly, our study reveals the contribution of mutant htt in glial cells to HD neurological symptoms even when it is not overexpressed, providing strong evidence for the critical and pathogenic role of mutant htt in glial cells.
The expression levels of transgenic htt, which are controlled by the promoters, the polyQ repeat numbers, and the size of N-terminal mutant htt determine the severity of neuropathology in various HD mouse models (13). For example, R6/2 mice, in which exon1 htt is abundant in both glial and neuronal cells, show more severe phenotypes (21), whereas mice expressing the same exon1 protein only in their neurons live normally and show much milder neurological phenotypes (28). Such differences raised the interesting issue of whether glial mutant htt could contribute to HD pathology. Overexpression of N-terminal htt in neuronal cells by the neuronal prion promoter also elicits severe neurological phenotypes in N171-82Q mice (19). As N171-82Q mice overexpress mutant htt transcripts at a much higher level than other HD mouse models (13), it is unclear if transgenic htt RNA toxicity could also contribute to the neuropathology in N171-82Q mice. Thus, different expression levels of transgenes in various types of cells in HD mice make it difficult to define the role of mutant htt in glial cells. Because mutant htt accumulates more slowly in glial cells than in neurons when mutant htt is expressed at the endogenous level in HD knockin (KI) mice, we chose to investigate the role of mutant htt in glial cells when it is not overexpressed.
The low level of transgenic mutant htt in astrocytes in mice could be due to the lower expression of transgenic htt or the greater capacity of glial cells to clear misfolded proteins, or both. Indeed, we have found that there is higher activity of the ubiquitin-proteasome system in glial cells than in neurons (14). The age-dependent accumulation of mutant htt in astrocytes in our transgenic mice further supports the idea that age-related decreases in the ubiquitin-proteasome system can promote htt accumulation and cytotoxicity.
Smaller htt fragments with larger polyQ repeats are known to be more prone to aggregation and misfolding. Notably, our transgenic mice expressing N-terminal htt (208 aa) with 160Q show early death, a phenotype not seen in HD mice that express full-length htt in both neurons and glial cells. Because we see the neurological phenotypes in htt-160Q, but not htt-23Q, transgenic mice, it is clear that these phenotypes are dependent on the length of the polyQ repeat. In addition, we know that protein context can modulate htt toxicity, as HD mice expressing smaller htt fragments often show more severe neurological phenotypes than HD mice expressing full-length htt (31). Taken together, the expression levels of mutant htt, its protein context, and its expression in different types of cells all contribute to the neurological phenotypes of HD transgenic mice.
Despite the relatively low levels of mutant htt in astrocytes in our transgenic mice, this model allows us to investigate HD pathology that is not the result of overexpressed mutant htt in glial cells. Although previous findings have shown that mutant htt reduces GLT-1 expression in transgenic HD mice (10, 22, 32) and decreases GLT-1 mRNA and glutamate uptake in the brains of HD patients (33, 34), the mechanism underlying this phenomenon has not been revealed.
We have demonstrated that mutant htt in astrocytes binds more Sp1 and reduces Sp1 occupancy of the GLT-1 promoter. This finding provides some molecular insight into the phenomenon of GLT-1 expression being decreased in various HD mouse models expressing toxic N-terminal htt fragments (10, 21). Miller et al. have recently shown that up-regulation of GLT-1 transporter attenuates some of the behavioral alterations in the R6/2 transgenic model (35), and recent findings reveal that decortication of glutamatergic projections to the striatum significantly lowered striatal glutamate levels and reduced behavioral phenotypes of R6/2 mice (36). Defective glial glutamate uptake as a meaningful pathological change has also been found in ALS, and neuronal dysfunction can be induced by mutant astrocytes in the absence of neurodegeneration (18, 22). Although our study focused on the effect of glial htt on GLT-1, we should point out that the exacerbating effect of glial htt can also be attributed to other cellular dysfunctions. Astrocytes produce neurotrophic factors and cytokines to regulate the synaptic function and morphology of neurons, and recent studies suggest that astrocytes secrete a substance that kills motor neurons in ALS (8, 37). Establishing HD models that express mutant htt in glial cells will allow these possibilities to be tested.
Materials and Methods
Antibodies and Plasmids.
The sources of all reagents can be found in the SI Text.
HD Mice.
N171-82Q mice and Hdh CAG (150) knockin mice, which express a 150Q repeat, were bred and maintained in the animal facility at Emory University under specific pathogen-free conditions in accordance with institutional guidelines of The Animal Care and Use Committee at Emory University. To generate GFAP-HD mice, cDNA encoding N-terminal human htt 208 aa containing 23Q, or 160Q was subcloned into the eukaryotic expression vector pGfa2 at the BamHI restriction site. This vector uses the 2.2-kb fragment of astrocyte-specific human glial fibrillary acidic protein (GFAP) promoter (15, 16). Microinjection of GFAP-HD vectors into the pronucleus of fertilized oocytes from FVB mice was conducted by the Emory University transgenic mouse core facility. Genomic DNA was isolated from mouse tails, and the PCR genotyping method was used for screening transgenic mice. Primers with sequences flanking the polyglutamine repeat were used for PCR. Sequences of the primers are: the forward primer (5′-ATGAAGGCCTTCGAGTCCCTCAAGTCCTTC-3′) and reverse primer (5′-AAACTCACGGTCGGTGCAGCGGCTCCTCAG-3′) were used for PCR. All of the positive founders and their corresponding lines carry the expected length (23Q and 160Q) of the polyQ repeat in transgenic htt.
Western Blot Analysis, Immunohistochemistry, and Mouse Behavioral Analysis.
See SI Text.
RT-PCR.
Total RNA from mouse cortex was isolated using the RNeasy Lipid Tissue Mini kit (QIAGEN #74804). First strand cDNA was obtained using the Invitrogen SuperScript First Strand Synthesis System for RT-PCR (11904-018). RT-PCR of mouse brain htt and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers has been described by our laboratory (13).
Four- to 6-week-old cultured astrocytes were treated overnight with 0.25 mM dBcAMP in serum-free media to increase glutamate transporter expression for RT-PCR. Total astrocyte RNA was collected using the Qiagen RNeasy Mini Kit (74104), and cDNA was produced using the same method as the brain lysate. Primers for GLT-1, GLAST (10), GAPDH, and htt (13) have been previously described in our early studies.
Glial Culture, Glutamate Uptake Assay, GLT-1 Transcription Reporter Assay, and ChIP.
See SI Text.
Statistical Analysis.
All values are expressed as means ± SE. We assessed statistical significance using Student's t test and considered a P value of <0.05 significant.
Supplementary Material
Acknowledgments.
We thank Zhihui Fang for her technical assistance. We thank Michael Brenner at the University of Alabama at Birmingham for providing the pGfa2 vector and Jeffrey Rothstein at Johns Hopkins University for providing rabbit anti-GLT1 antibody and the GLT-1 promoter reporter. This work was supported by National Institutes of Health Grants NS36232 (to X.-J.L.), AG19206 (to X.-J.L.), NS045016 (to S.L.), and AG031153 (to S.L.) and CHDI foundation, Inc. (X.-J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911503106/DCSupplemental.
References
- 1.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Calabresi P, et al. Striatal spiny neurons and cholinergic interneurons express differential ionotropic glutamatergic responses and vulnerability: Implications for ischemia and Huntington's disease. Ann Neurol. 1998;43:586–597. doi: 10.1002/ana.410430506. [DOI] [PubMed] [Google Scholar]
- 3.Beal MF. Huntington's disease, energy, and excitotoxicity. Neurobiol Aging. 1994;15:275–276. doi: 10.1016/0197-4580(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 4.Cepeda C, et al. NMDA receptor function in mouse models of Huntington disease. J Neurosci Res. 2001;66:525–539. doi: 10.1002/jnr.1244. [DOI] [PubMed] [Google Scholar]
- 5.Zeron MM, et al. Increased sensitivity to N-methyl-d-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 6.Maragakis NJ, Rothstein JD. Glutamate transporters in neurologic disease. Arch Neurol. 2001;58:365–370. doi: 10.1001/archneur.58.3.365. [DOI] [PubMed] [Google Scholar]
- 7.Lobsiger CS, Cleveland DW. Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat Neurosci. 2007;10:1355–1360. doi: 10.1038/nn1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebb MO, Denovan-Wright EM, Robertson HA. Expression of the Huntington's disease gene is regulated in astrocytes in the arcuate nucleus of the hypothalamus of postpartum rats. FASEB J. 1999;13:1099–1106. doi: 10.1096/fasebj.13.9.1099. [DOI] [PubMed] [Google Scholar]
- 10.Shin JY, et al. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171:1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lievens JC, Rival T, Iche M, Chneiweiss H, Birman S. Expanded polyglutamine peptides disrupt EGF receptor signaling and glutamate transporter expression in Drosophila. Hum Mol Genet. 2005;14:713–724. doi: 10.1093/hmg/ddi067. [DOI] [PubMed] [Google Scholar]
- 12.Custer SK, et al. Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nat Neurosci. 2006;9:1302–1311. doi: 10.1038/nn1750. Epub 2006 Aug 1327. [DOI] [PubMed] [Google Scholar]
- 13.Wang CE, et al. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington's disease. Hum Mol Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tydlacka S, Wang CE, Wang X, Li S, Li XJ. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J Neurosci. 2008;28:13285–13295. doi: 10.1523/JNEUROSCI.4393-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner M, Kisseberth WC, Su Y, Besnard F, Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. J Neurosci. 1994;14:1030–1037. doi: 10.1523/JNEUROSCI.14-03-01030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner M, Messing A. GFAP Transgenic Mice. Methods. 1996;10:351–364. doi: 10.1006/meth.1996.0113. [DOI] [PubMed] [Google Scholar]
- 17.Hackam AS, et al. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka K, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schilling G, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 20.Casper KB, McCarthy KD. GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol Cell Neurosci. 2006;31:676–684. doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 22.Lievens JC, et al. Impaired glutamate uptake in the R6 Huntington's disease transgenic mice. Neurobiol Dis. 2001;8:807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- 23.Su ZZ, et al. Insights into glutamate transport regulation in human astrocytes: Cloning of the promoter for excitatory amino acid transporter 2 (EAAT2) Proc Natl Acad Sci USA. 2003;100:1955–1960. doi: 10.1073/pnas.0136555100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu ZX, et al. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornett J, et al. Context-dependent dysregulation of transcription by mutant huntingtin. J Biol Chem. 2006;281:36198–36204. doi: 10.1074/jbc.M607839200. [DOI] [PubMed] [Google Scholar]
- 26.Chou SY, et al. Expanded-polyglutamine huntingtin protein suppresses the secretion and production of a chemokine (CCL5/RANTES) by astrocytes. J Neurosci. 2008;28:3277–3290. doi: 10.1523/JNEUROSCI.0116-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singhrao SK, et al. Huntingtin protein colocalizes with lesions of neurodegenerative diseases: An investigation in Huntington's, Alzheimer's, and Pick's diseases. Exp Neurol. 1998;150:213–222. doi: 10.1006/exnr.1998.6778. [DOI] [PubMed] [Google Scholar]
- 28.Gu X, et al. Pathological cell–cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 29.Maragakis NJ, Rothstein JD. Mechanisms of Disease: Astrocytes in neurodegenerative disease. Nat Clin Pract Neurol. 2006;2:679–689. doi: 10.1038/ncpneuro0355. [DOI] [PubMed] [Google Scholar]
- 30.Choi SH, et al. Non-cell-autonomous effects of presenilin 1 variants on enrichment-mediated hippocampal progenitor cell proliferation and differentiation. Neuron. 2008;59:568–580. doi: 10.1016/j.neuron.2008.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Li XJ. Multiple pathways contribute to the pathogenesis of Huntington disease. Mol Neurodegener. 2006;1:19. doi: 10.1186/1750-1326-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behrens PF, Franz P, Woodman B, Lindenberg KS, Landwehrmeyer GB. Impaired glutamate transport and glutamate-glutamine cycling: Downstream effects of the Huntington mutation. Brain. 2002;125:1908–1922. doi: 10.1093/brain/awf180. [DOI] [PubMed] [Google Scholar]
- 33.Arzberger T, Krampfl K, Leimgruber S, Weindl A. Changes of NMDA receptor subunit (NR1, NR2B) and glutamate transporter (GLT1) mRNA expression in Huntington's disease—An in situ hybridization study. J Neuropathol Exp Neurol. 1997;56:440–454. doi: 10.1097/00005072-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 34.Hassel B, Tessler S, Faull RL, Emson PC. Glutamate uptake is reduced in prefrontal cortex in Huntington's disease. Neurochem Res. 2008;33:232–237. doi: 10.1007/s11064-007-9463-1. [DOI] [PubMed] [Google Scholar]
- 35.Miller BR, et al. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington's disease phenotype in the R6/2 mouse. Neuroscience. 2008;153:329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stack EC, et al. Neuroprotective effects of synaptic modulation in Huntington's disease R6/2 mice. J Neurosci. 2007;27:12908–12915. doi: 10.1523/JNEUROSCI.4318-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.