Abstract
The cyanobacterium Synechococcus elongatus PCC 7942 exhibits oscillations in mRNA transcript abundance with 24-h periodicity under continuous light conditions. The mechanism underlying these oscillations remains elusive—neither cis nor trans-factors controlling circadian gene expression phase have been identified. Here, we show that the topological status of the chromosome is highly correlated with circadian gene expression state. We also demonstrate that DNA sequence characteristics of genes that appear monotonically activated and monotonically repressed by chromosomal relaxation during the circadian cycle are similar to those of supercoiling-responsive genes in Escherichia coli. Furthermore, perturbation of superhelical status within the physiological range elicits global changes in gene expression similar to those that occur during the normal circadian cycle.
Keywords: circadian clock, gene expression, supercoiling, cyanobacteria
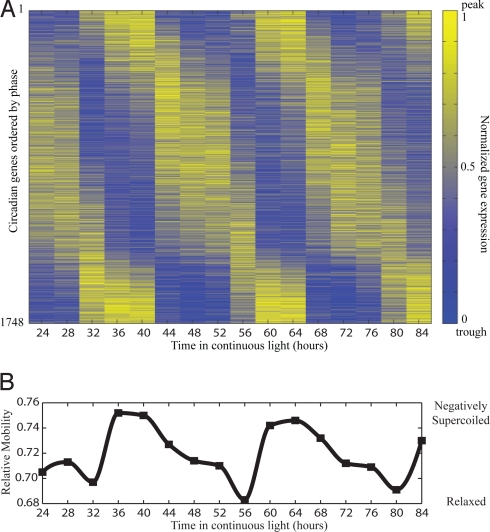
Circadian rhythms in gene expression have been identified in many organisms. In general, 5–15% of an organism's transcriptome oscillates with 24-h periodicity in the absence of external cues such as light to dark or dark to light transitions (1). These transcriptional rhythms are controlled by an endogenous biological clock and allow organisms to schedule processes at appropriate times during the day and night cycle. The cyanobacterium Synechococcus elongatus PCC 7942 (hereafter, S. elongatus) is particularly striking because the majority of its gene expression is under circadian control in continuous light conditions. A “promoter trap” analysis using a bacterial luciferase reporter integrated at approximately 30,000 random loci showed circadian oscillations in bioluminescence at all 800 locations where bioluminescence signal was detected (2). A recent measurement of mRNA levels by microarray analysis demonstrated that at least 30% of transcript levels oscillated in circadian fashion (3). The discrepancy between promoter trap and microarray analysis is not surprising and is at least partially attributable to a combination of: (i) the limited time-resolution of microarrays and; (ii) the time-averaging of transcript levels observed in the bioluminescence output of promoter trap studies due to finite luciferase protein lifetime. The actual percentage of circadian transcripts in S. elongatus is likely between 30 and 100%; here we observe that 64% of transcripts oscillate with circadian periodicity (Fig. 1A).
Fig. 1.
Circadian gene expression and topology in S. elongatus. (A) Heat map of circadian gene expression (1748/2724 = 64% of total predicted ORFs) ordered by phase. Each gene is normalized between 0 and 1 such that yellow (1) and blue (0) indicate a peak and trough of expression, respectively. Circadian genes were identified on the chromosome and both endogenous plasmids. (B) Oscillations in the superhelicity of an endogenous plasmid (pANS) from same time-course as expression analysis. Each superhelical state corresponds to a unique state of gene expression suggesting that oscillations in supercoiling can drive oscillations in gene expression.
In S. elongatus, circadian oscillations in transcriptional activity require three genes, kaiA, kaiB, and kaiC, whose products comprise the core circadian oscillator. The proteins encoded by the kai genes interact with one another to generate circadian rhythms in KaiC phosphorylation at serine and threonine residues (4). Amazingly, an in vitro mixture of the Kai proteins and ATP reproduces in vivo oscillations in the phosphorylation state of KaiC (5). Inactivation of any kai gene abolishes circadian oscillations in both transcription and phosphorylation, and when kaiC is mutated such that phosphorylation oscillations occur with periods other than 24 h, the period of transcriptional oscillation is similarly affected (3, 4, 6). Although the link between KaiC phosphorylation and transcription is strong, it is still conditional—when S. elongatus is exposed to constant darkness, oscillations in KaiC phosphorylation persist for several cycles, but transcriptional activity is greatly reduced and no longer appears circadian (3, 7).
Although a role for the Kai proteins in circadian gene expression is evident, the mechanism of promoter control is unclear. Circadian gene expression in S. elongatus is primarily divided into two phases—one subset of transcripts peaks at subjective dawn and the other subset at subjective dusk (2, 3). Systematic analysis of single promoters has yet to identify specific cis-elements controlling expression phase (8, 9), and neither random mutagenesis nor transposon insertion screens have identified trans-factors responsible for expression phase (10). Interestingly, several heterologous noncircadian promoters from Escherichia coli can drive circadian expression when integrated in S. elongatus, suggesting that a specific cis-sequence is not necessary for circadian promoter activity (11). The elusiveness of cis and trans factors has prompted the “oscilloid model” for circadian control of gene expression (12). In this model, circadian rhythms in the topology of the S. elongatus chromosome are thought to impart circadian gene expression patterns by modulating the affinity of the transcription machinery for promoters during the circadian cycle. After this model was proposed, two studies demonstrated circadian oscillations in the compaction and superhelical status of the S. elongatus chromosome and an endogenous plasmid, respectively (13, 14). Although circadian oscillations in the topology of the chromosome are evident, it is unclear whether these oscillations cause circadian changes in gene expression. To test whether changes in chromosome topology are responsible for generating circadian gene expression, we concurrently measured gene expression and endogenous plasmid topology. We demonstrate that each topological state corresponds to a distinct state in gene expression. We also observe that DNA sequence features of promoters and coding regions of S. elongatus genes that appear monotonically relaxation repressed or monotonically relaxation activated during the circadian cycle are similar to features found in relaxation sensitive genes in E. coli. Furthermore, we show that perturbation of topological status results in rapid and predictable changes in global transcript levels.
Results
Measurement and Analysis of Circadian Gene Expression in S. elongatus.
We analyzed changes in transcript abundance in S. elongatus using whole-genome microarrays. Cells were entrained by two consecutive alternating 12-h light–dark cycles and subsequently released into continuous light (T = 0 h). Samples were collected every 4 h for 60 h from T = 24 to T = 84 h (Fig. S1A). Our analysis revealed that 1,748 of 2,724 predicted ORFs, equivalent to 64%, oscillated with 22- to 26-h period (Fig. 1A, GSE18902, and Dataset S1). Hereafter, we refer to these ORFs as circadian genes.
We observed that the majority of circadian genes either peaked in the subjective dawn or subjective dusk, with approximately 30% more genes peaking in the subjective dawn (Figs. S1B and S2) (3). Circadian genes existed with amplitudes as high as a 32-fold change in expression, but approximately 90% of oscillating genes had amplitudes <2-fold (Fig. S1C). Surprisingly, a few genes exhibited oscillations with periods <24-h. For example, rpaA—encoding a two component transcriptional regulator—has transcript levels that oscillate with a 12-h period. Genes with a 12-h period may be inherently 24-h period genes whose expression is controlled (activated or repressed) by another 24-h period gene.
To investigate whether chromosomal location influences gene expression we explored the location dependence of gene expression profiles. We found that genes located on the same operon had almost identical gene expression profiles. For example, the Pearson correlation of the temporal profiles of kaiB and kaiC, two genes known to exist on the same operon, is 0.99 (6). In the extreme case of the ribosomal protein cluster, a contiguous 8.5-kb segment featuring 18 genes showed remarkable coregulation with mean Pearson correlation between neighbors of 0.89 (3). Despite the highly correlated expression profiles of neighboring genes, the overall transcription architecture of the chromosome appears random (Fig. S3A) (3). In fact, when the spatial organization of expression profiles is conditioned on putative operon structure, the correlation between neighboring operons is close to random (Fig. S3 B and C). That is, genes on the same mRNA transcript are highly correlated, but neighboring transcripts show close to no correlation. Similarly, the amplitude distribution along the chromosome appears random after conditioning on putative operon structure (Fig. S4). In addition to an approximate 2.7 Mb chromosome, S. elongatus contains two endogenous plasmids, pANL and pANS. We see several circadian genes on both endogenous plasmids. The existence of circadian genes on both the chromosome and plasmids suggests that circadian gene expression is regulated by a global mechanism.
To explore the function of circadian gene expression, we investigated the distribution of subjective dawn and subjective dusk genes in various cellular and metabolic pathways (3). Interestingly, we found that 41 of 46 circadian genes involved in photosynthesis were most highly expressed in the subjective dawn. Since protein levels have been shown to lag transcript abundance by 4–6 h (15, 16), these photosynthesis proteins are likely up-regulated during the subjective day and down-regulated during the subjective night. Several other metabolic and cellular pathways show significant enrichment for subjective dawn or subjective dusk genes (Dataset S1 and Fig. S5). In total, 70% of genes annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (17) exhibit circadian periodicity in their expression. Although it is clear that circadian expression has major metabolic and cellular consequences in continuous light, it is unclear whether and how this control is used in more physiologically relevant conditions where light and dark periods alternate.
Correlation Between Topological State and Gene Expression.
The oscilloid model suggests that circadian change in the topology of the chromosome drives circadian gene expression. If this model is true, we expect each topological state to map to a single gene expression state. As a proxy for the topological status of the S. elongatus chromosome, we quantified the superhelical status of the endogenous pANS plasmid (Fig. 1B) sampled from the same time-course as the expression analysis (Fig. S6) (14). A comparison of the topological analysis with the microarray data suggests that each topological state corresponds to a unique state of gene expression (Fig. 1 A and B). That is, circadian times with similar superhelical states generally have similar gene expression states. Interestingly, a dramatic increase in superhelicity occurs 8 h into the circadian cycle (CT 8), allowing each superhelical state to be sampled only once during the circadian cycle. If each gene requires a particular level of supercoiling for maximal transcription, then this increase in superhelicity could explain the predominance of sinusoidal 24-h period oscillations over oscillations with significant components at smaller periods.
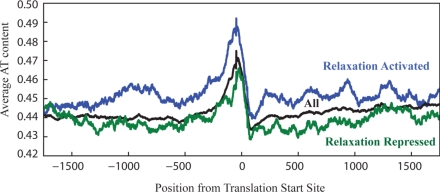
Studies of global expression changes due to chromosomal relaxation in E. coli have identified general sequence characteristics of relaxation activated and relaxation repressed genes (18, 19). To investigate if supercoiling plays a role in circadian gene expression we analyzed the sequence content of genes that appear activated and repressed by relaxation in S. elongatus. To identify genes whose expression was highly correlated (relaxation repressed) or anticorrelated (relaxation activated) with relaxation, we computed the Pearson correlation coefficient between each temporal expression profile and the supercoiling waveform (Dataset S1). The 250 most strongly correlated genes were designated monotonically relaxation repressed and the 250 most strongly anticorrelated genes were designated monotonically relaxation activated. By selecting the most correlated and most anticorrelated genes, we eliminate the genes whose relationship with supercoiling is nonlinear, including the genes for which maximal expression occurs at an intermediate physiological supercoiling state. Analysis of global expression changes due to relaxation in E. coli demonstrated that genes monotonically repressed in response to relaxation have a lower than expected AT content in both the coding region and promoter, whereas genes monotonically activated by relaxation have a higher than expected AT content (18, 19). Similarly, we see a significant difference in the AT content of genes that appear monotonically repressed versus monotonically activated by relaxation (Fig. 2A). This difference in AT content is highly significant in both the promoter region and coding region (all P < 0.05) (Fig. S7).
Fig. 2.
Sequence characteristics of genes suggest supercoiling mediated control. Genes that are monotonically relaxation activated (blue) and monotonically relaxation repressed (green) have increased and decreased AT content (in promoter and coding region), respectively, compared to all of the genes in the genome. Genes were aligned by translational start site and the mean AT content along −2,000–2,000 was calculated with a smoothing window of 100 nucleotides.
Perturbation of Topology Results in Predictable Expression Changes.
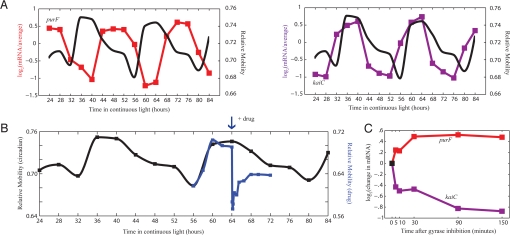
If superhelical state does in fact regulate circadian gene expression, then artificial manipulation of supercoiling should result in a predictable change in gene expression. In particular, if we are able to rapidly change the superhelical status of the chromosome from its most supercoiled to most relaxed state, then the instantaneous change in gene expression should be very similar to the change in gene expression caused by normal circadian relaxation. That is, the expression of genes that are expressed most strongly during supercoiled circadian times (Fig. 3A, purple) should immediately decrease, whereas the expression of genes that are expressed most strongly during relaxed circadian times (Fig. 3A, red) should immediately increase.
Fig. 3.
Manipulation of supercoiling via gyrase inhibition results in expected changes in gene expression. (A) Canonical subjective dawn (purF, red) and subjective dusk (kaiC, purple) genes oscillate with opposite phase in gene expression. Superimposed is the circadian oscillation in superhelicity of the endogenous pANS plasmid (black). Relaxed supercoiling states have lower relative mobility than negatively supercoiled states. KaiC expression is decreased during circadian relaxation while purF expression is increased. (B) Quantification of pANS plasmid supercoiling before and after addition of novobiocin at T = 64 h (blue) superimposed on supercoiling changes during the circadian cycle (black). Supercoiling was measured 5, 10, 15, 30, 60, 90, 150, 240, 330, and 480 min after novobiocin addition. Novobiocin addition immediately relaxes the pANS plasmid to a level similar to the most relaxed state during the circadian cycle. (C) Gyrase inhibition at T = 64 h in the circadian cycle results in an immediate change in gene expression due to drug-induced relaxation. Genes that have higher expression during relaxed circadian times immediately increase in gene expression (purF, red) whereas genes that have lower expression during relaxed circadian times immediately decrease in gene expression (kaiC, purple).
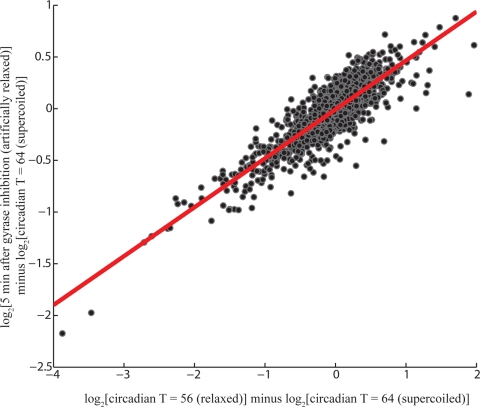
To test this hypothesis, we treated cells with the gyrase inhibitor novobiocin (0.1 μg/mL novobiocin sodium salt) during the most supercoiled state in the circadian cycle (CT 16, T = 64 h). At this concentration of novobiocin, the growth rate is not significantly affected and supercoiling resumes the circadian course after a brief excursion. Within 5 min of novobiocin treatment, the pANS plasmid reached a supercoiling state similar to the most relaxed state observed during the normal circadian cycle (Fig. 3B and Fig. S8 A and B, GSE18902). As expected, expression from the well characterized supercoiling sensitive promoters in E. coli—gyrA, gyrB, and topI—exhibited a homeostasis-generating response upon relaxation (20) (Fig. S8C). That is, since gyrase induces negative supercoils and topoisomerase I induces positive supercoils (relaxation), the expression of gyrase increases and the expression of topoisomerase I decreases in response to relaxation to regain the original superhelical state. Amazingly, genes whose expression is decreased or increased with circadian relaxation took opposite and predictable trajectories after novobiocin-induced relaxation (Fig. 3C). In fact, there is high correlation (Pearson correlation coefficient 0.85) between the change in gene expression after only 5 min of drug-induced relaxation and the change in gene expression due to relaxation in the circadian cycle (Fig. 4). Of the 1,748 circadian genes, 1,380 (79%) moved in the expected direction 5 min after novobiocin treatment. Of the genes whose expression differed by >2-fold between relaxed and supercoiled circadian time-points, none moved in the direction contrary to that expected. These results demonstrate that direct modulation of supercoiling within the physiological regime elicits the expected changes in expression, suggesting that oscillations in superhelicity may be sufficient to impart circadian gene expression.
Fig. 4.
Genome-wide changes in expression due to gyrase inhibition induced relaxation. Scatter plot of the change in gene expression due to circadian relaxation versus drug-induced relaxation for circadian genes. The normal relaxation induced change in gene expression (x axis) is highly correlated with the rate of gene expression due to drug-induced relaxation (y axis) suggesting that supercoiling plays a primary role in dictating circadian gene expression. The linear best fit line is shown in red with Pearson correlation coefficient 0.85.
Discussion
By measuring genome-wide gene expression and superhelicity of an endogenous plasmid, we demonstrate that each superhelical state corresponds to a unique state of expression. In addition, we show that genes monotonically repressed and monotonically activated by relaxation are depleted and enriched for AT content, respectively, similar to observations made for relaxation sensitive genes in E. coli (18, 19). Further, we show that the instantaneous change in gene expression after drug-induced relaxation closely correlates with the change resulting from relaxation during the normal circadian cycle.
But what is controlling the rhythmicity in supercoiling? Since we know that circadian oscillations in gene expression and supercoiling are dependent on KaiC (3, 6, 13, 14), the primary hypothesis is that KaiC either directly or indirectly controls supercoiling. KaiC sequence analysis suggests that it belongs in the RecA/DnaB superfamily (21) and KaiC has been shown to have DNA-binding activity in vitro (22), allowing for the possibility that KaiC may be able to directly change the superhelicity of DNA (14). An alternative model is that KaiC controls the ratio of negative versus positive supercoiling generating activity (14). For example, KaiC or an interacting protein may transcriptionally or post-transcriptionally control an enzyme that modulates supercoiling or affects the [ATP]/[ADP] ratio. In E. coli, changes in supercoiling are often accompanied by changes in the [ATP]/[ADP] ratio (23–25), and in vitro, the [ATP]/[ADP] ratio has been shown to modulate the activity of gyrase (26).
Supercoiling has been shown to play an important role in transcription regulation in E. coli (27). Superhelicity imposes torsional stress on DNA, which affects transcription by modulating the stability of interactions between RNA polymerase, nucleoid-like proteins, and transcription factors with the promoter or coding region of genes (27). Changes in the level of supercoiling have been observed during a variety of stress responses—temperature, peroxide, and osmotic—as well as growth conditions (23–25, 28), and changes in supercoiling have been shown to elicit a global transcriptional response (29). A role for supercoiling in mediating changes in expression is not limited to E. coli and has been observed in a variety of bacteria including Salmonella typhimurium, Vibrio cholera, Bacillus subtilis, and Synechocystis PCC 6803 (30–33).
Supercoiling mediated control of circadian gene expression poses several advantages over other modes of regulation: (i) supercoiling can globally affect all promoters, enabling genome-wide oscillations in transcription; (ii) promoters can have differential sensitivity to supercoiling, allowing different phases of circadian oscillation; and (iii) supercoiling is already under feedback control in bacteria, so evolution of an oscillator requires only a circadian perturbation of supercoiling levels since the return to homeostasis is already preprogrammed.
In S. elongatus, the extent of supercoiling mediated circadian gene expression, natural variation in superhelical status, and robust outputs—amplitude and phase—of supercoiling sensitivity make it an ideal system for understanding how supercoiling affects transcription. Although several mechanisms have been proposed for how supercoiling affects the transcription of individual promoters in E. coli, genome-wide analysis in S. elongatus may reveal the common mechanistic and sequence motifs of supercoiling mediated transcriptional control. By studying how sequence determines phase and how the association of transcription machinery with promoters and genes is modulated during the circadian cycle, we may be able to gain a better understanding of how supercoiling can affect transcription.
Materials and Methods
Continuous Culture of Cyanobacteria.
A continuous culture apparatus was developed to keep cells in constant conditions and provide real-time bioluminescence readings. S. elongatus (stain AMC 408 (8): psbAI::luxCDE fusion in NS1 (34) (spectinomycin) and purF::luxAB fusion in NSII (34) (chloramphenicol)) was grown in a 6-L cylindrical spinner flask (Corning) at a volume of 4.5 L. Cells were grown in modified BG-11 medium (35) with the following modifications: 0.0010 g/L FeNH4 citrate was used instead of 0.0012 g/L FeNH4 citrate and citric acid was supplemented at 0.00066 g/L. Cells were initially inoculated in the presence of antibiotics (5 μg/mL spectinomycin and 5 μg/mL chloramphenicol), and subsequently diluted with modified BG-11 only. Cells were exposed to surface flux of approximately 25 μmol photons m−2 s−1 white light, bubbled with 500 mL/min 1% CO2 in air, maintained at 30 °C, and stirred at 1 rotation per second. Constant optical density (OD750 0.15) and volume are achieved via a two state controller. OD does not fluctuate >8% during an experiment.
Cells are exposed to two light–dark cycles for entrainment before release into continuous light, collected at designated times by vacuum filtration, snap frozen in liquid nitrogen, and stored in −80 °C. Duplicate 120-mL cultures were harvested at each time-point—one for the supercoiling assay and one for the expression analysis.
Expression Microarrays.
RNA was extracted from frozen cells in two steps. First, cells were lysed in 65 °C phenol/SDS (sodium dodecyl sulfate) by vortexing and the total RNA was purified by phenol/chloroform extraction. Second, total RNA was subjected to DNase I (Promega) treatment followed by a second phenol/chloroform extraction. Total RNA was analyzed on agarose gel and Agilent Bioanalyzer for integrity.
Expression was measured using custom designed two-color 8 × 15 k microarrays (Agilent, Array ID 020846). Microarrays were designed using genome and plasmid sequences from GenBank CP000100, CP000101, and S89470. Four separate melting temperature matched probes (80 °C) for each predicted ORF were designed using eArray (Agilent). All probes <60 nucleotides were appended with the Agilent standard linker to reach 60 nucleotides. Probes against luxA through luxE and Arabidopsis spike-in controls (Ambion) were also included on the microarray.
cDNA was prepared for each individual time-point (foreground channel) as well as for a pool of all time-points (background channel). Spike-in RNA was introduced at different concentrations and ratios to the foreground and background channels before reverse transcription to ensure proper ratio detection in a wide dynamic range. Five micrograms total RNA (plus spike-ins) was reverse-transcribed with random 15-mer primers (Operon) and a 2:3 ratio of amino allyl-UTP:dTTP (Sigma) using SuperScript III reverse-transcriptase (Invitrogen) without amplification. RNA was hydrolyzed and cDNA was purified using Microcon 30 spin column (Millipore).
cDNA was labeled with N-hydroxysuccinimide-ester cyanine 3 (Cy3, foreground) or cyanine 5 (Cy5, background) (GE Biosciences) in 0.1 M sodium bicarbonate pH 9.0 for 6 h. Labeled cDNA was purified (Microcon 30, Millipore) in preparation for hybridization. Each array was hybridized with 150–300 ng Cy3 and 150–300 ng Cy5 labeled cDNA and rotated (5 rotations min−1) at 60 °C for 17 h in SureHyb chambers (Agilent). Arrays were subsequently washed in 6.7× SSPE and 0.005% N-lauryl sarcosine buffer for at least 1 min, 0.67× SSPE and 0.005% N-lauryl sarcosine buffer for 1 min, and then Agilent drying and ozone protection wash for 30 s at room temperature (1× SSPE = 0.15 M NaCl, 10 μM sodium phosphate, 1 mM EDTA, pH 7.4). The arrays were immediately scanned using an Axon 4000B scanner at 5-μm resolution. The average intensity of the Cy3 and Cy5 florescence at each spot was extracted using the GenePix software (Molecular Devices) and loaded into MATLAB (Mathworks). Loess and quantile normalization were performed using the MATLAB bioinformatics toolbox. All subsequent analysis was performed in MATLAB. Spike-in ratios were compared from array to array to ensure proper normalization and hybridization. Constant spike-in signal also ensures that rRNA to mRNA ratio is constant during all experimental conditions.
Identification and Classification of Circadian Genes.
The 4 melting temperature matched probes per predicted ORF were normalized and averaged. A modified Cosiner method was used to identify cycling genes and their period, phase, and amplitude (36). Briefly, the expression data for each predicted ORF was linearly de-trended and the first Fourier component was calculated assuming periods between 12 and 36 with increment of 0.1. The period that minimized the Euclidian distance between the first Fourier component and the experimental signal was designated the period of the predicted ORF. If the period was between 22 and 26, the ORF was considered circadian and its phase, period, and amplitude were given by the first Fourier component. To separate subjective dawn and subjective dusk genes as well as genes that peaked at particular times during the circadian cycle, the circadian genes were subjected to K-means clustering by Euclidean distance. Clustering with K = 6, generated clusters that peaked at unique times during the circadian cycle (CT 0, 4, 8, 12, 16, and 20). Genes peaking at CT 20, 0, and 4 were designated subjective dawn genes and genes peaking at CT 8, 12, and 16 were designated subjective dusk genes (Dataset S1 and Fig. S2).
Fig. S9 compares the data from this study with Ito et al. (3). Here, we identify as circadian 86% of the genes identified as circadian in Ito et al. (Fig. S9A). In addition, for the genes which both studies identified as circadian, the phase of expression was almost identical (Fig. S9B).
Isolation of Plasmid DNA and Chloroquine Agarose Gel Electrophoresis (CAGE).
Plasmid isolation and CAGE technique were performed similarly to (14). Endogenous S. elongatus plasmid (pANS) was isolated using QIAprep miniprep kit (Qiagen). Approximately 150 ng of isolated plasmid was run on a gel containing 0.8% agarose, 0.5× TBE, and 15 μg/mL chloroquine diphosphate (1× TBE = 90 mM Tris, 64.5 mM boric acid, and 2.5 mM EDTA, pH 8.3). Plasmid topoisomers were separated by electrophoresis at 50 volts/14 cm for 24 h in the dark at room temperature with buffer exchange. Gels were incubated for 1 h in 0.5× TBE and 1× SybrGold stain (Invitrogen). Gels were imaged on Typhoon Imager (GE). Chloroquine titration and gyrase inhibition (novobiocin treatment) were used to determine the position of relaxed and supercoiled forms on gel. Southern blot analysis was used to prove that topoisomers visualized by SybrGold staining were from the pANS plasmid. Blotting was performed by upward capillary transfer (37) onto Biodyne B nylon membrane (Pierce) and visualized by horseradish peroxidase-streptavidin chemiluminesence assay (Pierce). Probe template was synthesized by amplification with PCR primers pANSorfAfor (14), 5′-GGAAGTAGAAGGCTT-3′, and pANSorfArev (14), 5′-GGCAATGGCCAGCATCG-3′. The 468 nucleotide PCR product was amplified with random heptanucleotide primers in the presence of biotin-11-dUTP using random priming kit (Pierce). Gels were analyzed using ImageJ and MATLAB software. From densitometry traces, the relative mobility was calculated by the position of the mean of the Boltzmann distribution of topoisomers relative to the position of the open circular (oc) and relaxed (rel) forms. Relative Mobility (RM) = 1 − (mean − oc)/(oc − rel).
Supplementary Material
Acknowledgments.
We thank members of the O'Shea laboratory for discussion and commentary. We thank Dr. Susan Golden for the S. elongatus strain AMC 408. This work was supported by the Howard Hughes Medical Institute, National Defense Science and Engineering (V.V.) and National Science Foundation Graduate Research Fellowships (V.V.).
Footnotes
The authors declare no conflict of interest.
Data Deposition: The microarray data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE18902).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912673106/DCSupplemental.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology—Biological Timekeeping. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 2.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 3.Ito H, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 6.Ishiura M, et al. Expression of a gene cluster KaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 7.Tomita J, Nakajima M, Kondo T, Ishiura M. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 8.Min H, Liu Y, Johnson CH, Golden SS. Phase determination of circadian gene expression in Synechococcus elongatus PCC 7942. J Biol Rhythms. 2004;19:103–112. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Tsinoremas NF, Golden SS, Kondo T, Johnson CH. Circadian expression of genes involved in the purine biosynthetic pathway of the cyanobacterium Synechococcus sp. strain PCC 7942. Mol Microbiol. 1996;20:1071–1081. doi: 10.1111/j.1365-2958.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 10.Min H, Golden SS. A new circadian class 2 gene opcA, whose product is important for reductant production at night in Synechococcus elongatus PCC 7942. J Bacteriol. 2000;182:6214–6221. doi: 10.1128/jb.182.21.6214-6221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu Rev Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 12.Mori T, Johnson CH. Circadian programming in cyanobacteria. Semin Cell Dev Biol. 2001;12:271–278. doi: 10.1006/scdb.2001.0254. [DOI] [PubMed] [Google Scholar]
- 13.Smith SM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo T, et al. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Golden SS, Kondo T, Ishiura M, Johnson CH. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995;177:2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–44. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter BJ, et al. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong KS, Xie Y, Hiasa H, Khodursky AB. Analysis of pleiotropic transcriptional profiles: A case study of DNA gyrase inhibition. PLoS Genet. 2006;2:1464–1476. doi: 10.1371/journal.pgen.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: Homeostatic control of DNA supercoiling. Cell. 1983;35:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 21.Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 22.Mori T, et al. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA. 2002;99:17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hseih LS, Rouviere-Yaniv J, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: Changes associated with salt shock. J Bacteriol. 1991;12:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hseih LS, Burger RM, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J Mol Bio. 1991;219:443–450. doi: 10.1016/0022-2836(91)90185-9. [DOI] [PubMed] [Google Scholar]
- 25.Camacho-Carranza R, et al. Topoisomerase activity during the heat shock response in Escherichia coli K-12. J Bacteriol. 1995;177:3619–3622. doi: 10.1128/jb.177.12.3619-3622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westerhoff HV, O'Dea MH, Maxwell A, Gellert M. DNA supercoiling by DNA gyrase. A static head analysis. Cell Biophys. 1988;12:157–181. doi: 10.1007/BF02918357. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Martin J, Rojo F, Lorenzo V. Promoters responsive to DNA bending: A common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rui S, Tse-Dinh YC. Topoisomerase function during bacterial responses to environmental challenge. Front Biosci. 2003;8:256–263. doi: 10.2741/984. [DOI] [PubMed] [Google Scholar]
- 29.Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003;13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins CF, et al. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 31.Parsot C, Mekalanos JJ. Structural analysis of the acfA and acfD genes of Vibrio cholerae: Effects of DNA topology and transcriptional activators on expression. J Bacteriol. 1992;174:5211–5218. doi: 10.1128/jb.174.16.5211-5218.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alice AF, Sanchez-Rivas C. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr Microbiol. 1997;35:309–315. doi: 10.1007/s002849900260. [DOI] [PubMed] [Google Scholar]
- 33.Prakash JS, et al. DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium Synechocystis. Mol BioSyst. 2009 Sep 10; doi: 10.1039/B903022k. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Andersson CR, et al. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 35.Bustos SA, Golden SS. Expression of the psbDII fene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kucho K, et al. Global analysis of circadian gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2005;187:2190–2199. doi: 10.1128/JB.187.6.2190-2199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Russell DW. Molecular Cloning—A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.