Abstract
Auxin regulates most aspects of plant growth and development. The hormone is perceived by the TIR1/AFB family of F-box proteins acting in concert with the Aux/IAA transcriptional repressors. Arabidopsis plants that lack members of the TIR1/AFB family are auxin resistant and display a variety of growth defects. However, little is known about the functional differences between individual members of the family. Phylogenetic studies reveal that the TIR1/AFB proteins are conserved across land plant lineages and fall into four clades. Three of these subgroups emerged before separation of angiosperms and gymnosperms whereas the last emerged before the monocot-eudicot split. This evolutionary history suggests that the members of each clade have distinct functions. To explore this possibility in Arabidopsis, we have analyzed a range of mutant genotypes, generated promoter swap transgenic lines, and performed in vitro binding assays between individual TIR1/AFB and Aux/IAA proteins. Our results indicate that the TIR1/AFB proteins have distinct biochemical activities and that TIR1 and AFB2 are the dominant auxin receptors in the seedling root. Further, we demonstrate that TIR1, AFB2, and AFB3, but not AFB1 exhibit significant posttranscriptional regulation. The microRNA miR393 is expressed in a pattern complementary to that of the auxin receptors and appears to regulate TIR1/AFB expression. However our data suggest that this regulation is complex. Our results suggest that differences between members of the auxin receptor family may contribute to the complexity of auxin response.
Keywords: plant hormone, plant development
The plant hormone auxin influences virtually every developmental program in plants (1, 2). Auxin acts by regulating transcription through the action of at least three protein families called the TIR1/AFB F-box proteins (3, 4), the Aux/IAA transcriptional repressors (5, 6), and the ARF transcription factors (7, 8). The TIR1 F-box protein acts as an auxin receptor and directly links auxin perception to degradation of the Aux/IAA proteins (9, 10). Structural studies recently revealed that auxin acts as a “molecular glue” to increase the strength of the interaction between TIR1 and the Aux/IAA protein, thus promoting their ubiquitination and degradation (11–14).
In Arabidopsis there are 29 Aux/IAA genes (5, 6). The only known function of Aux/IAA proteins is to repress transcription of the auxin-regulated genes. According to the current model, Aux/IAAs recruit the corepressor TPL to promoters through interactions with both TPL and ARF proteins (15–18). The presence of auxin, and the subsequent degradation of the Aux/IAA proteins, releases this repression and permits ARF-dependent transcription (19).
TIR1 is a member of a small gene family that contains 5 additional AFB proteins (3). Previous studies have shown that TIR1, AFB1, AFB2, and AFB3 all function as auxin receptors and that the respective genes are broadly transcribed throughout the plant (3). Genetic experiments indicate that reducing the number of functional TIR1/AFB proteins in the plant results in increasing resistance to exogenous auxin. Further, the analysis of triple and quadruple tir1/afb mutants demonstrated that these genes have overlapping functions and collectively are essential for Arabidopsis growth and development (3). However many questions remain concerning the specific function of each gene. In this study, we show that individual members of the family make an unequal contribution to the auxin response and that they exhibit important differences in expression, biochemical activity, and function.
Results
Evolutionary History of the TIR1/AFB Protein Family.
To learn more about the evolutionary history of the TIR1/AFB proteins, we performed a phylogenetic analysis using the available sequences from land plants. Our results show that the proteins are conserved across land plants and have radiated during vascular plant evolution (Fig. S1). The seed plant TIR1/AFB proteins are more closely related to each other than to those from the nonvascular plant Physcomitrella patens, indicating that the genome of an ancestor to vascular plants likely encoded a single TIR1/AFB protein. Two duplication events established three distinct lineages—“TIR1/AFB2,” “AFB4,” and “AFB6”—before the angiosperm and gymnosperm lineages diverged roughly 300 million years ago (Mya). The duplication event distinguishing the TIR1 and AFB2 clades occurred early within the angiosperm lineage before the split between monocot plants and eudicot plants, possibly coinciding with a proposed whole-genome duplication event at the base of the angiosperm lineage (20). AFB6 homologs were lost independently early in the Brassicaceae and Poaceae families. Finally, the α-duplication event at the base of the Brassicaceae family ≈34 Mya (21) resulted in the three pairs of paralogs present in the Arabidopsis thaliana genome.
The TIR1 and AFB Proteins Do Not Contribute Equally to Auxin Response in the Root.
In a previous study we characterized a series of single and higher order mutants deficient in the TIR1, AFB1, AFB2, and AFB3 genes (3). However, many of the mutant combinations were mixed Ws and Col-0 backgrounds, complicating the analysis. We have now obtained Col-0 alleles for each gene. The tir1–10, afb1–3, afb1–5, afb2–3, and afb3–4 alleles carry T-DNA insertions, whereas the afb2–5 allele was recovered in a screen for enhancers of tir1–1 (Fig. S2) (22). The effects of each new mutation on RNA levels were determined by RT-PCR (Fig. S2). The results show that the tir1–10, afb1–3, afb1–5, and afb3–4 alleles do not produce full-length transcript. The tir1–10 and afb1 alleles are likely to be nulls because the T-DNA insertion is within the coding region near the 5′ end of the gene. In contrast the afb2–3 allele has a T-DNA insertion 37 bp upstream of the transcriptional start site that results in reduced levels of AFB2 transcript (Fig. S2C). The afb2–5 allele has a nucleotide substitution that results in substitution of glycine 70 with an arginine. Thus the afb2 alleles may retain some function. Similarly, because the T-DNA insertion in afb3–4 is near the end of the gene, it is possible that this gene retains some function. We generated a full series of double, triple, and quadruple mutants using these alleles and determined the effects of 2,4-D on root elongation in each genotype. Of the single mutants, only tir1–1 is clearly resistant to 2,4-D (Fig. S3 A and B). However the tir1–1 afb2 double mutant is more resistant to auxin than tir1–1 alone, clearly showing that AFB2 contributes to auxin response in the root. Similar results were obtained when tir1–1 was combined with the Ws allele afb2–1 (3). In contrast, the response of the tir1afb1, tir1afb3, and tir1afb1afb3 mutants is similar to that of tir1 seedlings, indicating that the loss of AFB1 and/or AFB3 does not affect the response to auxin when the seedling is already lacking TIR1 (Fig. S3 A and B). Further, the response of afb1afb2, afb1afb3, and afb2afb3 roots is similar to wild type, indicating that TIR1 is sufficient for normal response to 2,4-D, even in the absence of any two of the three AFB proteins (Fig. S3A). We used qRT-PCR to examine the relative expression of each TIR1/AFB gene in the root tissue of each single mutant background and did not observe any evidence for compensatory regulation (Fig. S4A).
The relative contribution of AFB1 and AFB3 to auxin response in the root is revealed by the analysis of higher order mutants. The tir1afb2afb3 line but not tir1afb1afb2 triple mutant is more resistant to 2,4-D than the tir1afb2 double mutant, implying that AFB3 contributes to auxin response in the root but AFB1 does not (Fig. S3B). However, AFB1 does appear to have a role later in plant development because tir1afb1afb2 plants are smaller in stature than tir1afb2 plants (Fig. S3H). The tir1afb2afb3 has a more severe phenotype than tir1afb1afb2. A significant proportion of these plants are embryo or seedling lethal (Fig. S5). This phenomenon was previously observed in the mixed ecotype triple mutant lines (3). Thus our data indicate that AFB3 plays a more significant role than AFB1 in auxin response.
The quadruple tir1–1afb1–3afb2–3afb3–4 mutant displays the same range of phenotypes observed in tir1–1afb2–3afb3–4 (Fig. S5). However in this case a higher proportion of the seedlings demonstrated the severe phenotype (Fig. S5). This phenotype is similar to that observed in the previously characterized tir1–1afb1–1afb2–1afb3–1 quadruple mutant (3) and demonstrates that each tir1/afb plays a role in seedling development.
TIR1, AFB2, and AFB3 Exhibit Posttranscriptional Regulation.
Previously we showed that TIR1, AFB1, AFB2, and AFB3 are broadly transcribed throughout development (3). To further explore expression of these genes, we created a series of translational GUS fusion proteins. The GUS enzyme was fused to the C-terminal region of each protein and introduced into plants under the control of the same promoters as were used to make the transcriptional fusion lines (3). To determine whether the TIR1-GUS fusion was functional, we introduced this transgene into tir1–1 plants and showed that the fusion protein restores normal auxin response to mutant seedlings (Fig. 3H). At least 10 independent transgenic lines were identified for each of pTIR1:cTIR1-GUS, pAFB1:cAFB1-GUS, pAFB2:cAFB2-GUS, and pAFB3:cAFB3:GUS. Homozygous lines with representative levels of expression were selected for further experimentation. We found that in young seedlings and in flowers containing the pTIR1:cTIR1-GUS, pAFB2:cAFB2-GUS, and pAFB3:cAFB3-GUS transgenes, GUS staining was much more restricted than in the equivalent promoter:GUS fusion lines (Fig. 1, Fig. S6). In contrast, the GUS expression pattern was similar in seedlings containing either pAFB1-GUS or pAFB1:cAFB1-GUS transgenes.
Fig. 3.
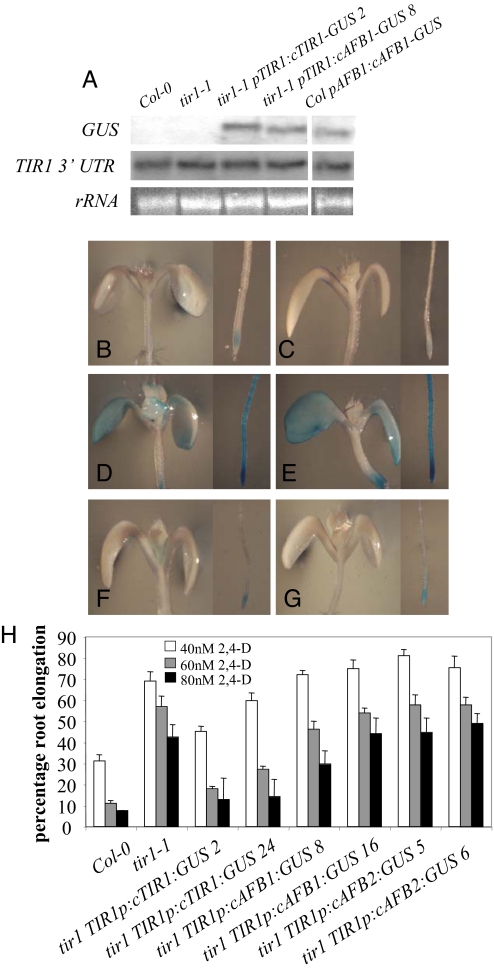
AFB1 and AFB2 cannot replace TIR1 in tir1–1 plants. (A) Northern blot of RNA isolated from 9-day-old Col-O, tir1–1, tir1 pTIR1:cTIR1:GUS, tir1 pTIR1:cAFB1:GUS, and col pAFB1:cAFB1:GUS seedlings. Expression evaluated using P32-labeled probes that bind the GUS coding region or the 3′-UTR of the TIR1 gene. rRNA levels are stained using EtBr. (B-G) GUS expression in 8-day-old seedlings containing tir1 TIR1p-cTIR1:GUS line 2 (B), tir1 TIR1p-cTIR1:GUS line 24 (C), tir1 TIR1p-cAFB1:GUS line 8 (D), tir1 TIR1p-cAFB1:GUS line 16 (E), tir1 TIR1p-cAFB2:GUS line 5 (F), and tir1 TIR1p-cAFB2:GUS line 6 (G) transgenes. (H) Root elongation as performed in Fig. 2A with 4-day-old seedling grown in the presence of 2,4-D. Numbers denote independent lines.
Fig. 1.
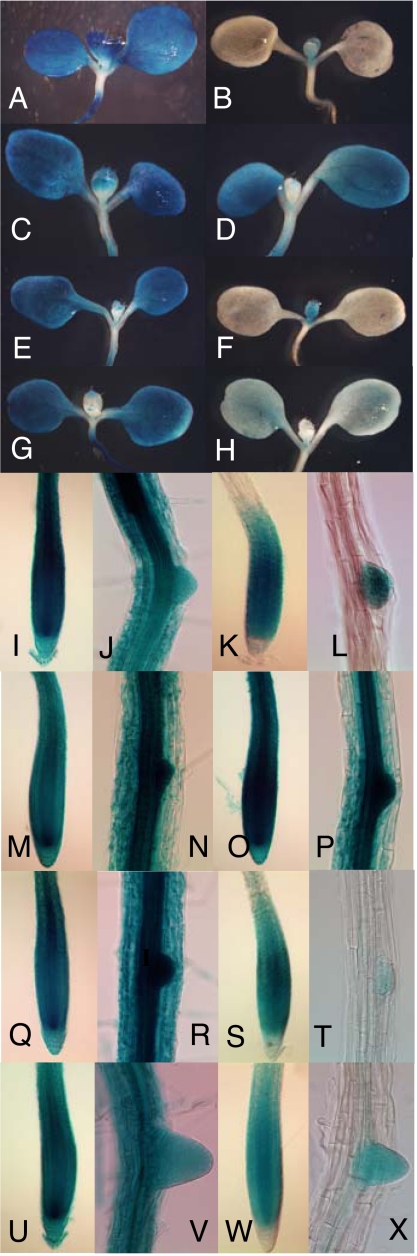
TIR1/AFB gene expression involves posttranscriptional regulation. GUS expression in the cotyledons, young leaves (A–H), primary root tip, and young lateral roots (I–X) of 8-day-old transgenic seedlings. X-Gluc staining is representative of expression levels in seedlings containing pTIR1:GUS (A, I, and J), pTIR1:cTIR1:GUS (B, K, and L), pAFB1:GUS (C, M, and N), pAFB1:cAFB1:GUS (D, O, and P), pAFB2:GUS (E, Q, and R), pAFB2:cAFB2:GUS (F, S, and T), pAFB3:GUS (G, U, and V), and pAFB3:cAFB3:GUS (H, W, and X). Samples A–H, J–L, N–P, and R–T X-Gluc were stained overnight. Samples I, M, Q, and U–X were stained for 5 h.
GUS staining in the TIR1-, AFB2-, and AFB3- GUS translational fusion lines is restricted to the primary and lateral root tips, young leaves, and the young flower buds, regions of the plant that are actively growing. However the AFB1:GUS fusion protein is found throughout the plant (Fig. 1, Fig. S6). We extracted RNA from flowers of pTIR1:GUS and pTIR1:cTIR1-GUS transgenic lines and showed by RT-PCR that RNA levels are similar in these lines even though the GUS staining is higher in pTIR1:GUS flowers (Fig. S6).
These data show that the TIR1, AFB2, and AFB3 genes undergo significant posttranscriptional regulation. Based on the translational GUS fusions, these proteins are found in regions of cell division and/or expansion consistent with their role in auxin-dependent processes. Notably, differences in the apparent distribution of TIR1 and the AFB proteins do not appear to explain the differences in their relative contribution to auxin response.
The TIR1/AFB Genes Are Not Rapidly Auxin Regulated.
Expression of many hormone-related genes is subject to negative and positive regulatory loops. For example transcription of the Aux/IAA genes, encoding negative regulators of auxin response, is rapidly induced by auxin. To determine whether auxin regulates expression of the TIR1/AFB genes in the root we performed qRT-PCR experiments using RNA isolated from auxin-treated roots. Somewhat surprisingly we did not detect any changes in TIR1/AFB RNA level even after treatment with high concentration of IAA (Fig. S4B). These data indicate that transcription of the auxin receptor genes does not change rapidly in response to auxin.
The microRNA miR393 Regulates Auxin Response.
Previous studies demonstrated that miR393 plays a role in the regulation of TIR1/AFB expression (23). This RNA is produced from two loci called miR393a and miR393b. To assess the effect of increased levels of miR393 we used the previously characterized 35S:miR393a transgenic line (23) and also generated transgenic lines containing a 35S:miR393b construct. We tested the response to 2,4-D in these transgenic seedlings by root elongation assay and showed that the roots of both 35S:miR393a and 35S:miR393b lines are auxin resistant. The level of resistance is similar to that observed in tir1–1 seedlings (Fig. 2A).
Fig. 2.
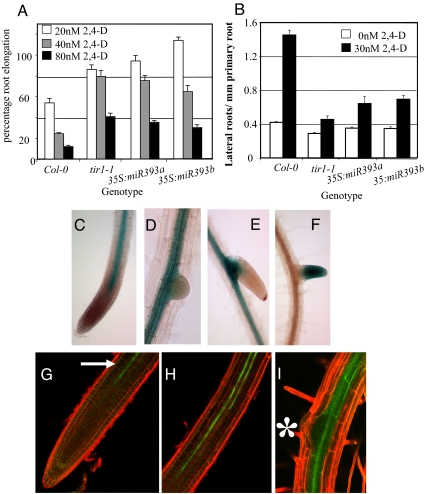
miR393 influences the auxin response in the root. (A) Four-day-old seedlings grown on MS media were transferred to media containing 2,4-D. Root elongation was measured after an additional 4 days and expressed as a proportion of growth in the absence of auxin (Scale bars, SE). (B) Lateral root assay. Four-day-old seedlings grown on MS media were transferred onto media −/+ 2,4-D for an additional 9 days. The number of emerged lateral roots/mm primary root length was measured. (Scale bars, SE.) (C–E) Expression of pmiR393:GUS in the primary root tip (C) and lateral roots (D and E) of 10-day-old seedlings. (F) Expression of pTIR1:cTIR1:GUS in an emerging lateral root in a 10-day-old seedling. (G–I) Expression of pimR393a:GFP in the primary root tip region (G, arrow), root elongation zone (H), and beneath an emerging lateral root (I, asterisk) in 10-day-old seedlings.
Growth of seedlings on auxin stimulates lateral root formation. The number of lateral roots in wild-type seedlings increases over threefold when grown on 30 nM of 2,4-D. However the roots of tir1–1 and 35S:miR393 seedlings did not respond to auxin in the same manner with only a modest increase in the number of lateral roots (Fig. 2B). These results demonstrate that overexpression of miR393 produces an auxin-resistant phenotype. However the phenotype is relatively weak when compared to that of the higher order tir1afb mutants. Further, neither overexpression line displays any of the severe growth defects exhibited by these mutants, indicating that increased levels of miR393 has only a modest effect on TIR1/AFB levels.
To investigate the expression pattern of miR393a and miR393b we constructed promoter fusions lines in which 2.5 kb upstream of each gene was used to drive expression of the GUS reporter. Expression is identical in both pmiR393a:GUS and pmiR393b:GUS seedlings. In the primary root, GUS staining is observed throughout the central stele but not in the overlying tissues (Fig. 2D). Interestingly, the stelar GUS expression domain stops ≈0.5 mm from the root tip and is restricted from the root apical meristem (Fig. 2C). Similarly, expression is also restricted from lateral roots until later stages of development when expression is once again only observed in the stele (Fig. 2E). Strikingly, the pattern of expression of both genes in the roots is complementary to that seen in pTIR1:cTIR1-GUS roots (Fig. 1 K–L, Fig. 2F). pTIR1:cTIR1-GUS is also expressed in young leaves whereas pmiR393a:GUS and pmiR393b:GUS expression is absent from this region (Fig. S7 A and B). However in older leaves there is strong expression of pmiR393a:GUS and pmiR393b:GUS but not of pTIR1:cTIR1:GUS (Fig. S7 C–E).
We also examined pmiR393a:GFP and pmiR393b:GFP transgenic lines (23). Both reporters display the same expression patterns as that observed in the pmiR393:GUS fusion lines. In the stele of the primary root, GFP signal is restricted to two strands of expression and is not found in either the root apical meristem or the emerging lateral roots (Fig. 2 G–I).
To determine whether miR393 plays a role in TIR1 regulation, we generated a pTIR1:cTIR1-GUS construct that includes four silent nucleotide changes within the miR393 recognition site predicted to make the transgene resistant to miR393-directed regulation (Fig. S8A) (24). We evaluated over 20 independent T2 lines containing the pTIR1:mTIR1-GUS transgene and found that the introduction of the miR393 target site mutations had no effect on the pattern of GUS staining (Fig. S8 B–G). Similar results were observed for pAFB2:mAFB2-GUS and pAFB3:mAFB3-GUS transgenic lines.
AFB1 contains a single nucleotide mismatch in the miR393 recognition site that is thought to cause resistance to miR393 directed mRNA cleavage (23). This might explain why pAFB1:AFB1-GUS is expressed through the entire seedling. To test this hypothesis we created a pAFB1:AFB1-GUS transgenic line that included a nucleotide change that alters the miR393 recognition site in AFB1 to be identical to that in TIR1 (Fig. S8H). These pAFB1:AFB1TIR1-GUS transgenic lines exhibited the same expression pattern as in the wild-type pAFB1:AFB1-GUS seedlings (Fig. S8 I and J).
TIR1 and AFB1 Have Distinct Functions in Vivo.
TIR1 and AFB1 are 70% identical at the amino acid level. To test whether these similar proteins have equivalent functions in planta we generated a promoter swap translational GUS fusion construct in which the AFB1 coding region, fused to GUS, was cloned downstream of the TIR1 promoter and introduced into tir1–1 plants. We also generated a similar line in which the TIR1 promoter was fused to the AFB2 coding region. We assessed the GUS staining pattern in at least 10 independent lines and continued our work with lines that were representative.
RNA blot analysis showed that the level of GUS RNA was similar in each line (Fig. 3A). However when we examined GUS staining we found that levels were much higher in tir1–1 pTIR1:cAFB1-GUS than in tir1–1 pTIR1:cTIR1-GUS seedlings (Fig. 3 B–E), indicating that the higher level of expression in the pAFB1:AFB1-GUS line is related to the AFB1 coding region and not the AFB1 promoter (Fig. 1). In tir1 pTIR1:cAFB2-GUS seedlings we observed similar GUS expression to that in tir1 pTIR1:cTIR1-GUS (Fig. 3 F and G). This is reminiscent of the level of GUS expression in pAFB2:cAFB2-GUS seedlings (Fig. 1) indicating that AFB2-GUS undergoes the same mechanisms of posttranscriptional control as TIR1-GUS.
We next evaluated whether the various constructs were able to restore wild-type 2,4-D response to tir1–1 mutant roots. The tir1–1 pTIR1:cTIR1:GUS lines displayed a wild-type level of 2,4-D response showing that the TIR1-GUS protein is able to rescue the mutant phenotype. However neither AFB1-GUS nor AFB2-GUS rescued the tir1–1 root growth phenotype even though AFB1-GUS is expressed at a high level throughout the seedling (Fig. 3H). These experiments demonstrate that AFB1 and AFB2 are unable to substitute for TIR1 protein even when regulated by the TIR1 promoter. These results indicate that TIR1, AFB1, and AFB2 do not have identical activities and may have specialized function.
TIR1 and AFB2 Exhibit a Stronger Interaction with Selected Aux/IAAs Than AFB1 and AFB3.
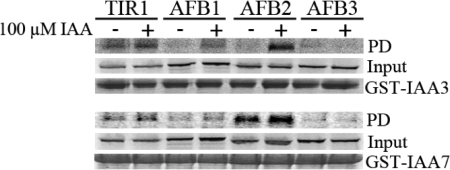
It is possible that differences in the function of TIR1/AFB family members are related to their binding to Aux/IAA substrates. In previous studies we showed that each of these proteins interact with IAA7 in an auxin-dependent manner (3). However, because some of the experiments were done using plant extracts and others with protein synthesized in TNT extracts, it was not possible to compare the strength of the interaction between different TIR1/AFB proteins and IAA7. To address this issue, we tested the ability of the Aux/IAA proteins IAA3 and IAA7 to bind each F-box protein in the absence and presence of auxin. The receptors were synthesized in TNT extracts and pulldown assays were performed with GST-tagged Aux/IAA proteins. Both TIR1 and AFB2 interact with each Aux/IAA protein in an auxin-dependent way (Fig. 4). In contrast, Aux/IAA binding to AFB1 and AFB3 was low or undetectable with this assay (Fig. 4). These results suggest that AFB1 and AFB3 may have a reduced affinity for Aux/IAA proteins compared to TIR1 and AFB2.
Fig. 4.
TIR1 and AFB2 exhibit a stronger interaction with Aux/IAA proteins than AFB1 and AFB3 in vitro. TIR1, AFB1, AFB2, and AFB3 proteins were synthesized in vitro and incubated with GST-IAA3 and GST-IAA7 proteins in the presence or absence of 100 μM of IAA. After pulldown reactions (PD), recovery of the F-box proteins was assessed by autoradiography (Top panels). Middle panel shows the input of in vitro synthethized TIR1, AFB1, AFB2, and AFB3 in the pulldown reactions, whereas Lower panels show the coomassie staining of entire pulldown reactions as loading control for GST-Aux/IAA proteins.
Discussion
The TIR1 protein has a critical role in auxin signaling. In Arabidopsis, TIR1 is a member of a small group of related proteins that includes AFB1 through AFB5 and the jasmonic acid receptor, COI1. Previous studies suggested that AFB1, AFB2, and AFB3 function together with TIR1 to regulate most aspects of auxin response throughout plant growth and development (3). However, the precise role of each member of the family is unclear. In this study we present evidence that the TIR1/AFB proteins have distinct biochemical and biological functions and that the regulation of these genes is complex.
Our phylogenetic studies indicate that the TIR1/AFB proteins are conserved among all land plants. The TIR1/AFB2, AFB4, and AFB6 lineages have been distinct since before the division of angiosperms and gymnosperms. Later, but before the separation of monocot and eudicot plants, the TIR1/AFB2 clade divided into two distinct clades (AFB1 and AFB2). The fact that these four clades have been maintained since these early events in plant evolution strongly suggests that the function of each clade is distinct. Consistent with this, our genetic studies indicate that TIR1 and AFB2 play a greater role in auxin response in the root than either AFB1 or AFB3. On the basis of the phenotype of single mutants, TIR1 appears to make the largest contribution followed by AFB2. As we reported in an earlier analysis of Ws alleles, both AFB1 and AFB3 contribute to auxin response, but this contribution is only apparent in higher order mutant combinations.
Interestingly, the phenotype of the tir1–1 afb2–2 afb3–4 and tir1–1 afb1–3 afb2–2 afb3–4 quadruple mutants is quite variable. Phenotypic variability was also observed in a previous study with a different combination of mutant alleles, indicating that this behavior is not related to the specific alleles involved (3). It seems likely that homeostatic mechanisms, perhaps related to regulation of auxin levels or response, contribute to this variability.
It is possible that the differences in relative contribution to auxin response exhibited by each TIR1/AFB protein are related to their expression level. However, an examination of transcript and protein levels indicates that this is not the case. AFB1 is expressed at a much higher level than TIR1, AFB2, or AFB3. Further, we find that AFB1 and AFB2 do not rescue the tir1 mutant even when regulated by the TIR1 promoter. These results suggest that despite their close relationship, these proteins are biochemically distinct.
There are 29 Aux/IAA proteins in Arabidopsis, 23 of which have the conserved domain II required for interaction with TIR1. It is possible that differences in TIR1/AFB function are related to the way that these protein interact with the Aux/IAAs. Consistent with this, we find that TIR1 and AFB2 exhibit a stronger interaction with several AuxIAA proteins than AFB1 or AFB3. At this point, the basis for this difference is unclear. Most of the residues believed to be important for IAA and Aux/IAA binding are conserved between TIR1 and AFB1. However, both AFB1 and AFB3 contain several nonconservative substitutions within loop 12 of these proteins, a region thought to be important for interaction with the Aux/IAA proteins (11). Further experiments will be required to determine whether these differences are responsible for changes in biochemical activity.
Regulation of TIR1/AFB Expression.
Our results demonstrate that the TIR1, AFB2, and AFB3 genes exhibit posttranscriptional regulation. The microRNA miR393 is a prime candidate for this regulation. Previous studies indicate that miR393 negatively regulates TIR1, AFB2, and AFB3 in response to pathogen attack and that overexpression of miR393 results in decreased levels of TIR1 mRNA (23). We have extended this work by showing that miR393 overexpression results in auxin-resistant root growth. Further, we show that expression of miR39a and miR393b is complementary to that of pTIR1:TIR1-GUS consistent with the hypothesis that miR393 negatively regulates TIR1 expression. On the basis of these results we were surprised to find that the introduction of mutations into the miR393 target sequence of TIR1, AFB2, and AFB3 did not affect expression of the respective fusion protein.
These results suggest that miR393 may not contribute to the developmental regulation of the TIR1/AFB genes. However, recent studies of ARGONAUTE 1 (AGO1) regulation suggest another possibility. Earlier work had shown that AGO1 is negatively regulated by miR168 (25). In addition to this regulation, recent studies show that miR168 slicing results in the generation of numerous 21-nucleotide siRNAs that map in the vicinity of the slice site (26). These siRNAs result in further silencing of the AGO1 locus. Strikingly, examination of the Arabidopsis small RNA project (ASRP) database reveals the presence of numerous 21-nucleotide siRNAs in the vicinity of the miR393 target site in TIR1, AFB2, and AFB3, but not in AFB1 (27). Thus it is possible that silencing of the pTIR1:mTIR1-GUS transgene involves production of siRNAs from the endogenous tir1 gene. Further experiments will explore this hypothesis.
The complementary expression pattern in the root of TIR1/AFB and miR393 suggests that miR393 might act in the formation of lateral roots. Interestingly the stelar expression of miR393:GFP is reminiscent of that observed in the GAL4 enhancer trap line J0121 (28). Transgenic lines that have an attenuated auxin response in this J0121 expression domain do not form lateral roots (28). Therefore in future work it will be interesting to discover whether the expression of miR393 in these cell files represents a mechanism to control lateral root formation by modulating TIR1/AFB expression.
Methods
The details of plant growth assays, generation of transgenic lines, PCR, protein expression and pulldowns, and phylogentic analysis are described in the SI Materials and Methods. Briefly, plant growth assays were performed as described previously (3). RNA for RT-PCR experiments was isolated using QIAGEN RNeasy plant kit or Tri-reagent (Sigma). Biochemical studies of the TIR1/AFB protein were performed as described (9). Phylogenetic studies were conducted using MRBAYES 3.1.2 (29) with the parameters aamodelpr = mixed, nst = 6, and rates = invgamma.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Institutes of Health (GNM43644 to M.E. and GM067203 to W.M.G.), National Science Foundation (MCB-0519970 to M.E.), Department of Energy (DE-FG02–02ER15312 to M.E.), and Marie Curie Intra-European Fellowship within the 7th European Community Framework Program (PIEF-GA-2008–220506 to B.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911967106/DCSupplemental.
References
- 1.Benjamins R, Scheres B. Auxin: The looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- 2.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot (Lond) 2005;95(5):707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 5.Overvoorde PJ, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17(12):3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135(3):1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 10.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 11.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 12.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13(10):2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenser N, Dreher KA, Edwards SR, Callis J. Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J. 2003;35(3):285–294. doi: 10.1046/j.1365-313x.2003.01801.x. [DOI] [PubMed] [Google Scholar]
- 14.Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319(5868):1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 16.Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16(2):533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13(12):2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weijers D, et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24(10):1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui L, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16(6):738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18(5):1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 23.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312(5772):436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 24.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18(10):1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallory AC, Vaucheret H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009;10(5):521–526. doi: 10.1038/embor.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backman TW, et al. Update of ASRP: The Arabidopsis Small RNA Project database. Nucleic Acids Res. 2008;36:D982–985. doi: 10.1093/nar/gkm997. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134(4):681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 29.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.