Abstract
Translational control plays an important role in cell growth and tumorigenesis. Cap-dependent translation initiation of mammalian mRNAs with structured 5′UTRs requires the DExH-box protein, DHX29, in vitro. Here we show that DHX29 is important for translation in vivo. Down-regulation of DHX29 leads to impaired translation, resulting in disassembly of polysomes and accumulation of mRNA-free 80S monomers. DHX29 depletion also impedes cancer cell growth in culture and in xenografts. Thus, DHX29 is a bona fide translation initiation factor that potentially can be exploited as a target to inhibit cancer cell growth.
Initiation is a tightly regulated rate-limiting step in the translation of eukaryotic mRNAs. Ribosome recruitment to the mRNA commences with binding of translation initiation factor 4F (eIF4F) to the 7-methyl guanosine cap structure, which is present at the 5′ end of all nuclear-encoded eukaryotic mRNAs (1). eIF4F (comprising the cap-binding protein eIF4E, the DEAD-box RNA helicase eIF4A and eIF4G, a scaffold for binding eIF4E and eIF4A) binds to the cap, unwinds (with the aid of eIF4A) the cap-proximal region of the mRNA, and, through interaction with the ribosome-bound eIF3, recruits the 40S ribosomal subunit to the mRNA (2–4). The 40S subunit then scans the 5′ UTR in a 5′ to 3′ direction until it encounters an initiation codon. A subsequent joining of the 60S ribosomal subunit and release of eIFs result in formation of an elongation-competent 80S ribosome.
Secondary structures in 5′UTRs of mRNAs are thought to become unwound to allow ribosomal complexes to move along the mRNA in search of the initiation codon. Thus, in addition to its role in the initial attachment of ribosomal complexes to mRNA, eIF4A is believed to assist ribosomal complexes during scanning (5). Recent observations suggest that the process of eukaryotic initiation requires additional members of the DEAD/DExH-box protein family; for instance, a DEAD-box protein, yeast Ded1, and its mammalian homologue, DDX3, are biochemically and genetically implicated in translation initiation on long structured 5′UTRs (6), and another DExH-box protein, DHX29, strongly stimulates cap-dependent initiation on mRNAs with structured 5′UTRs in vitro (7). Here we studied the importance of DHX29 for translation in vivo and characterized it as a novel factor required for cell proliferation.
Results
DHX29 Is a Ubiquitously Expressed Cytoplasmic Protein That Associates with the 40S Ribosomal Subunit.
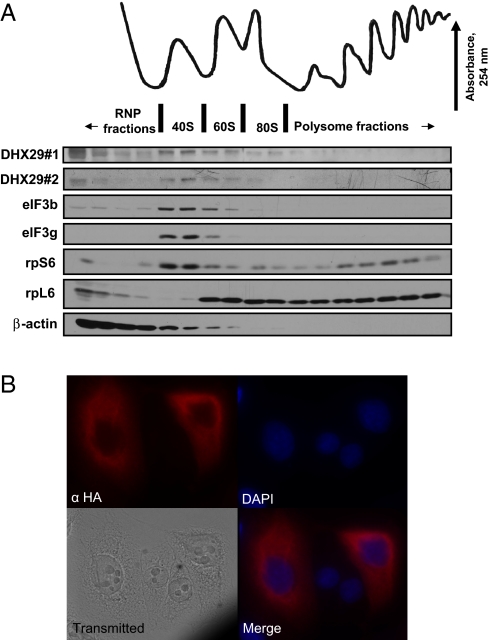
DHX29 has been found to interact with the 40S ribosomal subunit in vitro and to associate with 40S ribosomal complexes in a rabbit reticulocyte lysate (7). To determine how general these findings are, we examined the distribution of DHX29 in polysome preparations from HeLa cells using 2 commercial DHX29 antibodies (Fig. 1A). The specificity of these antibodies to human DHX29 was confirmed by immunoblotting against the purified native protein [supporting information (SI) Fig. S1] and recombinant DHX29, as well as by the loss of immunoreactivity following DHX29 RNAi depletion in HeLa cells (see below). DHX29 was enriched in 40S fractions (Fig. 1A). A low level of DHX29 was associated with 60S and 80S ribosomes. The eIF3b and eIF3g subunits of eIF3 showed the same distribution as DHX29. As expected, the 40S subunit ribosomal protein, rpS6, was associated with 40S, 80S, and polysome fractions, whereas the 60S subunit ribosomal protein, rpL6, was associated predominantly with 60S, 80S, and polysome fractions. β-actin protein was found primarily in the top gradient fractions.
Fig. 1.
DHX29 is a cytoplasmic protein that associates with the 40S ribosomal subunit. (A) Distribution of DHX29 in 10%–50% sucrose density gradient fractions of HeLa S3 cells as measured by immunoblotting. Polysomal profile distributions of eIF3b, eIF3g, rpS6, rpL6, and β-actin are shown for comparison. DHX29#1 and DHX29#2 represent antibodies against DHX29, from Bethyl Laboratories and Santa Cruz Biotechnologies, respectively. (B) Immunofluorescent detection of HA-DHX29 protein in HeLa cells using an anti-HA antibody (α-HA).
The distribution of DHX29 was similar to that of the eIF3 subunits. eIF3 is a canonical initiation factor that interacts with the 40S subunit (8, 9). Enrichment of DHX29 in 40S fractions in vivo is in agreement with the in vitro data (7) and consistent with the suggested function of DHX29 at the translation initiation stage.
As a component of the translation machinery, DHX29 would be expected to be expressed ubiquitously. Analysis of DHX29 mRNA expression using several databases [i.e., SOURCE, http://smd.stanford.edu/cgi-bin/source/sourceSearch (10); BioGPS, http://biogps.gnf.org; and ONCOMINE, http://www.oncomine.org (11)] supports this notion. To validate these data, we examined DHX29 expression in cell lines. Like eIF3b, DHX29 was expressed at similar levels in all cell lines examined (Fig. S2). The ubiquitous expression of DHX29 is consistent with its general physiological role in translation.
We next examined the subcellular localization of DHX29 by immunofluorescence (IF) staining of cells. Despite the good quality of the commercial antibodies for DHX29 detection by immunoblot analysis, these antibodies lack the specificity required for IF analysis. Thus, we transfected HeLa cells with an HA-tagged DHX29 expression vector and performed IF analysis using anti-HA antibodies. Consistent with its suggested role in translation, DHX29 was localized to the cytoplasm (Fig. 1B). The specificity of the IF staining by anti-HA antibodies was demonstrated by the absence of a fluorescent signal on staining with anti-Flag antibody used as a nonspecific isotype control (Fig. S3A), and also by the absorption of anti–HA-specific antibodies with the HA peptide (Fig. S3B).
DHX29 Down-Regulation Disrupts Polysomes and Inhibits Translation.
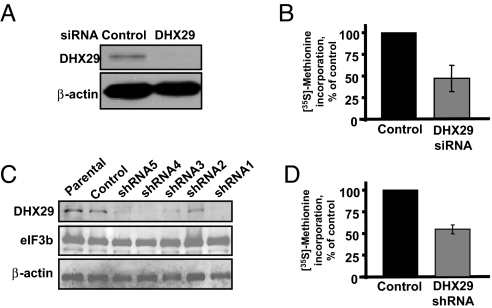
The function of several translation initiation factors in vivo has been studied previously using RNA interference (12–15). To investigate the role of DHX29 in translation in vivo, we reduced its amount in the cells by RNA interference. Transfection of HeLa cells with DHX29 siRNA resulted in a >90% reduction in DHX29 expression (Fig. 2A). The effect of reduced DHX29 expression on translation was determined using metabolic protein labeling experiments, in which siRNA-mediated knockdown of DHX29 led to a 2-fold reduction in [35S]-methionine incorporation (Fig. 2B).
Fig. 2.
Protein synthesis is impaired in DHX29-silenced cells. (A) Western blot analysis of DHX29 expression in HeLa cells transfected with nontargeting control or DHX29 siRNAs. (B) Effects of DHX29 silencing on protein synthesis. [35S]-methionine labeling of cells transfected with DHX29 or control siRNAs. The y-axis represents counts per minute (cpm) obtained from DHX29-silenced cells divided by cpm obtained from control cells. (C) Expression levels of DHX29 in parental HeLa cells and cells expressing 5 different DHX29-silencing (shRNA 1–5) and nontargeting control (control) shRNAs. (D) [35S]-methionine incorporation in cells expressing nontargeting control or DHX29 shRNA. The y-axis is as in (B).
To rule out the possibility that the results obtained with siRNA were due to “off-target” effects, and to study the long-term effects of DHX29 down-regulation on cell physiology, we established stable cell lines using lentiviral shRNAs against DHX29. We used several DHX29 shRNAs and a nontargeting shRNA control. Five different DHX29 shRNAs were introduced into HeLa cells. Various degrees of DHX29 silencing were achieved (Fig. 2C). Two stably infected cell lines (shRNA1 and shRNA4) that showed >90% reduction in DHX29 levels were used for subsequent experiments (Fig. 2C). Because of the great similarity of the data obtained from the 2 shRNA clones, here we present the results for shRNA1 only. In agreement with the results obtained with siRNA, DHX29 shRNA-expressing cells exhibited an average 2-fold reduction in [35S]-methionine incorporation into proteins (Fig. 2D).
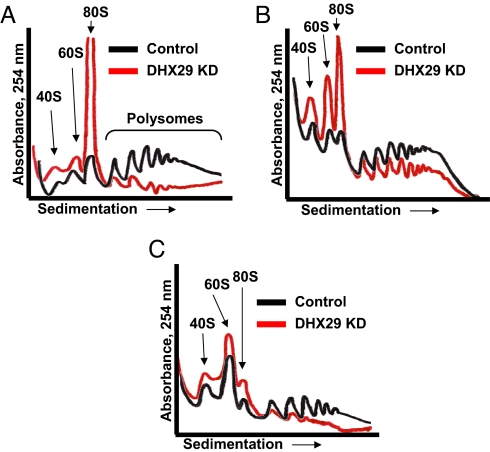
To determine the step at which translation is inhibited, we examined polysome profiles from DHX29 and control siRNA-transfected cells. DHX29 down-regulation caused dissociation of polysomes, with a concomitant increase in the abundance of 80S ribosomes as well as 40S and 60S subunits (Fig. 3A). Light polysomes were relatively less affected by DHX29 silencing compared with heavy polysomes. Notably, silencing of DHX29 resulted in dissociation of polysomes and an increase in the 80S peak. These effects on the polysomal profile are the hallmark of inhibited translation initiation (16). In DHX29 shRNA-expressing cells, polysome profiles showed a reduction in polysomes with concomitant increases in 40S, 60S, and 80S ribosome fractions (Fig. 3B). These data recapitulate those observed with siRNA and confirm that DHX29 functions in translation initiation.
Fig. 3.
DHX29 silencing disrupts polysomes and inhibits translation. (A) Polysomal profiles of HeLa cells transfected with DHX29 and control siRNAs. (B) Polysomal profiles from cells expressing control and DHX29 shRNA. (C) Polysomal profiles of siRNA DHX29-silenced and control cell lysates analyzed by sucrose density gradient centrifugation in buffer containing 0.5 M KCl (see Materials and Methods).
To determine whether the 80S ribosomes that accumulate on silencing of DHX29 are translation-competent monosomes assembled on the mRNA or mRNA-free ribosomes, we analyzed polysome profiles in the presence of high salt (0.5 M KCl), which disrupts mRNA-free 80S complexes that are not engaged in translation (17, 18). Polysome profiles from DHX29-silenced cells showed a dramatically reduced abundance of 80S ribosomes, due to their dissociation into individual 40S and 60S subunits (Fig. 3C). This result demonstrates that DHX29 silencing strongly reduces the assembly of 80S ribosomal complexes on mRNA.
DHX29 Is Required for Translation Initiation on mRNAs with Structured 5′UTRs.
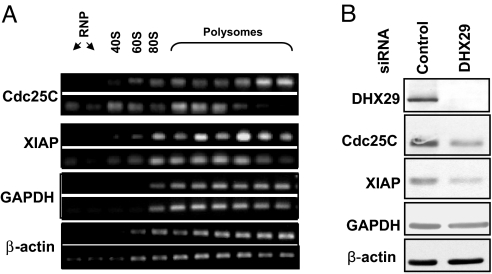
DHX29 plays a prominent role in the translation of mRNAs with highly structured 5′UTRs (7). The addition of DHX29 to an in vitro reconstituted translation system strongly stimulated 48S complex formation on mRNAs containing stable secondary structures in their 5′UTRs, such as Cdc25C mRNA (ΔG = −85 kcal/mol) (7). To examine the effects of DHX29 on the translation of Cdc25C mRNA in vivo in DHX29-silenced and control cells, we determined Cdc25C mRNA distribution in sucrose density gradients. In control cells, Cdc25C mRNA sedimented predominantly with heavy polysomes, whereas in DHX29-silenced cells, it was shifted to light polysomes, indicating decreased translation initiation of this mRNA (Fig. 4A). In agreement with these findings, Cdc25C protein levels decreased by more than 3-fold on DHX29 silencing (Fig. 4B).
Fig. 4.
DHX29 stimulates translation of mRNAs with structured 5′UTR. (A) Effects of DHX29 reduction on distribution of endogenous mRNAs in sucrose density gradients from control (Top) and DHX29-silenced (Bottom) cells. (B) Protein expression of DHX29, Cdc25C, XIAP, GAPDH, and β-actin in control and DHX29-silenced cells.
To examine the effect of DHX29 silencing on translation of mRNAs that have 5′UTRs with less-stable secondary structures than Cdc25C mRNA, we analyzed the polysome distribution of XIAP mRNA (ΔG = −47 kcal/mol) in sucrose density gradients. DHX29 depletion caused a shift in XIAP mRNA from heavy to light polysomal and subpolysomal fractions (Fig. 4A). XIAP protein expression was at least 3-fold repressed on DHX29 silencing (Fig. 4B). Together, these data demonstrate that DHX29 silencing inhibits translation of mRNAs containing moderately to extensively structured 5′UTRs.
To study the effect of DHX29 depletion on mRNAs with weak secondary structures, we assayed the polysomal association of β-actin and GAPDH mRNAs (ΔG = −16 and −22 kcal/mol, respectively). DHX29 depletion had a minimal effect on the polysomal distribution of β-actin and GAPDH mRNAs (Fig. 4A). Consistent with this result, no significant changes in protein levels for GAPDH and β-actin were observed on DHX29 silencing (Fig. 4B). In agreement with the in vitro results (7), these data indicate that in vivo, DHX29 predominantly stimulates translation of structured mRNAs. Analysis of all human 5′UTRs extracted from the RefSeq database shows that 66% of mRNAs have ΔG < −40 kcal/mol and thus can be considered moderately to extensively structured (Fig. S4). The prevalence of structured mRNAs in the eukaryotic cells could explain the global effects of DHX29 silencing on polysome distribution and decreased protein synthesis.
DHX29 Silencing Inhibits Cancer Cell Proliferation.
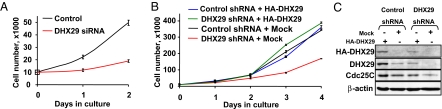
The inhibition of translation initiation impedes cell growth and proliferation (19). In addition, several translation factors are implicated in tumorigenesis (19–22). Consistent with a reduction in translation, HeLa cell proliferation was inhibited by almost 3-fold in DHX29 siRNA-silenced cells (Fig. 5A). Cell proliferation also was significantly (3-fold) inhibited by shRNA-mediated silencing of DHX29 after 4 days, compared with a nontargeting shRNA control (Fig. 5B). These results demonstrate that DHX29 is required for cell proliferation.
Fig. 5.
DHX29 promotes cell proliferation. (A) Proliferation of DHX29-silenced and control HeLa cells transfected with DHX29 siRNA and control siRNA, respectively. (B) Proliferation of DHX29-silenced and control HeLa cells transfected with HA-DHX29 and mock plasmid. (C) Expression of HA-tagged and endogenous DHX29 protein in DHX29-silenced and control cells transfected with HA-DHX29. Expression of Cdc25C and β-actin is also shown.
To exclude off-target effects of the DHX29 shRNA, we transfected DHX29-silenced HeLa cells with a plasmid encoding HA-tagged DHX29. This resulted in a 2-fold increase in proliferation compared with DHX29-silenced mock transfected cells (Fig. 5B). Control cells transfected with HA-DHX29 showed no significant change in cell proliferation compared with the mock-transfected control. Immunoblot analyses were performed to determine DHX29 expression levels. In control cells, the expression of HA-tagged DHX29 increased total DHX29 protein levels by about 3-fold (Fig. 5C; DHX29, compare lanes 1 and 2). In DHX29-silenced cells, the level of DHX29 was restored after transfection with HA-DHX29, compared with controls (Fig. 5C; DHX29 compare lanes 3 and 2). Consistent with the finding that DHX29-silencing causes the inhibition of Cdc25C mRNA translation (Fig. 4A), Cdc25C protein levels were restored to those of controls on rescue of DHX29 expression (Fig. 5C; Cdc25C, compare lanes 3 and 2). These findings indicate that rescue of DHX29 expression results in restoration of cell proliferation to control nonsilenced levels, underscoring the specificity of the observed DHX29-silencing effects.
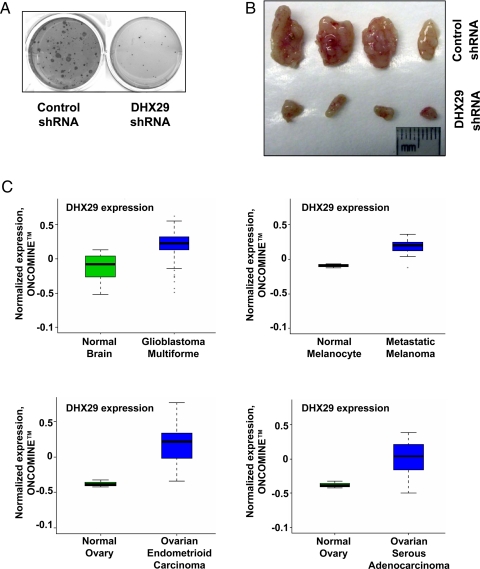
Given the strong effects of DHX29 on cell proliferation, we wished to determine whether DHX29 plays a role in tumorigenicity. We explored this issue using a HeLa cell–based system used previously to assess cell proliferation and tumorigenicity (23–30). First, anchorage-independent cell growth was assessed in shRNA-mediated DHX29-silenced and control cells. The DHX29-silenced cells exhibited at least a 10-fold decrease in the number of colonies formed in soft agar (Fig. 6A). Second, DHX29's role in tumor formation was assessed in xenograft experiments. DHX29-silenced and control HeLa cells were injected s.c. into nude mice, and after 4 weeks, tumors were excised. The DHX29-silenced cells formed significantly (P < .05) smaller tumors (average, 25 mg) compared with the nonsilenced control cells (average, 106 mg) (Fig. 6B). These findings demonstrate that silencing of DHX29 is sufficient to impair tumorigenicity.
Fig. 6.
DHX29 promotes tumorigenesis. (A) Soft-agar assay with DHX29-silenced and control cells. A total of 5,000 cells were plated and assayed 4 weeks later. (B) Tumors isolated from nude mice injected with 106 cells of the indicated type. Tumors were excised and photographed at 4 weeks postinjection. (C) Analysis of DHX29 mRNA expression in various cancers using the ONCOMINE database (11). ONCOMINE was searched for data sets in which DHX29 mRNA levels exhibited significant differences (t-test P value <10−5) in cancer versus normal tissue. Four such data sets were identified, and the normalized (by ONCOMINE) data were obtained. The graphs show boxplots of the 4 data sets comparing the cancer and normal samples. The 25th–75th percentiles are indicated by a closed box, with the median indicated by a line; different degrees of outliers are indicated by the whiskers and the points, as defined for standard boxplots.
Our query of the ONCOMINE database revealed significant overexpression of DHX29 in various types of cancers, including glioblastoma multiforme (P = 1.91e−07), metastatic melanoma (P = 6.25e−07), ovarian endometrioid carcinoma (P = 4.1e−13), and ovarian serous adenocarcinoma (P = 7.86e−11) (Fig. 6C). This suggests that translational inhibition through DHX29 silencing might present a means by which cancer cell growth could be inhibited.
Discussion
Our data demonstrate that DHX29 exhibits properties of a bona fide translation initiation factor. This is supported by several lines of evidence, including (i) the similar polysome distribution of DHX29 and other canonical translation initiation factors, (ii) the transient association of DHX29 with the 40S subunit, (iii) the decreased protein synthesis, and most importantly, (iv) the shift of heavy polysomes to light polysomes and free ribosomal subunits on RNAi-mediated reduction of DHX29 cellular content.
Early studies to identify eIFs using in vitro reconstituted initiation systems (31, 32) used β-globin mRNA, which has a relatively unstructured 5′UTR. More recent studies have confirmed that eIFs 2/3/1/1A/4A/4B/4F are sufficient to promote efficient 48S complex formation on β-globin mRNA and on model synthetic mRNAs with minimally structured 5′UTRs (5, 7, 33). The use of mRNAs with minimally structured 5′UTRs in previous biochemical assays is most likely the reason why DHX29 was identified only very recently. However, it is also noteworthy that preparations of factors used in the late 1970s contained substantial amounts of impurities, and that initiation factors such as eIF4F and eIF5, known to be required for initiation on β-globin mRNA, likely were present as contaminants and were not discovered until improved purification strategies became available. Thus, it is possible that DHX29 also may have been present as a contaminant in early preparations of initiation factors.
Both in vitro and in vivo studies show that DHX29 affects the initiation of translation of mRNAs with moderately to extensively structured 5′UTRs. Many mRNAs harboring extensive secondary structures have been shown to encode mainly for proteins involved in regulating growth, proliferation, and apoptosis (19, 34, 35); thus, it is conceivable that the decreased cell proliferation on DHX29 silencing is mediated through reduced translation of these mRNAs. In this regard, DHX29 protein behaves similarly to the canonical translation initiation factor eIF4E, which primarily controls translation of mRNAs with an extensive 5′UTR secondary structure and is strongly implicated in tumorigenesis (19, 35). Indeed, many of the eIF4E-regulated mRNAs are those involved in controlling cell proliferation and apoptosis (34, 35). But eIF4E does not exhibit helicase activity, and its modulatory role in the translation of structured mRNAs is thought to be exerted through eIF4G-mediated recruitment of the eIF4A helicase that unwinds 5′UTR secondary structures (5).
Although in silico DHX29 has a conserved helicase domain, it does not exhibit helicase activity in vitro (7). Our data indicate that DHX29 is associated with the 40S ribosomal subunit. The 43S ribosomal complex has intrinsic RNA scanning activity (5), which could be enhanced by DHX29 through remodeling of the 40S subunit. Data from Pisareva et al. (7) also suggest this possibility. Advancement in our understanding of the eukaryotic ribosome structure and its conformational changes during the process of translation would help to answer this question, as well as other important questions.
Materials and Methods
Cells.
HeLa S3 cells were obtained from the ATCC and maintained in DMEM (Sigma-Aldrich) containing 10% FBS.
Immunoblotting and Antibodies.
SDS/PAGE and Western blot analysis were performed as described previously (36), using antibodies against human Cdc25C (H-6), rpL6, eIF3b, DHX29, and rpS6 (Santa Cruz Biotechnologies); DHX29 (Bethyl Laboratories); eIF3g (from T. K. Tang, Academia Sinica, Taipei, Taiwan); eIF4B (37), rabbit anti-HA, and XIAP (Cell Signaling Technology); β-actin (Sigma-Aldrich); mouse anti-HA (HA11; Covance Research); and mouse anti-Flag (M2; Sigma-Aldrich).
Transfections.
Transfections were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
siRNA.
The anti-human DHX29 ON-TARGETplus SMARTpool and Non-Targeting Pool siRNA system was purchased from Dharmacon.
Lentivirus Packaging and Infections.
Lentiviruses for shRNA-silencing experiments were prepared as described previously (38), with some modifications. The MISSION shRNA bacterial glycerol stock containing 5 anti-DHX29 shRNA sequences and SHC002 shRNA Control were purchased from Sigma-Aldrich. Results from shRNA TRCN0000051238 (Sigma-Aldrich) are presented in this paper. Stable shRNA-expressing cell lines were established and selected by puromycin. Analyses of the protein synthesis and the polysome profile were conducted on day 6 after infection with anti-DHX29 and nontargeting control shRNA-expressing viruses.
[35S]-Methionine Metabolic Labeling.
Cells were seeded in 24-well plates, and [35S]-methionine labeling was performed as described previously (39).
Polysome Analysis.
Polysome analysis was performed as described previously (35). Polysomal fractionation experiments using 500 mM KCl were done as described previously (18).
RT-PCR.
Sucrose gradient fractions were subjected to RNA extraction using TRIzol (Invitrogen). Reverse transcription was performed using a SuperScript III Reverse-Transcriptase Kit (Invitrogen) and Random Hexamers (Invitrogen) according to the manufacturer's instructions. PCRs were carried out in a Mastercycler Realplex2 (Eppendorf) or a CFX96 (Bio-Rad) RT-PCR system using iQ Sybr Green Supermix (Bio-Rad) according to the manufacturer's instructions. The linear amplification range was determined by quantitative PCR. PCR was performed using 1 μL of RT reaction in a total reaction volume of 10 μL containing reverse and forward primers (0.5 μL of 10 μM solutions). The amplification conditions consisted of an initial denaturation step of 2 min at 95 °C, followed by up to 40 cycles of 15 s at 95 °C, 15 s at 55 °C, and 20 s at 68 °C. PCR products were analyzed in 1.8%–2% agarose gels. Primers for β-actin were as described previously (40). Primers for Cdc25C were: forward, 5′-TGGGATGAATCATGACCAGCACCT; reverse, 5′-TCTCTTTCTATGGCCACGGTCCAA. Primers for XIAP were: forward, 5′-TGTTTCAGCATCAACACTGGCACG; reverse, 5′-TGCATGACAACTAAAGCACCGCAC. Primers for GAPDH were: forward, 5′-TCGACAGTCAGCCGCATCTTCTTT; reverse, 5′-ACCAAATCCGTTGACTCCGACCTT.
Cell Proliferation.
HeLa cells were plated at 12,000 cells per well in a 6-well plate. For siRNA-silenced cells, they were plated 1 day after siRNA transfection as described above. Cells were trypsinized and counted using a hemocytometer or a Beckman Coulter counter.
Plasmids.
Human DHX29 cDNA (NCBI reference sequence NM_019030.2) was cloned into pcDNA3-HA using standard RT-PCR procedures. The empty vector was used as a control for possible effects caused by the transfected plasmid.
Soft Agar Assays and Injection in Nude Mice.
Soft agar assays were done as described previously (21). CD1 male nude mice (6–8 weeks old; Charles River Laboratories) were injected s.c. with 1 × 106 cells in 100 μL of PBS. Tumor growth was monitored according to McGill University guidelines.
Immunofluorescence.
IF experiments were performed as described previously (41). Cells were incubated for 3 hours with mouse anti-HA or anti-Flag antibodies at 1:2,000 dilution. IF images were obtained on an Axio Imager M1 (Carl Zeiss) using DAPI, rhodamin, and phase contrast settings.
Bioinformatics.
All human 5′UTRs were extracted from the RefSeq database downloaded on September 1, 2008. When multiple alternative 5′UTRs for a single gene existed, the longest 5′UTR was used for the analysis. The minimal free energy from each 5′UTR was estimated using UNAfold 3.6 with default settings (42). The minimal ΔG was set at −500.
Statistical Analysis.
Error bars for all data represent SDs from the mean. P values were calculated using t-tests.
Supplementary Material
Acknowledgments.
We thank Colin Lister and Pam Kirk for technical support, Thomas Duchaine and Nam-Sung Moon for the access to the PCR equipment, and Christos Gkogkas and Lian Wee Ler for help with IF. A.P. is an award holder from the Fonds de la Recherche en Santé Québec. This work was supported by grants from the National Cancer Institute of Canada to N.S. N.S. is a Howard Hughes Medical Institute International Scholar. O.L. is supported by a fellowship from the Knut and Alice Wallenberg Foundation.
Footnotes
Conflict of interest statement: C.U.H. and T.V.P. have filed a patent on DHX29 and its use to identify therapeutic regulators of gene expression.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909773106/DCSupplemental.
References
- 1.Shatkin AJ. mRNA cap-binding proteins: Essential factors for initiating translation. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 2.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E- inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pestova TV, Lorsch JR, Hellen CU. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–129. [Google Scholar]
- 5.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser CS, et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J Biol Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- 9.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehn M, et al. SOURCE: A unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes DR, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svitkin YV, et al. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site–mediated translation. Mol Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida M, et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher JW, Kubica N, Kimball SR, Jefferson LS. Reduced eukaryotic initiation factor 2Bepsilon-subunit expression suppresses the transformed phenotype of cells overexpressing the protein. Cancer Res. 2008;68:8752–8760. doi: 10.1158/0008-5472.CAN-08-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews MB, Sonenberg N, Hershey JW. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 1–41. [Google Scholar]
- 17.Vournakis J, Rich A. Size changes in eukaryotic ribosomes. Proc Natl Acad Sci USA. 1971;68:3021–3025. doi: 10.1073/pnas.68.12.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groppo R, Palmenberg AC. Cardiovirus 2A protein associates with 40S but not 80S ribosome subunits during infection. J Virol. 2007;81:13067–13074. doi: 10.1128/JVI.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider RJ, Sonenberg N. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 401–433. [Google Scholar]
- 20.Eberle J, Krasagakis K, Orfanos CE. Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro. Int J Cancer. 1997;71:396–401. doi: 10.1002/(sici)1097-0215(19970502)71:3<396::aid-ijc16>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 21.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 22.Silvera D, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura K, et al. Down-regulation of the insulin-like growth factor I receptor by antisense RNA can reverse the transformed phenotype of human cervical cancer cell lines. Cancer Res. 2000;60:760–765. [PubMed] [Google Scholar]
- 24.Wu Y, Pan S, Luo W, Lin SH, Kuang J. Hp95 promotes anoikis and inhibits tumorigenicity of HeLa cells. Oncogene. 2002;21:6801–6808. doi: 10.1038/sj.onc.1205849. [DOI] [PubMed] [Google Scholar]
- 25.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 26.An J, et al. Inactivation of the CYLD deubiquitinase by HPV E6 mediates hypoxia-induced NF-kappaB activation. Cancer Cell. 2008;14:394–407. doi: 10.1016/j.ccr.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, An H, Mayo MW, Baldwin AS, Yarbrough WG. LZAP, a putative tumor suppressor, selectively inhibits NF-kappaB. Cancer Cell. 2007;12:239–251. doi: 10.1016/j.ccr.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Huang JJ, et al. Ectopic expression of a COOH-terminal fragment of the human telomerase reverse transcriptase leads to telomere dysfunction and reduction of growth and tumorigenicity in HeLa cells. Cancer Res. 2002;62:3226–3232. [PubMed] [Google Scholar]
- 29.Tartour E, et al. Interleukin 17, a T cell–derived cytokine, promotes tumorigenicity of human cervical tumors in nude mice. Cancer Res. 1999;59:3698–3704. [PubMed] [Google Scholar]
- 30.Kato T, et al. Characterization of the HDAC1 complex that regulates the sensitivity of cancer cells to oxidative stress. Cancer Res. 2009;69:3597–3604. doi: 10.1158/0008-5472.CAN-08-4368. [DOI] [PubMed] [Google Scholar]
- 31.Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis, II: The assembly of the initiation complex with purified initiation factors. J Mol Biol. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- 32.Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 33.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 34.Larsson O, et al. Apoptosis resistance downstream of eIF4E: Posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 2006;34:4375–4386. doi: 10.1093/nar/gkl558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mamane Y, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahbazian D, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Methot N, Song MS, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase–dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–10273. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 40.Martineau Y, et al. Poly(A)-binding protein–interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rong L, et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA. 2008;14:1318–1327. doi: 10.1261/rna.950608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markham NR, Zuker M. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.