Abstract
Despite long-standing interest, the molecular mechanisms underlying the establishment of anterior–posterior (AP) polarity remain among the unsolved mysteries in metazoans. In the planarians (a family of flatworms), canonical Wnt/β-catenin signaling is required for posterior specification, as it is in many animals. However, the molecular mechanisms regulating the posterior-specific induction of Wnt genes according to the AP polarity have remained unclear. Here, we demonstrate that Hedgehog (Hh) signaling is responsible for the establishment of AP polarity via its regulation of the transcription of Wnt family genes during planarian regeneration. We found that RNAi gene knockdown of Dugesia japonica patched (Djptc) caused ectopic tail formation in the anterior blastema of body fragments, resulting in bipolar-tails regeneration. In contrast, RNAi of hedgehog (Djhh) and gli (Djgli) caused bipolar-heads regeneration. We show that Patched-mediated Hh signaling was crucial for posterior specification, which is established by regulating the transcription of Wnt genes via downstream Gli activity. Moreover, differentiated cells were responsible for the posterior specification of undifferentiated stem cells through Wnt/β-catenin signaling. Surprisingly, Djhh was expressed in neural cells all along the ventral nerve cords (along the AP axis), but not in the posterior blastema of body fragments, where the expression of Wnt genes was induced for posteriorization. We therefore propose that Hh signals direct head or tail regeneration according to the AP polarity, which is established by Hh signaling activity along the body's preexisting nervous system.
Keywords: body polarity, regeneration, anteroposterior axis, neuron, stem
Planarians are one of the most useful animals for investigation of the molecular and cellular mechanisms by which the anterior–posterior (AP) polarity of the body is established during regeneration. It is well known that planarians regenerate a head or a tail from the anterior and posterior ends, respectively, of the amputated stump, with maintenance of the original AP polarity. More than a century ago, T.H. Morgan reported one of the earliest descriptions of “polarity” in planarian regeneration (1), based on his findings that very thin cross-sectional fragments made by cutting along the AP axis regenerated bipolar two-headed planarians (2), termed “Janus-heads” (an allusion to the Roman god Janus; 3, 4). Since then, much effort has been focused on this fascinating maintenance of the original polarity, and its disruption, such as the induction of Janus-heads formation by treatment with colchicine or colcemid (5, 6), although the molecular mechanisms still remain unknown.
Recently, canonical Wnt/β-catenin signaling was shown to be required for posterior specification in planarians. Wnt/β-catenin signaling is evolutionarily well conserved and plays a role in establishing the AP axis during development in several animal species. In the planarian Schmidtea mediterranea, gene knockdown by RNA interference (RNAi) of Smed-βcatenin-1 causes Janus-heads formation during regeneration (4, 7, 8). RNAi of posterior-specific Smed-wntP-1 also causes the Janus-heads phenotype, as well as tail-less regeneration in Smed-wntP-1 or wnt11–2 RNAi regenerants (9). These findings provided insight into the molecular mechanisms directing the AP polarity during planarian regeneration. However, it remained necessary to discover the regulatory system for the posterior-specific activation of Wnt signals to fully understand the mechanism of the establishment of AP polarity in planarians.
Here, we investigated the upstream regulator(s) of Wnt/β-catenin signaling. We found that Hedgehog (Hh) signaling plays a crucial role for posterior specification in planarians by regulating the transcription of posterior-specific Wnt genes. In addition, we discuss the mechanism of the establishment of AP polarity during planarian regeneration.
Results
“Janus-Tails” Formation in Djptc RNAi Planarians.
To get a better understanding of the molecular mechanisms directing AP polarity, we screened a planarian Dugesia japonica EST library (10) for genes robustly responsible for AP polarity. Body fragments (head, trunk, and tail fragments) from transversely amputated planarians were examined for regeneration at both the anterior and posterior ends after RNAi by dsRNA injection (11). A homology search and the RNAi screen revealed that D. japonica patched (Djptc) is required for head regeneration in planarians (Fig. 1). Djptc, which is expressed ubiquitously (Fig. S1), codes for a Patched (Ptc) -like receptor, and possesses a conserved sterol-sensing domain characteristic of membrane proteins such as Ptc and NPC-1 (Fig. S2).
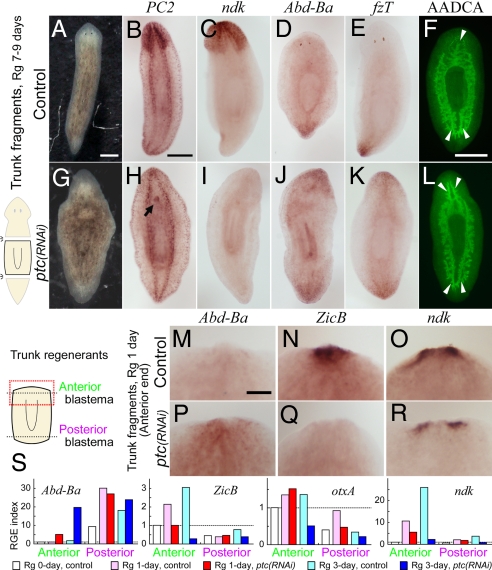
Fig. 1.
“Janus-tails” phenotype is caused by Djptc RNAi. Anterior end is placed at the top in all panels. Rg, regeneration. (A–L) Djptc(RNAi) caused abnormal anterior blastema formation in trunk fragment regenerants at 7–9 days postamputation. (A and G) Live animals. (B–F and H–L) Characterization by in situ detection of DjPC2 (CNS marker; B and H), Djndk (a head marker; C and I), DjAbd-Ba (a tail marker; D and J), and DjfzT (a tail marker; E and K), and by immunostaining for DjAADCA (digestive gut branch staining; F and L). Arrowheads indicate the single anterior and two posterior gut branches (F), and the bifurcated branching of the main gut tract in the anterior region as posterior fate (L). Arrow indicates an extra pharynx (H). (Scale bars, 500 μm.) (M–R) Posteriorized anterior end in Djptc(RNAi) trunk regenerants 1 day after amputation. Expression patterns of DjAbd-Ba (M and P), DjZicB (anterior marker; N and Q), and Djndk (O and R) in the anterior ends. (Scale bar, 200 μm.) (S) Time course of relative gene expression (RGE) level reveals gradual posteriorization of anterior blastema after Djptc(RNAi) during early regeneration (1 and 3 days). The expression levels of target genes (DjAbd-Ba, DjZicB, DjotxA, and Djndk) in anterior or posterior blastemas were normalized and expressed relative to the level in the anterior end of control trunk regenerants at day 0 taken as 1.0 (white column of “Anterior” bars; indicated by dashed line). Mean of quadruplicates.
To characterize the defect in head regeneration of Djptc RNAi [Djptc(RNAi)] planarians, we examined the expression patterns of anatomical and molecular markers for AP identity. The expression of DjPC2, a central nervous system (CNS) marker (12), showed that a cephalic brain did not develop in the “anterior blastema” (a mass of undifferentiated cells at the anterior cut edge of a stump) of Djptc(RNAi) trunk regenerants (i.e., regenerating fragments from the middle part of amputated planarians), although ventral nerve cords (VNCs) did regenerate (Fig. 1H). Expression of Djndk, a head marker (13), was absent from the anterior portion of Djptc(RNAi) trunk regenerants (Fig. 1I). Moreover, two tail marker genes, DjAbd-Ba (14) and Dj frizzled-T (DjfzT), a tail-specific Frizzled receptor gene (4), were ectopically induced in anterior blastemas by Djptc RNAi (Fig. 1 J and K). Regarding gut morphology (the digestive system, consisting of a single anterior and two posterior gut branches; Fig, 1F), staining of the gut tract with anti-DjAADCA antibody (15) revealed that Djptc(RNAi) regenerants showed posterior fate in the anterior region, as indicated by their bifurcated branching (Fig. 1L). These results show that not only is the head-identity lost upon Djptc(RNAi), but the polarity is reestablished to yield two oppositely oriented posterior ends, leading to the generation of the “Janus-tails” formation in Djptc(RNAi). Actually, Djptc(RNAi) regenerants occasionally developed an oppositely oriented pharynx, anterior to the original one (arrow in Fig. 1H), suggesting the intercalation of an extra pharynx according to the reestablished polarity.
Gradual Transformation to Posterior Polarity after Djptc RNAi.
To gain more insight into the polarization, we analyzed the expression patterns of relevant marker genes and their expression levels by reverse transcription and quantitative PCR (RT-qPCR) during early regeneration of trunk fragments after Djptc RNAi. DjAbd-Ba expression was induced ectopically and increased gradually in the anterior end of Djptc(RNAi) fragments during regeneration (Fig. 1 P and S). In contrast, the expression of the anterior-specific gene DjZicB (16) was not induced in the anterior blastema at regeneration day 1, but rather decreased to the same level as that in the posterior blastema at day 3 in Djptc(RNAi) (Fig. 1 Q and S). However, Djptc(RNAi) showed no effect on the relative gene expression (RGE) level of brain marker DjotxA (17) at day 1, but showed a decrease by day 3 of regeneration (Fig. 1S). Also, Djndk expression showed moderate induction in the anterior blastema of Djptc(RNAi) fragments at day 1, but thereafter decreased gradually (Fig. 1 R and S). These findings indicate that Djptc RNAi does not cause a complete reversal of polarity from the beginning of regeneration. Rather, the initial anterior polarity of body fragments is gradually transformed into posterior polarity during regeneration.
Djptc Functions Upstream of Wnt Signaling.
Wnt/β-catenin signaling is crucial for posterior specification in planarians (4, 7–9), as it is in many animals. β-catenin is a key mediator of the canonical Wnt/β-catenin cell-to-cell signal transduction cascade in animals (18). To examine the possible relationship between Djptc and Wnt/β-catenin signaling in planarians, we conducted double gene knockdown by coinjection of dsRNAs. We confirmed Janus-heads formation in trunk regenerants after RNAi of Djβ-cateninB (Fig. 2C), an ortholog of Smed-βcatenin-1 (4, 7, 8). Simultaneous Djptc and Djβ-cateninB RNAi also caused Janus-heads formation (Fig. 2D), indicating that Djptc functions upstream of Djβ-cateninB signaling.
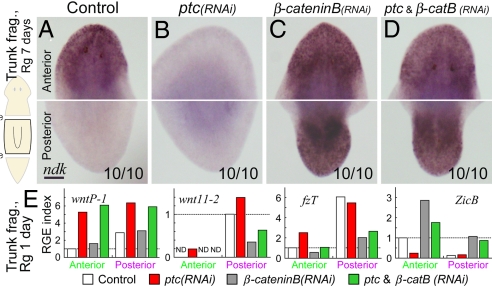
Fig. 2.
Djptc functions upstream of Wnt/β-catenin signaling. (A–D) Djndk expression in anterior (Upper) and posterior (Lower) regions of RNAi trunk regenerants at day 7. Djptc(RNAi) caused Janus-tails formation (B), whereas Djβ-cateninB(RNAi) caused Janus-heads formation (C) with or without simultaneous Djptc(RNAi) (D). Numbers represent the fraction of tested regenerants showing the indicated phenotype. (Scale bar, 200 μm.) (E) RGE levels of Wnt genes in anterior and posterior blastemas of RNAi trunk regenerants at day 1, with the anterior (DjZicB) and posterior (DjfzT) marker genes shown as positive controls for RNAi phenotype. ND, not detected.
To examine the Djptc/Wnt relationship, we analyzed the transcription of posterior-specific Wnt genes (7, 9), DjwntP-1 and Djwnt11–2 in RNAi trunk regenerants 24 h after amputation (Fig. 2E). Quantification of the RGE level at both ends of Djptc(RNAi) trunk regenerants revealed that Djptc RNAi increased the expression of DjwntP-1 and Djwnt11–2 ectopically in the anterior blastema. We found that this ectopic induction of Djwnt11–2, but not DjwntP-1, was abrogated by simultaneous Djβ-cateninB RNAi. The same tendencies were also observed in the posterior end. These results indicate that Djptc-mediated signaling regulates the transcription of posterior-specific Wnt genes, as is thought to occur in flies (19), and that transcription of Djwnt11–2 is further regulated by downstream Djβ-cateninB. These results suggest that Djptc regulates the transcription of DjwntP-1, which may act as a primary stimulus to activate Wnt/β-catenin signaling.
Hh Signaling Is Responsible for Posteriorization.
In general, Ptc is known to be a receptor in the evolutionarily well-conserved Hh signaling pathway (20–24). The basic machinery of the Hh signaling pathway functions via a series of repressive interactions: the ligand Hh, a transmembrane transducer Smoothened (Smo) and an activated form of the transcription factor Gli, act as positive regulators, whereas other components such as the Hh receptor Ptc and a Gli-interacting protein, Suppressor of Fused (SuFu), act as negative regulators. We identified planarian homologs of hedgehog (Djhh), suppressor of fused (Djsufu), and gli (Djgli) from our EST database. A smoothened homolog was not identified from our EST database, although one can be found in an S. mediterranea genome database (http://smedgd.neuro.utah.edu/). Djsufu and Djgli were expressed nearly ubiquitously in planarians as similarly to Djptc (Fig. S1). In contrast, Djhh expression was specifically localized in a subset of cells in intact planarians (Fig. S1).
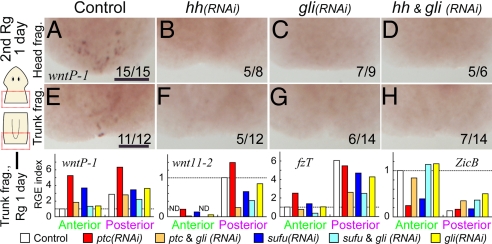
To determine whether Hh signaling is involved in AP polarity during regeneration, we performed RNAi analyses for Hh signaling component genes. In regenerants with RNAi of the positive regulators Djhh and Djgli, weak RNAi treatment caused tail malformation, while strong RNAi treatment and a second round of regeneration caused Janus-heads formation (Fig. 3). In the head fragments after the first round of regeneration, RNAi of Djhh and Djgli caused Janus-heads formation with the generation of ectopic eyes in the posterior blastemas, as well as loss of the tail identity, as shown by the loss of DjfzT expression (Fig. 3 B and C). Almost half of RNAi trunk regenerants finally showed loss of the tail identity after the second round of regeneration (Fig. 3 B′ and C′), although they showed no clear defect during the first round of regeneration. Simultaneous RNAi for Djhh and Djgli enhanced this phenotype, including ectopic eye formation around the posterior end of trunk regenerants (Fig. 3 D and D′). Thus, Djhh and Djgli are necessary for posterior specification.
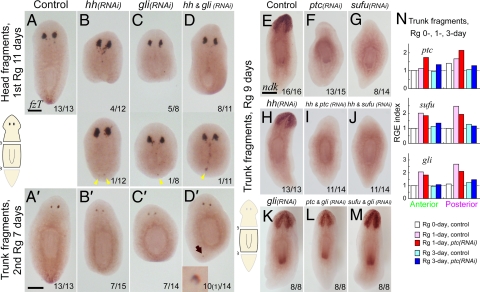
Fig. 3.
Hh signaling is involved in the establishment of AP polarity. (A–D) Loss of Hh signaling activity caused Janus-heads or loss of the tail identity in regeneration. DjfzT expression in head fragments at day 11 of the first round of regeneration (A–D), and trunk fragments at day 7 of the second round of regeneration (A′–D′) after strong inhibition of the positive regulators of the Hh signaling cascade (RNAi of Djhh or Djgli). Note that the generation of ectopic eyes in the posterior blastemas (arrowheads) and extra eye formation (arrow; Inset) near the posterior end (7%; 1/14, D′). (Scale bars, 300 μm.) (E–M) Janus-tails formation after RNAi of negative regulators of Hh signaling cascade (Djptc or Djsufu) and its suppression by simultaneous RNAi of positive regulators. Djndk expression in trunk fragments at regeneration day 9. (Scale bar, 300 μm.) (N) RGE index of Hh signaling component genes in anterior and posterior blastemas after Djptc(RNAi) as in Fig. 1S.
In the case of RNAi of the negative regulator Djsufu, as assessed by Djndk expression in trunk regenerants, Janus-tails formation was also observed similarly to that in Djptc(RNAi) (Fig. 3 F and G and Fig. S3), a finding that accords with the similarity of the phenotypes seen in ptch1 and sufu mouse mutants (25, 26). Furthermore, simultaneous RNAi of Djgli was sufficient to suppress this ectopic anterior-tail formation (Fig. 3 L and M), but not simultaneous Djhh RNAi (Fig. 3 I and J). In addition, quantification of the RGE level after Djptc RNAi revealed that the expression of Djptc itself tended to increase (Fig. 3N), presumably due to the autoinhibitory regulation of Djptc, which has been described in flies and mice (27, 28). In contrast, the RGE levels of Djsufu and Djgli were not affected in the anterior blastema in Djptc(RNAi) (Fig. 3N). Together with the conservation of the Hh signaling cascade among animal species (20–24), these findings strongly suggest that conserved Hh signaling is responsible for posterior specification in planarian regeneration, and Janus-tails formation was caused by activation of downstream DjGli ectopically in the anterior blastema.
Hh Signaling Regulates Transcription of Wnt Genes.
To examine the possibility that the transcription of Wnt genes is a primary output of the Hh signaling, we analyzed the expression of Wnt genes in RNAi animals for Hh signaling component genes at regeneration day 1. Actually, the transcription of DjwntP-1 was repressed at the posterior end of fragments as a result of the loss of Hh signaling activity (Fig. 4 A–H). DjwntP-1 expression was absent in the posterior ends of regenerants from both head and trunk fragments after RNAi of Djhh (Fig. 4 B and F) or Djgli (Fig. 4 C and G), as well as simultaneous Djhh and Djgli RNAi (Fig. 4 D and H). Conversely, RGE quantification after loss of negative regulators of the Hh signaling pathway indicated that the transcription of DjwntP-1 and Djwnt11–2 was aberrantly up-regulated in the anterior blastema of trunk regenerants in Djptc(RNAi) or Djsufu(RNAi) (Fig. 4I). Also, simultaneous Djgli(RNAi) suppressed the ectopic up-regulation of these posterior Wnt genes in the anterior blastema in Djptc or Djsufu (RNAi) (Fig. 4I). These results indicated that Hh signaling specifies posterior identity through regulating the transcription of Wnt genes in planarian regeneration.
Fig. 4.
Hh signaling regulates transcription of Wnt genes. (A–H) DjwntP-1 expression was lost by RNAi of Djhh or Djgli in the posterior end of both head (A–D) and trunk (E–H) regenerants on day 1 of the second round of regeneration. (Scale bars, 200 μm.) (I) RGE quantification of Wnt genes in anterior and posterior blastemas of trunk fragments at regeneration day 1 after RNAi of Hh signaling components genes, with the anterior (DjZicB) and posterior (DjfzT) marker genes as positive controls for RNAi phenotype. ND, not detected.
Differentiated Cells Provide Signal Transduction for Posterior Specification.
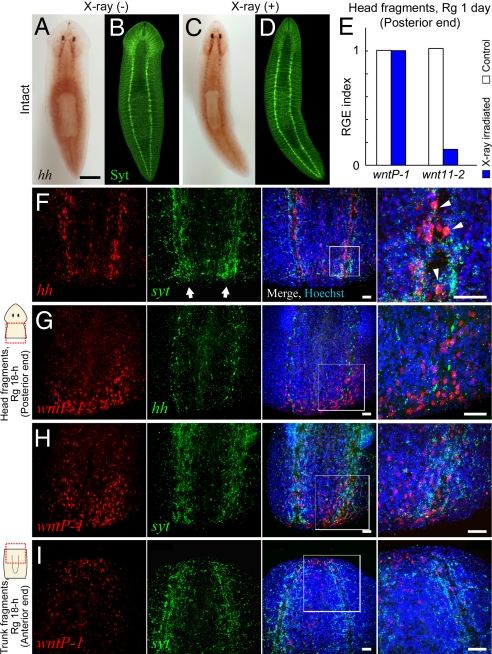
The ability of planarians to regenerate missing parts relies on pluripotent stem cells distributed throughout the body (3, 29, 30), and the surrounding differentiated cells might provide appropriate cues for the differentiation of stem cells (31). To investigate how differentiated cells contribute to the regulation of Wnt signaling for the establishment of AP polarity, we analyzed X-ray-irradiated planarians, in which stem cells are specifically eliminated. Djhh was apparently expressed in the X-ray-resistant cells within the VNCs, which were stained with anti-Dj Synaptotagmin (DjSyt) antibody (Fig. 5 A–D and Fig. S4), and Djptc expression was detected in X-ray-resistant cells along the VNCs (Fig. S4). Interestingly, the expression of DjwntP-1 was induced exclusively in X-ray-resistant cells at the posterior end of head regenerants 24 h after amputation, whereas Djwnt11–2 induction occurred mainly in X-ray-sensitive cells (Fig. 5E). These findings imply that posterior identity is established in differentiated cells by the induction of DjwntP-1 expression even in the absence of stem cells in the posterior end.
Fig. 5.
Posteriorizing signal originates in differentiated cells (neuron). (A–D) Expression of Djhh was observed in X-ray-resistant differentiated cells of the VNCs. In situ detection of Djhh in nonirradiated (A) and irradiated (C) planarians, followed by labeling of the CNS with anti-DjSynaptotagimin antibody (neural marker; Syt; B and D). (Scale bar, 1 mm.) (E) RGE index of Wnt genes in the posterior blastema of head fragments from X-ray-irradiated animals 1 day after amputation. (F-I) Confocal images of expression patterns of Djhh, DjwntP-1, or Djsyt by dual in situ detection in the posterior (F–H) and anterior (I) end in regenerants from head and trunk fragments, respectively, at 18 h after amputation. Panels show individual expression patterns (red or green color) and their merged image with the image of nuclear staining (Hoechst; blue). The rightmost panel is a magnified view of each boxed area. Arrow indicates VNC (F). Note that Djhh was expressed in neural cells of the VNCs, which were labeled with Djsyt probe (arrowheads; F), but not in the posterior blastema (F and G). Also, DjwntP-1 was predominantly expressed in the posterior end surrounding the VNCs (G and H), but was also weakly expressed in the anterior end (I). (Scale bars, 50 μm.)
To further identify the cell type in differentiated cells involved in the cross-talk between Hh and Wnt signaling for posteriorization, we conducted dual-color in situ detection for the expression pattern of Djhh and DjwntP-1 at an early stage of regeneration. Djhh expression was significantly induced in the anterior blastema of regenerants, although this expression was absolutely derived from stem cells (Fig. S4). Thus, this anterior-expression of Djhh has no contribution to the induction of DjwntP-1 expression (Fig. 5E). At 18 h of regeneration, the stage before the induction of Djhh expression in the anterior blastema (Fig. S1), Djhh expression was not induced in the posterior blastema of regenerants, but was detected in neural cells of the preexisting VNCs, as indicated by the colocalization of Djsyt mRNA (32) (Fig. 5 F and G). In contrast, DjwntP-1 expression was predominantly induced in cells along and between the VNCs at the posterior end of regenerants (Fig. 5 G and H), but it was also weakly detected in the anterior region (Fig. 5I). Especially, expression of Djhh and DwntP-1 was not colocalized in the same cells at the posterior end of regenerating fragments (Fig. 5G). These findings suggest that de novo expression of Djhh is not required for the induction of DjwntP-1 expression during regeneration.
Discussion
Hh Signaling Triggers Posterior Specification.
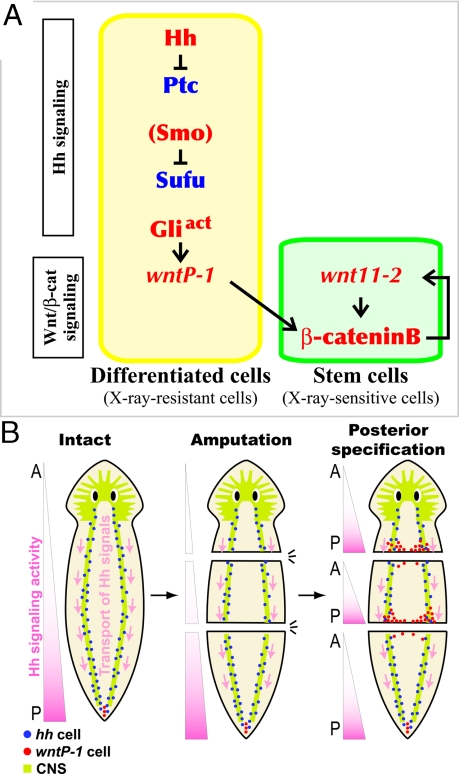
Based on the findings in this report, we propose a model of the signaling cascade for the posterior specification in planarians (Fig. 6A). First, Hh signaling triggers posterior specification by activating the transcription of DjwntP-1 in X-ray-resistant differentiated cells at the posterior end, and then the posterior specification signal is further transmitted by the induction of Djwnt11–2 in X-ray-sensitive stem cells through β-catenin signaling. Finally, the resultant committed stem cells direct posterior-fate-specification of undifferentiated stem cells through Wnt/β-catenin signaling. Thus, cues for differentiation of stem cells are provided from not only differentiated cells but also stem cells during planarian regeneration.
Fig. 6.
Models for two aspect of the establishment of AP polarity by Hh signaling in planarians. (A) The signaling cascade model for posterior specification in planarian regeneration inferred from the present study. Hh signaling triggers posteriorization by inducing transcription of Wnt genes in differentiated cells (yellow-shaded box), from which the signal is transmitted to stem cells (green-shaded box) through Wnt/β-cateninB signaling for tail regeneration. Positive and negative regulators of posterior specification are written with red and blue letters, respectively. (B) A possible model for the establishment of AP polarity by Hh signaling in planarians. DjwntP-1-expressing cells (red dots) at the posterior end suggests that Hh signaling is exclusively activated there (pink gradients on the left), although Djhh-expressing cells (neural cells; blue dots) are localized all along the VNCs (CNS is indicated by light green). In this model, Djhh gene products would be transported posteriorly via VNCs (pink arrows), leading to the induction of posterior specification at the posterior end.
Hh signaling is thus the activator of a key molecular switch for posterior specification, and Wnt/β-catenin signaling acts as this switch. It is known that Hh signaling regulates the morphogenesis of various tissues and organs through canonical Wnt signaling in many animal species, including the morphogenesis of intrasegmental patterning and neural tube patterning; however, our findings reveal the role of Hh signaling to establish AP polarity throughout the body in planarians.
Putative Role of Hh Signaling in Intercalary Regeneration.
It has been shown that ectopic transplantation of a tail fragment into the anterior portion of a planarian caused intercalation of a newly formed ectopic pharynx in the region between the graft and the original pharynx (33, 34). We observed similar intercalation of an extra pharynx in Djptc(RNAi) animals (Fig. 1H). The anterior blastema of Djptc(RNAi) regenerants was transformed to become the posterior end and then it induced ectopic pharynx formation by intercalation between the induced posterior end and the anterior base of the pharynx. In other words, identification of the “posterior terminus” by the induction of DjwntP-1 in differentiated cells might be responsible for the intercalary regeneration of a tail in planarians. Differentiated cells provide the signal indicating the terminus (“distalization”), and missing parts are regenerated from stem cell populations between the initial tissue and the terminal cells (tissue) according to the positional information (“intercalation”), in accord with a process called “rearrangement of the body regionality” (refer to 35).
Possible Mechanisms for the Establishment of AP Polarity in Planarians.
The present study clearly demonstrated that Hh signaling functions as a regulator for posteriorization in planarians. How does Hh signaling establish AP polarity during planarian regeneration? Djhh expression itself is not induced in the posterior blastema of regenerating fragments when DjwntP-1 is expressed there. Rather, Djhh is expressed in differentiated neural cells of VNCs. Based on these observations, we propose a possible model in which Hh signaling present in the neural cells of the VNCs is activated exclusively at the posterior end, and induces DjwntP-1. That is, DjHh signals (protein or mRNA) might be transported posteriorly via the neural axons of the VNCs (Fig. 6B), in a similar way to that reported in photoreceptor neurons in fly (36).
These putative Hh signals transported uni-directionally in the VNCs might also maintain AP polarity in intact planarians, by directing the expression of DjwntP-1 to be strongest at the posterior tip (Fig. 6B) (7, 9). Thus, after amputation, the planarian regeneration system does not require a ‘reconstitution’ of AP polarity in the remaining tissue. Instead, planarians regenerate their body according to the initial AP polarity of the fragment along the VNCs. This scenario could account for our observation that anterior polarity was gradually lost and transformed into posterior identity during the course of regeneration in the anterior blastema after Djptc(RNAi). That is, regeneration in the anterior blastema of Djptc(RNAi) regenerants was initiated according to the initial AP polarity of the body as defined by Hh signaling activity, and subsequently, the newly formed tissue was gradually transformed to possess posterior identity. We speculate that ectopic activation of downstream DjGli progressively activates Wnt signaling in the anterior blastema, which gradually represses the anterior (head) identity and transforms it into tail fate. The transient expression of DjwntP-1 observed in the anterior end during regeneration (Fig. 5I) might be induced by the “leaky” activation of Hh signaling at the cut surface but then repressed by a resultant depletion of the Hh signaling activity via the uni-directional transport of Djhh gene products.
Some puzzling observations made in classical and recent planarian regeneration experiments can be explained by this model. For example, as originally reported by Morgan (2), the Janus-heads formation in regenerants from very thin cross-sectional fragments could be explained by the lack of a sufficient total amount of Hh signal available for transport and delivery in these thin tissues to transform the polarity of the end to a posterior fate. Disruption of microtubules by treatment with colchicine or colcemid causes Janus-heads formation after regeneration (5, 6), which our model suggests would be the result of the inhibition of Hh signal transport along the microtubules in neural cells. Similarly, Janus-heads formation has been shown in regenerants in which Ca2+ signaling was perturbed by chemical inhibition of voltage-operated calcium channels (37), implying loss of DjHh axonal transport and/or signal transduction in neural cells of the VNCs. Further studies to clarify whether uni-directional transport of Hh signals along the AP axis plays a crucial role in establishing the AP polarity in planarians should yield fruitful insights into this long-standing puzzle.
Materials and Methods
Planarians.
A clonal strain of the planarian D. japonica was used in all experiments. The colony was maintained in autoclaved tap water at room temperature in dim light. In all experiments, planarians of 8- to10-mm length that had been starved for at least 1 week were used. For regeneration studies, planarians were cut into three fragments (head, trunk, and tail) transversely anterior and posterior to the pharynx and allowed to undergo regeneration.
cDNA Clones.
cDNA clones encoding the respective proteins were identified in a previously constructed library of expressed sequence tags (ESTs) (10). Partial cDNA fragments encoding DjFzT, DjWntP-1, and DjWnt11–2 were cloned by PCR. To obtain the full-length cDNA, 5′ and 3′ rapid amplification of cDNA ends (RACE) were carried out using a SMART RACE cDNA amplification kit (Clontech).
X-Ray Irradiation.
Worms placed on wet filter paper on ice were irradiated with 12 R X-rays using an X-ray generator (SOFTEX B-4). Five days after irradiation, worms were subjected to the examinations indicated in the text.
Whole-Mount In Situ Hybridization and Immunohistochemistry.
Whole-mount in situ hybridization was performed with digoxigenin (DIG)-labeled riboprobes (Roche Diagnostics). Animals were treated with 2% HCl in 5/8 Holtfreter's solution for 5 min at 4 °C and fixed in 5/8 Holtfreter's solution containing 4% paraformaldehyde and 5% methanol for 3 h at 4 °C. Hybridization and detection of DIG-labeled RNA probes were carried out as described in ref. 38. After whole-mount in situ hybridization, specimens were subjected to immunohistochemistry using anti-DjAADCA or anti-DjSyt antibody essentially as described (15, 32). In brief, specimens were incubated with the primary antibody (1:1,000 dilution) overnight at 4 °C. They were visualized using a fluorescent microscope Biozero BZ-8000 (Keyence) with fluorescence-labeled secondary antibodies (Alexa Fluor 488, 1:500; Molecular Probes).
Dual-color detection of mRNA expression was performed essentially as described. For the detection of DIG- or fluorescein-labeled RNA probes, samples were incubated with specific antibodies conjugated with alkaline phosphatase or horseradish peroxidase (1:2,000; Roche Diagnostics). To develop fluorescent color, a TSA kit No. 2 (Molecular Probes) and an HNPP Fluorescent Detection Set (Roche Diagnostics) were used according to the manufacturer's instructions. Cell nuclei were labeled with Hoechst No. 33342. Fluorescence was detected with a confocal laser scanning microscope FV1000 (Olympus).
RNA Interference (RNAi).
Double-stranded RNA (dsRNA) was synthesized from appropriate cDNA clones, and was injected into planarians essentially as described (usual-dose RNAi; 11). Control animals were injected with distilled water (DW; the solvent for dsRNA). For regeneration studies, planarians were cut 3 days after the third injection. For the high-dose RNAi in single Djhh and Djgli RNAi experiments, the synthesized dsRNAs were dissolved in half the volume of DW compared to the volume used for usual-dose RNAi. Injected animals were allowed to undergo a second round of regeneration: the animals were cut transversely, and then both the anterior and posterior blastemas of the fragments were cut off at regeneration day 4.
Reverse Transcription and Quantitative PCR (RT-qPCR) Analysis.
Planarians were cut into three fragments and allowed to undergo regeneration for various numbers of days (e.g., day 0, just after the cutting; day 1, approximately 24 h after amputation) as indicated in the text. Pieces of the anterior or posterior ends from RNAi trunk fragments (n ≥ 20), or the posterior end from head fragments of X-ray-irradiated planarians (n = 10 each), which included regenerating blastemas, were then collected. Total RNA was extracted from each pool of blastemas using ISOGEN-LS (Nippon Gene), and cDNA was synthesized from 1 μg total RNA using a QuantiTect Reverse Transcription Kit (Qiagen). Quantitative analysis of the amount of each gene product was performed essentially as described in ref. 39 using the 7900HT Real-Time PCR System (Applied Biosystems). Each reaction (10 μL) contained 1× QuantiTect SYBR Green PCR Master Mix (Qiagen), gene-specific primer at 0.3 μM, and 1 μL diluted (1:20) cDNA template, and was subjected to PCR as follows: 95 °C for 15 min, 50 cycles of 95 °C for 15 s, 60 °C for 30 s, 72 °C for 1 min. PCR primers for each target gene are listed in Table S1. The primer set for the Djptc gene was not complementary to its dsRNA, rather, primers were designed to correspond to the 3′ end of the Djptc gene and the dsRNA was designed to correspond to the 5′ end. Measurements were performed in quadruplicate for technical replicates (coefficient of variation was 0.1–2.8% for all primer sets), and were normalized to the expression level of DjG3PDH (32). In the analyses of trunk regenerants, the expression level in the anterior end of the control fragment was taken as the standard for the relative gene expression (RGE) value, except in the case of Djwnt11–2, for which the level of the posterior end of the control fragment was taken as the standard because its expression was not detectable in the anterior end. In head regenerants, the expression level in the control fragment was taken as the standard. The RGE values were calculated according to Ogawa et al. (39). In brief, the level of the target gene transcript detected was compared with the baseline level detected in the standard, and the RGE index of the standard was set to 1.0 (dashed line in each figure). The RGE index reported here is the mean of quadruplicate measurements.
Supplementary Material
Acknowledgments.
We thank J. Tasaki for immunostaining of DjSyt, Y. Saito for drawing illustrations, J. C. Rink of the University of Utah for a personal communication informing us that a Smo homolog could be found in the S. mediterranea genome database, and E. Nakajima, L. Rouhana, and C. Lamy for helpful discussions and comments on the manuscript. This work was supported by Global Centers of Excellence Program A06 of Kyoto University and a Grant-in-Aid for Creative Research to K. A. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence data for the reported cDNAs of D. japonica have been deposited in the DDBJ/EMBL/GeneBank database under the accession codes AB504738 (Djptc), AB504739 (Djhh), AB504740 (Djsufu), AB504741 (Djgli), AB504743 (DjfzT), AB504744 (DjwntP-1), AB504745 (Djwnt11–2), AB505439 (Djβ-cateninB), and AB504746 (DjG3PDH).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907464106/DCSupplemental.
References
- 1.Morgan TH. An analysis of the phenomena of organic ‘polarity’. Science. 1904;20:742–748. doi: 10.1126/science.20.518.742. [DOI] [PubMed] [Google Scholar]
- 2.Morgan TH. The control of heteromorphosis in Planaria maculata. Arch Entw Mech Org. 1904;17:683–695. [Google Scholar]
- 3.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 4.Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McWhinnie MA. The effect of colchicine on reconstitutional development in Dugesia dorotocephala. Bio Bull. 1955;108:54–65. [Google Scholar]
- 6.Kanatani H. Formation of bipolar heads induced by demecolcine in the planarian, Dugesia gonocephala. J Fac Sci Tokyo Univ. 1958;8:253–270. [Google Scholar]
- 7.Petersen CP, Reddien PW. Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 8.Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-βcatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- 9.Adell T, Saló E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- 10.Mineta K, et al. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci USA. 2003;100:7666–7671. doi: 10.1073/pnas.1332513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agata K, et al. Structure of the planarian central nervous system (CNS) revealed by neuronal cell markers. Zoolog Sci. 1998;15:433–440. doi: 10.2108/zsj.15.433. [DOI] [PubMed] [Google Scholar]
- 13.Cebrià F, et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- 14.Nogi T, Watanabe K. Position-specific and non-colinear expression of the planarian posterior (Abdominal-B-like) gene. Dev Growth Differ. 2001;43:177–184. doi: 10.1046/j.1440-169x.2001.00564.x. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura K, et al. Reconstruction of dopaminergic neural network and locomotion function in planarian regenerates. Dev Neurobiol. 2007;67:1059–1078. doi: 10.1002/dneu.20377. [DOI] [PubMed] [Google Scholar]
- 16.Aruga J, et al. A wide-range phylogenetic analysis of Zic proteins: Implications for correlations between protein structure conservation and body plan complexity. Genomics. 2006;87:783–792. doi: 10.1016/j.ygeno.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Umesono Y, Watanabe K, Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev Genes Evol. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- 18.Schneider SQ, Finnerty JR, Martindale MQ. Protein evolution: Structure-function relationships of the oncogene beta-catenin in the evolution of multicellular animals. J Exp Zoolog B Mol Dev Evol. 2003;295:25–44. doi: 10.1002/jez.b.6. [DOI] [PubMed] [Google Scholar]
- 19.Martinez Arias A, Baker NE, Ingham PW. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich LV, Scott MP. Hedgehog and patched in neural development and disease. Neuron. 1998;21:1243–1257. doi: 10.1016/s0896-6273(00)80645-5. [DOI] [PubMed] [Google Scholar]
- 21.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 22.Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 23.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, McMahon AP, Allen BL. Shifting paradigms in Hedgehog signaling. Curr Opin Cell Biol. 2007;19:159–165. doi: 10.1016/j.ceb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Cooper AF, et al. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 26.Svärd J, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Hidalgo A, Ingham P. Cell patterning in the Drosophila segment: Spatial regulation of the segment polarity gene patched. Development. 1990;110:291–301. doi: 10.1242/dev.110.1.291. [DOI] [PubMed] [Google Scholar]
- 28.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 29.Saló E. The power of regeneration and the stem-cell kingdom: Freshwater planarians (Platyhelminthes) Bioessays. 2006;28:546–559. doi: 10.1002/bies.20416. [DOI] [PubMed] [Google Scholar]
- 30.Agata K, et al. Two different evolutionary origins of stem cell systems and their molecular basis. Semin Cell Dev Biol. 2006;17:503–509. doi: 10.1016/j.semcdb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Kato K, Orii H, Watanabe K, Agata K. Dorsal and ventral positional cues required for the onset of planarian regeneration may reside in differentiated cells. Dev Biol. 2001;233:109–121. doi: 10.1006/dbio.2001.0226. [DOI] [PubMed] [Google Scholar]
- 32.Takano T, et al. Regeneration-dependent conditional gene knockdown (Readyknock) in planarian: Demonstration of requirement for Djsnap-25 expression in the brain for negative phototactic behavior. Dev Growth Differ. 2007;49:383–394. doi: 10.1111/j.1440-169X.2007.00936.x. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi C, Nogi T, Watanabe K, Agata K. Ectopic pharynxes arise by regional reorganization after anterior/posterior chimera in planarians. Mech Dev. 1999;89:25–34. doi: 10.1016/s0925-4773(99)00192-6. [DOI] [PubMed] [Google Scholar]
- 34.Agata K, Tanaka T, Kobayashi C, Kato K, Saitoh Y. Intercalary regeneration in planarians. Dev Dyn. 2003;226:308–316. doi: 10.1002/dvdy.10249. [DOI] [PubMed] [Google Scholar]
- 35.Agata K, Saito Y, Nakajima E. Unifying principles of regeneration I: Epimorphosis versus morphallaxis. Dev Growth Differ. 2007;49:73–78. doi: 10.1111/j.1440-169X.2007.00919.x. [DOI] [PubMed] [Google Scholar]
- 36.Chu T, Chiu M, Zhang E, Kunes S. A C-terminal motif targets Hedgehog to axons, coordinating assembly of the Drosophila eye and brain. Dev Cell. 2006;10:635–646. doi: 10.1016/j.devcel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Nogi T, Zhang D, Chan JD, Marchant JS. A novel biological activity of praziquantel requiring voltage-operated ca channel β subunits: Subversion of flatworm regenerative polarity. PLoS Negl Trop Dis. 2009;3:e464. doi: 10.1371/journal.pntd.0000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa K, et al. Induction of a noggin-like gene by ectopic DV interaction during planarian regeneration. Dev Biol. 2002;250:59–70. doi: 10.1006/dbio.2002.0790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.