Abstract
Notch signaling regulates cell specification and homeostasis of stem cell compartments, and it is counteracted by the cell fate determinant Numb. Both Numb and Notch have been implicated in human tumors. Here, we show that Notch signaling is altered in approximately one third of non–small-cell lung carcinomas (NSCLCs), which are the leading cause of cancer-related deaths: in ≈30% of NSCLCs, loss of Numb expression leads to increased Notch activity, while in a smaller fraction of cases (around 10%), gain-of-function mutations of the NOTCH-1 gene are present. Activation of Notch correlates with poor clinical outcomes in NSCLC patients without TP53 mutations. Finally, primary epithelial cell cultures, derived from NSCLC harboring constitutive activation of the Notch pathway, are selectively killed by inhibitors of Notch (γ-secretase inhibitors), showing that the proliferative advantage of these tumors is dependent upon Notch signaling. Our results show that the deregulation of the Notch pathway is a relatively frequent event in NSCLCs and suggest that it might represent a possible target for molecular therapies in these tumors.
Keywords: γ-secretase inhibitors, NSCLC, NUMB
The Notch signaling pathway mediates a variety of context-dependent biological functions (1–4). In humans, there are four Notch receptors that, upon engagement by ligands of the DSL family, are proteolytically cleaved to release the intracellular domain of Notch (NICD), which translocates into the nucleus to modulate gene expression (1). The activity of Notch is counteracted by Numb (3, 5), through a mechanism that is not completely understood but that is underscored by the fact that loss of Numb function phenocopies Notch gain-of-function, in developmental systems (3).
The best-characterized function of Notch is to regulate cell fate; this has been linked to the homeostasis of stem cell compartments (1, 6–8). Not surprisingly, therefore, aberrant Notch signaling has been implicated in human cancer (7). The clearest example of cell-autonomous oncogenic activation of Notch occurs in T-cell acute lymphoblastic leukemia/lymphoma (T-ALL), where NOTCH-1 is activated through chromosomal translocations or mutations (7, 9). Deregulated expression of Notch receptors, ligands, or targets has also been reported in solid tumors [reviewed in (7, 10)], including breast (11) and lung cancers (12–15). To date, however, cell-autonomous activating mutations of Notch receptors have not been found in solid tumors. Finally, lack of attenuation of Notch signaling also plays a role in cancer, as loss of NUMB expression, in breast cancer, causes increased Notch activity and a Notch-dependent proliferative advantage (16, 17).
The inhibition of Notch signaling holds, therefore, promise for cancer therapies. A family of compounds, γ-secretase inhibitors (GSIs), is the object of intense scrutiny for this purpose. γ-Secretase is pivotal in the activation of Notch, as it executes the last proteolytic cleavage that releases the NICD from the plasma membrane (10). However, the clinical application of GSIs must still overcome important hurdles. First, GSIs display significant acute toxicity (7, 10). Second, we need patient stratification criteria, to determine eligibility for GSI treatment. In this framework, the identification of alterations in Notch signaling in major solid tumors might provide new impetus to clinical research in this area.
Here, we show that alterations of the Notch pathway are frequent in non–small-cell lung carcinomas (NSCLC). We identify two major alterations: loss of NUMB expression, and gain-of-function mutations of the NOTCH-1 gene. We also show that NSCLCs, harboring activation of the Notch pathway, depend upon Notch signaling for their growth potential. Thus, our results suggest that targeted interference with Notch activation represents a promising therapeutic avenue in NSCLC, and provide biomarkers for patient stratification.
Results
Deregulation of NUMB Expression in NSCLC.
In an initial survey of more than 200 NSCLCs performed by immunohistochemistry (IHC) on tissue microarrays, we observed frequent loss of NUMB expression. Normal lung parenchyma invariably showed moderate/intense homogeneous NUMB staining (Fig. 1A). In comparison, only ≈70% of NSCLCs displayed moderate/intense staining (class 2 and 3 tumors) (SI Text), whereas 30% of all NSCLCs showed absent or barely detectable NUMB (class 1 tumors) (Fig. 1A, Table S1). There was no correlation between loss of NUMB expression and tumor histotype (adenocarcinoma vs. squamous cell carcinoma, P = 0.69), indicating that the two major types of NSCLC harbor alterations of NUMB expression with similar frequency.
Fig. 1.
Loss of NUMB expression in NSCLC. (A) NUMB expression in NSCLCs detected by IHC. Entire TMA cores, from representative samples, are shown on the left at an original magnification ×20; the boxed areas are magnified on the right (original magnification ×40) to show the typical cytosolic and plasma membrane localization of Numb. (B) (Left) NUMB mRNA levels measured by Q-PCR in primary NSCLC cultures expressed relative to the reference cell line BEAS-2B (= 1). Results represent the mean mRNA level detected in cultures from four patients for each NUMB-class shown. (Right) NUMB protein levels in primary NSCLC cultures. Blots are representative of three independent experiments.
We established primary pure epithelial cultures from several NSCLCs. In these cultures, we detected comparable levels of NUMB mRNA in class 1 vs. class 3 tumors (Fig. 1B). This contrasts with the markedly different levels of NUMB protein in the same cultures (Fig. 1B). When class 1 cultures were treated with the proteasome inhibitor MG132, NUMB protein was restored to levels indistinguishable from those of class 3 cultures (Fig. 1B). Thus, loss of NUMB expression in NSCLCs is determined at the posttranslational level, through enhanced protein degradation, similarly to what was shown in breast cancers (16). Of note, we did not detect mutations in the NUMB coding sequence (cds) at the genomic level in 13 samples representative of the various classes of NUMB staining in NSCLC (see below).
Loss of NUMB Expression and Activation of Notch Signaling in NSCLCs.
We investigated whether loss of NUMB expression was associated with increased Notch signaling in a cohort of 49 NSCLC patients for whom both formalin-fixed paraffin-embedded (FFPE) and frozen specimens were available. We analyzed the levels of activated NOTCH-1 by IHC on FFPE specimens using an antibody (Ab) that specifically recognizes the activated version of the NOTCH-1 receptor (see SI Text and Fig. S1 for details). We observed a strong inverse correlation between the levels of NUMB and of activated–NOTCH-1 (Fig. 2 A and B). We then measured the expression levels of the Notch target gene, HES1 (4), in frozen samples. Also in this case, we observed a significant inverse correlation between HES1 mRNA levels and NUMB status (Fig. 2C, Table S2). Note that in Fig. 2 B and C, six patients harboring mutations of the NOTCH-1 gene (see below) were not included.
Fig. 2.
NUMB expression and Notch signaling in NSCLCs. (A and B) Inverse correlation between NUMB and activated-NOTCH-1 status in NSCLCs. (A) Representative IHC in serial sections (original magnification ×40). (B) Quantitative assessment of activated-NOTCH-1 in NSCLC as a function of NUMB-class. (C) HES1 mRNA levels in NSCLCs displaying different levels of NUMB. Data were obtained by Q-PCR on frozen specimens of the cohort of 49 patients and are expressed relative to the reference cell line BEAS-2B (= 1). The six patients harboring mutations in the NOTCH-1 gene (see Figs. 3A and 5A) were excluded from the results reported in panels (B) and (C). (D) NUMB-class (measured by IHC) and HES1 levels, measured as in (C), in 10 primary cultures from NSCLC patients.
We analyzed NUMB protein and HES1 mRNA levels in primary NSCLC cultures derived from 10 patients. Under these conditions, NUMB levels and Notch pathway activity could be analyzed in a pure epithelial tumor population. Again, we observed a strong inverse correlation between the levels of NUMB and of HES1 mRNA (Fig. 2D). We concluded that deregulation of NUMB is a frequent event in NSCLC, and that this correlates with the activation of the Notch signaling pathway.
Genetic Alterations of NOTCH-1 in NSCLC.
Despite the significant inverse correlation between NUMB protein and HES1 mRNA (or activated NOTCH-1) levels, there were a significant number of outliers. In particular, in some specimens (Table S2) we noted high levels of HES1 mRNA (> 2.0) together with high/intermediate levels of activated NOTCH-1, despite substantial levels of NUMB. We hypothesized that these patients might harbor activating NOTCH-1 mutations. Thus, we performed a mutation analysis of the 49 NSCLC cohort, by sequencing the entire C-terminal region of the NOTCH-1 cds (Fig. 3A). We found four different heterozygous NOTCH-1 mutations in six NSCLC samples (Fig. 3A, Fig. S2, Table S2). For patients 42 and 44, we confirmed the presence of mutations also in primary cultures (Fig. 6A). Of note, we also sequenced 45 breast cancers without detecting any mutations.
Fig. 3.
Mutations of the NOTCH-1 gene in NSCLCs. (A) Schematic representation of the NOTCH-1 protein with its domains: Ankyrin, ankyrin region; EC, extracellular; EGF-like, EGF-like repeats; HDC, heterodimerization domain C-terminal; HDN, heterodimerization domain N-terminal; LNR, Lin-12/Notch repeats; NICD, intracellular; N-N-TM, transmembrane; PEST, PEST region; RAM, RBP-J/Su(H)/CBF1-associated molecule; TAD, transactivation domain. Sequencing strategy indicating positions of the sequenced fragments (1–7) is shown (top). Locations of the detected mutations, confirmed in two independent experiments, are indicated by red dots (bottom). Note that mutations did not show evident specificity for tumor histotype, as they were detected both in adenocarcinomas (three of 34 cases) and in squamous cell carcinomas (three of 15 cases). (B) An example of a somatic mutation (D1643H) in NOTCH-1, in patient 40: chromatograms of normal and tumor cDNAs are shown (arrowheads indicate the position of the G to C mutation). The complete dataset is in Fig. S2. Also note that, in three cases (cases 36, 40, and 44), the mutations allowed restriction polymorphism analysis that confirmed the presence of WT alleles in the normal tissues and the presence of heterozygous mutations in the tumor samples (Fig. S2). (C) HES1 mRNA levels in frozen normal (N) and tumor (T) lung tissue specimens from patients harboring mutations of the NOTCH-1 gene. Data are expressed relative to the reference cell line BEAS-2B (= 1). Asterisks indicate a P value <0.01 in tumor vs. normal.
Fig. 6.
Inhibition of Notch signaling affects the growth potential of primary NSCLC cultures harboring alterations of the Notch pathway. A. Eight NSCLC primary cultures, representing 3 groups based on NOTCH-1/NUMB status, were tested for their sensitivity to the GSIs MRK-003 and DAPT. The characteristics of the primary cultures are summarized in the panel. (B and C) Survival of the primary NSCLC cultures, after treatment with MRK-003 (B) or DAPT (C) with respect to vehicle (DMSO). Results are expressed as the mean ± SEM of two independent experiments. (D) Summary of GSI experiments. Survival, with respect to vehicle-treated cells, is shown for the three types of NSCLC cultures described in (A): filled bars, “WT” cells (no alterations in the Notch pathway, see main text); empty bars, NUMB-negative cells; gray bars, NOTCH-1-mutated cells. Results for primary cultures belonging to the same group were combined and are expressed as the mean ± SEM of two independent experiments. Asterisks indicate a significant difference (P < 0.01) relative to vehicle-treated cells as well as to WT cells treated with MRK-003 (left) or DAPT (right).
Alterations in the NOTCH-1 cds might represent somatic mutations or polymorphisms. Thus, we sequenced the NOTCH-1 cds in matched normal samples, where these were available. For patients 36, 40, 42, 44, and 49 (Table S2), normal frozen lung tissue was used; for patient 42, normal lymphocytes were also tested. In all cases, wild-type (WT) NOTCH-1 alleles were detected in the normal tissues (Fig. 3B, Fig. S2).
Because of the limited availability of tumor tissues, we performed our analysis at the cDNA level. It was important, therefore, to confirm that the mutations in the NOTCH-1 cDNAs corresponded to mutations in the NOTCH-1 gene. In four cases (cases 36, 40, 42, and 49, representing the four different mutations), we had enough material to perform this analysis, which confirmed the presence of the mutations at the genomic level (Fig. S3).
We also performed a mutational analysis of the NUMB cds, using genomic DNA extracted from FFPE samples, from four NSCLCs with low NUMB expression (patients 34, 36, 39, and 46) and nine NSCLCs with intermediate/high NUMB levels (patients 4, 9, 13, 33, 38, 40, 42, 47, and 49, four of whom also displayed mutations of the NOTCH-1 gene). This analysis did not reveal any mutations in the NUMB cds (Table S2).
Finally, the availability of matched normal/tumor tissues allowed us to analyze the activation of the Notch pathway, as a function of the malignant conversion. The levels of HES1 mRNA were higher in tumor frozen tissues compared to the normal counterparts for all patients tested (Fig. 3C). Comparable results were also obtained for FFPE samples, which were used to test both HES1 and another Notch target gene, HEY2 (Fig. S4).
NOTCH-1 Mutations in NSCLC Are Gain-of-Function Mutations.
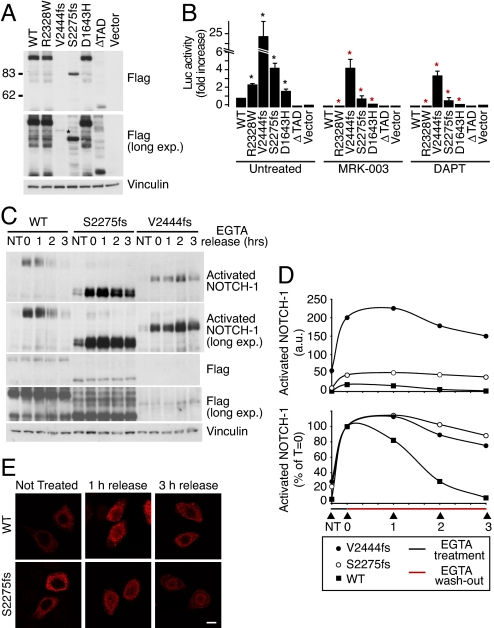
To prove that the NOTCH-1 mutations in NSCLC are gain-of-function mutations, we cloned them in the context of full-length NOTCH-1 (Fig. 4A), and tested their activity in a luciferase reporter assay. Increased luciferase levels were detected in cells transfected with NOTCH-1 mutants compared to WT transfectants (Fig. 4B). This increase was reversed by treatment with two different GSIs, MRK-003 (15, 18) and DAPT (Fig. 4B). Finally, by using the anti-activated–NOTCH-1 Ab, we directly demonstrated an increased basal level of activation for the NOTCH-1 mutants compared with WT NOTCH-1 (Fig. 4C, Fig. S5A, lanes NT).
Fig. 4.
NOTCH-1 mutations in NSCLCs are gain-of-function mutations. (A) Expression of flag-tagged NOTCH-1 constructs (shown on top) in HeLa cells. ΔTAD, inactive NOTCH-1 used as negative control. Note that the V2444fs mutant was expressed at very low levels and was detectable only in long exposures (asterisk). (B) Basal activity of NOTCH-1 mutants. HeLa cells were transfected with NOTCH-1 constructs (as indicated in (A) and a CBF1-Luc reporter. Luciferase activity was then determined in standard conditions (untreated) or after treatment with the GSIs, MRK-003 (1 μM) and DAPT (1 μM), and is reported as the fold increase vs. WT NOTCH-1, for each condition. In untreated cells, asterisks indicate a significant difference (P < 0.01) vs. WT control. In treated cells, red asterisks indicate a significant difference (P < 0.01) vs. the same transfectants in untreated conditions. (C) The effect of mutations on NOTCH-1 activation. HeLa transfectants were treated for 20 min with 5 mM EGTA or left untreated (NT). Cells were then washed to remove EGTA and returned to culture for an additional 0–3 h (EGTA release = 0, 1, 2 or 3 h). The levels of activated and total (FLAG) NOTCH-1 in total cell lysates were determined. A longer exposure of the blot for activated NOTCH-1 (long. exp.) is also shown to better visualize the kinetics of activated NOTCH-1 extinction in WT cells and V2444fs transfectants. (D) Densitometric analysis of the immunoblots shown in (C). (Top) amount of activated NOTCH-1 normalized to the NT sample in HeLa-WT NOTCH-1 cells (= 1). Data are expressed as arbitrary units (a.u.) relative to anti-FLAG signal. (Bottom) amount of activated NOTCH-1 expressed as a percentage of T 0 (= 100%) for each transfectant. (E) Subcellular localization of WT and S22275fs NOTCH-1. EGTA treatment and washout of HeLa cell transfectants was performed as in (C). The subcellular localization of NOTCH-1 was determined by immunofluorescence using an anti-FLAG antibody. Bar, 10 μM. Note how, in unstimulated cells, both WT and mutated NOTCH-1 displayed a predominantly extranuclear distribution. Upon EGTA treatment, a significant proportion of both WT and mutant NOTCH-1 translocated into the nucleus and was still visible in this location 1 h after washout. However, 3 h after washout, WT NOTCH-1 had completely disappeared from the nucleus, whereas persistent nuclear NOTCH-1 staining was detectable in S2275fs transfectants.
To increase our understanding of the impact of the mutations on NOTCH-1 activity, we measured the degree and duration of NOTCH-1 activation. NOTCH-1 activation was induced in HeLa transfectants by a short exposure to EGTA, which, by chelating calcium, activates the cleavage/activation sequence of NOTCH-1 (19). The levels of activated NOTCH-1, after treatment, were considerably more pronounced for the NOTCH-1 mutants compared with WT NOTCH-1 (Fig. 4 C and D, Fig. S5). EGTA was then washed away to measure the kinetics of extinction of NOTCH-1 activation. In WT transfectants, NOTCH-1 activation decayed with a half-life of ≈1/1.5 h. In contrast, in mutant transfectants, NOTCH-1 activation persisted for a longer time (≥3 h; Fig. 4 C and D and Fig. S5). This correlated with a persistence of NOTCH-1 in the nucleus, a hallmark of NOTCH-1 activation (Fig. 4E).
We concluded that the NOTCH-1 mutations in NSCLC are gain-of-function mutations.
Impact of Alterations of Notch Signaling in Lung Cancer.
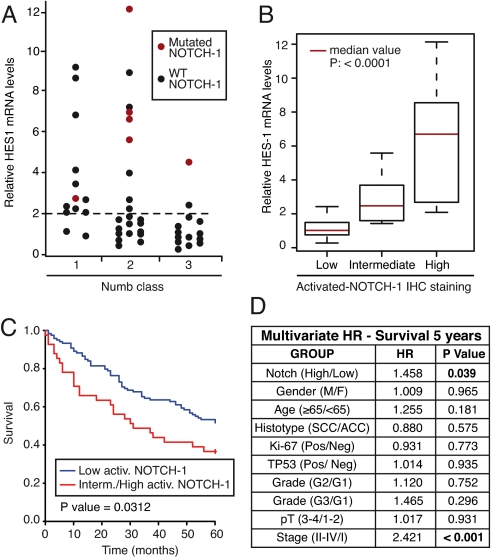
In the 49 NSCLC cohort, high levels of HES1 mRNA (≥2, an arbitrary threshold corresponding to twice the value of the reference sample) were observed in 22 (45%) cases (Fig. 5A). These 22 cases were distributed between the NUMB classes as follows: class 1, 11 of 13 samples (85%, note that in one case, 36, we also detected a NOTCH-1 mutation); class 2, 9 of 22 samples (41%), four of which harbored NOTCH-1 mutations (Fig. 5A and Table S2); class 3, 2 of 14 samples (14%, of which 40 also displayed a NOTCH-1 mutation).
Fig. 5.
Impact of alterations of Notch signaling in NSCLC. (A) Summary of the alterations of the Notch signaling pathway in the cohort of 49 NSCLC patients. HES1 mRNA levels are reported vs. NUMB status (as described in Fig. 2C). The six patients harboring NOTCH-1 mutations are indicated by red dots. (B) HES1 mRNA levels (expressed relative to the reference cell line BEAS-2B = 1) in NSCLCs displaying different levels of activated NOTCH-1 by IHC. mRNA levels were measured by Q-PCR on frozen specimens of the cohort of 49 patients. (C) The activation status of NOTCH-1 was used to predict overall survival in the subgroup of p53-negative NSCLC patients (i.e., patients without TP53 mutations, n = 176, 48.9% of the entire NSCLC cohort). Data are shown as the probability of survival, in Kaplan–Meier plots, as a function of low (Low) or intermediate-to-high (Interm./High) activated-NOTCH-1 levels. (D) The activation status of NOTCH-1 (low vs. intermediate-to-high activated-NOTCH-1 levels) was tested for prediction of survival in the same cohort of patients as in (C), in a multivariate comparative analysis using the indicated biological and biochemical parameters. HR, hazard ratio. A value of P < 0.05 was considered statistically significant.
Therefore, in 16 of 22 cases, high HES1 levels can be attributed to two distinct mechanisms: loss of NUMB expression or NOTCH-1 gain-of-function mutations (Fig. 5A). In addition, in the set of 49 tumors, there was very good correlation between HES1 mRNA levels and activated–NOTCH-1 (Fig. 5B). We note that, in three cases (cases 38, 45, and 47), we detected high levels of HES1 mRNA (≥4) together with intermediate/high levels of Notch activation, in the presence of intermediate levels of NUMB and in the absence of NOTCH-1 mutations. In these cases, the constitutive activation of the Notch pathway is not readily accounted for. It remains to be established whether additional, yet unidentified, molecular mechanisms of Notch activation are at play in these tumors.
To determine whether aberrant Notch signaling correlates with clinically relevant parameters, we analyzed an independent large case collection of NSCLCs for which clinical follow-up was available (the “clinical cohort”; Table S1 and Table S3). In this cohort, ≈26% of patients displayed intermediate/high levels of activated NOTCH-1 (Table S3). In the entire cohort, there was a trend correlating intermediate/high levels of activated NOTCH-1 with poorer clinical outcome, which, however, did not reach statistical significance (P = 0.09, Fig. S6). In the subgroup of patients without TP53 mutations (p53-negative patients; n = 176, 48.9% of the entire NSCLC cohort; Table S3), however, there was a significant correlation between Notch activation and poor prognosis (Fig. 5C). This correlation was maintained in multivariate analysis (Fig. 5D). Although these initial results need further corroboration, they suggest that Notch activation might impact on the natural history of NSCLC, at least in the subgroup of tumors without TP53 mutations.
A relevant question is whether alterations of the Notch pathway constitute the causal lesions, selected for during tumorigenesis, required for the maintenance of the malignant phenotype (driver mutations) or whether they are just part of the broad repertoire of molecular alterations occurring in the natural history of the tumor (passenger mutations). In the former case, one would expect NSCLCs harboring alterations of the Notch pathway to entirely rely on deregulated Notch signaling for sustained proliferative advantage. Thus, we tested the sensitivity of primary NSCLC cultures to the GSIs MRK-003 and DAPT. The selected primary NSCLC cultures belonged to three categories, based on the levels and modes of activation of the Notch pathway: WT, NUMB-Low, NOTCH-1-mutated (Fig.6A). The survival of WT cells was not significantly affected by GSIs (Fig. 6 B, C, and D). Conversely, the inhibitors selectively killed primary cells belonging to the NUMB-Low or to the NOTCH-1-mutated groups. We concluded that NSCLCs showing molecular alterations in the Notch pathway are dependent upon Notch signaling.
Discussion
We show that subversion of Notch signaling is frequent in NSCLC. Two types of alterations were detected: heterozygous mutations of the NOTCH-1 locus (≈10% of the cases), and loss of NUMB expression (≈30% of the cases).
In the case of NOTCH-1 mutations, we demonstrated that they represent gain-of-function mutations. We note that the NOTCH-1 mutations identified in this study occurred in the same regions previously identified as hot spots in T-ALL (1): the HD and the C-terminal/PEST region. It has been shown that mutations in the HD domain destabilize the NOTCH-1 heterodimer, prompting ligand-independent cleavage, whereas mutations within the PEST region result in the stabilization of the NICD and sustained NOTCH-1 signaling, probably because of alterations and/or removal of phosphodegron domains required for NICD degradation (1). Similar mechanisms are most likely at play in NSCLC. The “real” frequency of NOTCH-1 mutations in NSCLC remains to be determined. We found six mutations in 49 cases (≈12%). The limited size of the cohort does not allow definitive conclusions on the magnitude of the event. In a recent study by Ding et al. (20), performed in lung adenocarcinomas, NOTCH-1 mutations were not reported at high frequency. In that study, one mutation V1671I, which falls in the HD domain, was described, although it is not known whether it represented a polymorphism. Whether the differences between our study and that of Ding et al. are due to differences in study design (sequencing strategy) or to intrinsic differences in the patient cohorts remains to be established.
The second alteration of the Notch pathway in NSCLC is loss of NUMB expression resulting from enhanced NUMB degradation. It is possible that the instability of NUMB is caused by mutations in its cds. However, a mutational analysis of the NUMB gene, in tumors representative of the various NUMB subclasses, failed to evidence any such alteration. Moreover, in breast cancers, we reported a mechanism for loss of NUMB expression indistinguishable from that uncovered here for NSCLC, in the absence of any alteration of the NUMB locus (16). Thus, we rather favor the possibility that the primary lesion, responsible for loss of NUMB expression, affects a gene encoding for a protein involved in the control of NUMB stability: E3 ligases or de-ubiquitinating enzymes being the prime suspects.
We have previously proposed that NUMB is a tumor suppressor (16, 17). From our studies in breast cancer and in tumor cell lines, loss of NUMB expression can confer a proliferative advantage through two mechanisms: (i) lack of counteraction over Notch (16), and (ii) increased degradation of the tumor suppressor TP53 (17). In the case of NSCLC, a cogent case can be made that NOTCH-1 is activated in NUMB-defective tumors, with ensuing proliferative advantage, because (i) there was a very tight correlation between loss-of-NUMB and high levels of activated NOTCH-1 and of HES1 mRNA, and (ii) NUMB-negative primary NSCLC cultures were sensitive to GSIs.
It is more questionable whether NUMB control over TP53 plays a role in NSCLC. In the case of breast cancer, inactivating mutations of the TP53 gene, or alterations of circuitries regulating TP53 levels are relatively infrequent (21–28), thereby providing a strong rationale for how loss of NUMB and the ensuing increased TP53 degradation can confer a proliferative advantage (17). The situation is different in NSCLCs, which are characterized by a high frequency of mutations of the TP53 gene (29). Whether loss of NUMB plays a role in TP53-proficient NSCLC, through mechanisms similar to those described in breast cancer remains therefore to be established.
An important issue concerns whether alterations in Notch signaling in NSCLC extend beyond those reported here, as also supported by findings of overexpression of NOTCH-3 or of Notch ligands in this tumor (12–15). To this end, it is important to note that the anti-activated NOTCH Ab, that we used, will fail to detect tumors in which deregulated NOTCH signaling does not dependent on augmented production or stability of NICD of NOTCH-1. Indeed, in three cases of the 49-patient cohort, we detected high levels of the Notch target HES1, in the absence of alterations in the expression of NUMB or of activating mutations of NOTCH-1. Thus additional, yet undefined alterations of Notch signaling might be involved in NSCLC, although probably at low frequency.
Our results also suggest that altered Notch signaling affects the natural history of NSCLC. Indeed, we detected a correlation between the levels of activated NOTCH-1 and poor prognosis in the subset of NSCLC patients without TP53 mutations. Importantly, the correlation was maintained in multivariate analysis and was even independent of tumor stage, which is the strongest prognostic factor in NSCLC. We do not know why the correlation was only evident in the subgroup of patients in which TP53 is not mutated. One plausible explanation is that when TP53 is mutated, and therefore functionally subverted, the impact of aberrant NOTCH-1 activity on the natural history of the tumor might be masked by the plethora of genetic changes, and of their biological consequences, typically occurring in TP53-mutated cancers.
Finally, our data have therapeutic implications, as we show that primary NSCLC, harboring alterations in the Notch signaling pathway, are sensitive to GSIs. Because approximately one third of NSCLC cases display altered Notch signaling, in principle, these patients might benefit from treatment with GSIs. The prospective use of these inhibitors in the clinical setting has been plagued by their toxicity (7, 10). Recent developments, however, highlighted the possibility that combinatorial treatment with glucocorticoids might counteract the toxicity of GSIs (30, 31). The perspective development of combined GSI-based therapeutic protocols, with reduced toxicity, would constitute a major advancement in NSLC, in which the development of effective targeted therapies is still a largely unmet need (32). In this framework, the molecular alterations of the Notch pathway herein described might constitute an effective tool for the stratification of eligible patients.
Materials and Methods
NSCLC Specimens and Analyses.
All specimens were from lung cancer patients undergoing surgery at the European Institute of Oncology (IEO) in Milan, Italy. Ethics approval for tissue collection for research purposes was obtained from the IEO Institutional Review Board, after written informed consent had been obtained from all patients. Details on IHC analysis and NSCLC classification according to their NUMB or Activated-NOTCH status are given in SI Text. In SI Text, the methodology for mutational analyses of NOTCH-1 and NUMB, and for quantitative RT-PCR analysis of NOTCH-1 targets is also described.
Cell Lines, Expression Vectors for NOTCH-1 Mutants and Biochemical Studies.
Primary cultures were obtained as previously described (16) (SI Text). HeLa cells were transfected with Lipofectamine PLUS (Invitrogen). For luciferase assays (Dual Luciferase Kit, Promega), cells were transfected with the NOTCH-1 constructs together with a Notch-dependent CBF1-responsive luciferase reporter (6x-RBP-Jk-luc) (16) and a Renilla luciferase plasmid, and tested 48 h after transfection. GSIs were added to cells immediately after transfection. Luciferase activity was normalized to the Renilla transfection control and to NOTCH-1 expression levels. Results of three independent experiments performed in triplicate are shown. Immunofluorescence and immunoblotting were performed as described (17). Plasmids and antibodies are described in the SI Text.
Patients in Clinical Cohort and Statistical Analyses.
The clinical cohort of 420 consecutive NSCLC cases (Table S3) was constituted by patients who had undergone surgical resection at IEO between June 1998 and December 2002. Overall survival was defined as the interval between surgery and either death from any cause, or last contact. The median duration of follow-up was 62 months (range, 0–122 months). The 5-year survival rate was 52.0% (Stage I, 68.4%; Stage II–IV, 38.3%). During the 5-year follow-up period, a total of 200 (48%) events (deaths) were registered. Plots of the overall survival according to activated–NOTCH-1 expression were drawn using the Kaplan–Meier method. The statistical significance of differences in survival rates between groups was established by the log-rank test. Multivariate analyses were carried out using the Cox proportional hazards method to assess the prognostic value of activated–NOTCH-1 status before and after correction for different independent risk factors, including age at diagnosis of the tumor, pathological stage, tumor grade of differentiation, nodal status, TP53 status, and Ki-67. SAS statistical software was used for all of the analyses (SAS Institute, Inc.). A value of P ≤ 0.05 was considered significant.
Supplementary Material
Acknowledgments.
We are indebted to the following individuals: G. Veronesi for the acquisition of human tissue samples; G. Draetta, D. Bergstrom, and P. Strack at Merck Research Laboratories (MRL, Boston) for the MRK-003 compound and for sharing their unpublished results; G. Taliento, M. Simone, G. Paciucci, and M. Bianchi for technical assistance; the Biological Resource Center and the Tumor Registry at IEO and the Molecular Pathology Unit, the Real Time PCR and DNA Sequencing Service, the Imaging Service, the Monoclonal Service, and the tissue culture Service at the IFOM-IEO Campus, for technical support; and G. Goisis for statistical analyses. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC), Italian Ministry of Health, and MIUR (to S.P. and P.P.D.F.); the European Community (VI Framework), the CARIPLO foundation, the Ferrari Foundation, and the Monzino Foundation (to P.P.D.), and the G. Vollaro Foundation (to S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907781106/DCSupplemental.
References
- 1.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 3.Gonczy P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 4.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 5.Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Farnie G, Clarke RB. Mammary stem cells and breast cancer–role of Notch signalling. Stem Cell Rev. 2007;3:169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 7.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: Differentiation, cancer and adhesion. Curr Opin Cell Biol. 2008;20:171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 10.Shih Ie M, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 11.Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 12.Choi K, et al. Distinct biological roles for the notch ligands Jagged-1 and Jagged-2. J Biol Chem. 2009;284:17766–17774. doi: 10.1074/jbc.M109.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SM, et al. Expression of Notch 1 and 3 is related to inhibition of lymph node metastasis and progression in non-small lung carcinoma. Basic Applied Pathol. 2008;1:93–97. [Google Scholar]
- 14.Dang TP, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 15.Konishi J, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 16.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colaluca IN, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 18.Lewis HD, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Rand MD, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCann AH, et al. Amplification of the MDM2 gene in human breast cancer and its association with MDM2 and p53 protein status. Br J Cancer. 1995;71:981–985. doi: 10.1038/bjc.1995.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 23.Pharoah PD, Day NE, Caldas C. Somatic mutations in the p53 gene and prognosis in breast cancer: A meta-analysis. Br J Cancer. 1999;80:1968–1973. doi: 10.1038/sj.bjc.6690628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 25.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 26.Silva J, et al. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene. 2001;20:4586–4590. doi: 10.1038/sj.onc.1204617. [DOI] [PubMed] [Google Scholar]
- 27.Silva J, et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 28.Vestey SB, et al. p14ARF expression in invasive breast cancers and ductal carcinoma in situ—relationships to p53 and Hdm2. Breast Cancer Res. 2004;6:R571–R585. doi: 10.1186/bcr912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 30.Grosveld GC. Gamma-secretase inhibitors: Notch so bad. Nat Med. 2009;15:20–21. doi: 10.1038/nm0109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Real PJ, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.