Abstract
The pathogenesis of Huntington's disease (HD) remains elusive. The identification of increasingly early pathophysiological abnormalities in HD suggests the possibility that impairments of striatal medium spiny neuron (MSN) specification and maturation may underlie the etiology of HD. In fact, we demonstrate that HD knock-in (Hdh-Q111) mice exhibited delayed acquisition of early striatal cytoarchitecture with aberrant expression of progressive markers of MSN neurogenesis (Islet1, DARPP-32, mGluR1, and NeuN). Hdh-Q111 striatal progenitors also displayed delayed cell cycle exit between E13.5–15.5 (BrdU birth-dating) and an enhanced fraction of abnormal cycling cells in association with expansion of the pool of intermediate progenitors and over expression of the core pluripotency (PP) factor, Sox2. Clonal analysis further revealed that Hdh-Q111 neural stem cells (NSCs) displayed: impaired lineage restriction, reduced proliferative potential, enhanced late-stage self-renewal, and deregulated MSN subtype specification. Further, our analysis revealed that in addition to Sox2, the core PP factor, Nanog is expressed within the striatal generative and mantle regions, and in Hdh-Q111 embryos the fraction of Nanog-expressing MSN precursors was substantially increased. Moreover, compared to Hdh-Q18 embryos, the Hdh-Q111 striatal anlagen exhibited significantly higher levels of the essential PP cofactor, Stat3. These findings suggest that Sox2 and Nanog may play roles during a selective window of embryonic brain maturation, and alterations of these factors may, in part, be responsible for mediating the aberrant program of Hdh-Q111 striatal MSN specification and maturation. We propose that these HD-associated developmental abnormalities might compromise neuronal homeostasis and subsequently render MSNs more vulnerable to late life stressors.
Keywords: development, huntingtin, medium spiny neurons, neurodegeneration
Huntington's disease (HD) is caused by mutation in exon 1 of the gene that codes for huntingtin (Htt) (1). Although Htt is pan-neuronal (2), pathological changes in HD are selective, targeting mainly medium spiny neurons (MSNs) of the striatum (3). Research initiatives in pathological brain aging have traditionally focused on defining biological processes mediating neuronal dysfunction and death during adult life. However, there is increasing evidence that Htt has selective functions in the developing striatum (4, 5), suggesting that the nexus of HD pathogenesis may occur at earlier time periods. Accordingly, some reports have proposed that failure of normal brain development may lead to altered neuronal homeostasis and increased cellular vulnerability to late life stressors (6). In fact, cumulative reports set the beginning of abnormalities in HD before the occurrence of cell death and motor abnormalities (7–11). Some models even place the beginning of the disease at birth (12), when striatal neurogenesis is still occurring. Htt interacts with a wide spectrum of developmental factors and the mutation may compromise a subset of its functions resulting in subtle developmental defects that may have been overlooked. We therefore hypothesize that the HD mutation causes early molecular and cellular alterations that compromise the specification and maturation of MSNs and acquisition of the mature striatal chemoarchitecture. Our findings reveal that HD knock-in mice exhibited a series of developmental defects in striatal NSC-mediated MSN neurogenesis, from the stages of NSC maintenance and incipient MSN lineage specification to progressive neuronal maturation. Moreover, these HD-associated developmental deficits are linked to corresponding alterations in the deployment of the core PP factors, Sox2, and Nanog during sequential phases of striatal NSC maintenance and lineage restriction, and MSN lineage specification and maturation, thereby suggesting innovative molecular targets for therapeutic initiatives involving stem cell reprogramming.
Results
Impairment in Progressive Maturation of Striatal Medium Spiny Neurons and Deregulation of the Striatal Chemoarchitecture in Hdh-Q111 Mice.
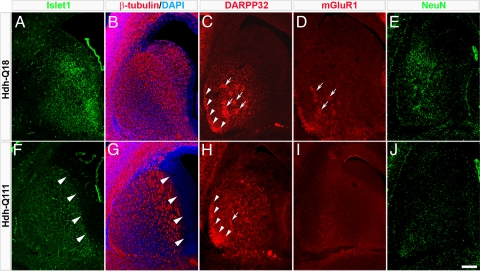
To define the maturational state of the striatum at E17.5, we examined the profiles of expression of markers of MSN specification and maturation (Islet1, β-tubulin, DARPP32, mGluR1, and NeuN). Compared to Hdh-Q18 embryos, expression of Islet1 and β-tubulin in paramedian germinative areas was substantially reduced in Hdh-Q111 embryos (Fig. 1 A–B and F–G, arrowheads). Further, in contrast to the patchy distribution of DARPP32 in the Hdh-Q18 striatum, DARPP32 was diffusely expressed throughout the Hdh-Q111 striatum (Fig. 1 C and H, arrows). Moreover, the Hdh-Q111 subcallosal streak, a murine subcompartment of the striosome, lacked the typical thin crescent moon shape, and instead appeared thickened and poorly defined (Fig. 1 C and H, arrowheads). Likewise, mGluR1 did not display the patchy distribution normally observed at this time (Fig. 1 D and I, arrowheads), and expression of NeuN was notably reduced in Hdh-Q111 embryos (Fig. 1 E and J). Conversely, at postnatal day 2 (PND2), the Hdh-Q111 striatum exhibited the expected striosome profile defined by patchy expression of μ-opioid receptor 1 (MOR1) (Fig. S1 A and C, arrowheads). In addition, expression of calbindin (CB), a matrix maturation marker, although detected in both models at PND7, did not display the typical CB mosaic pattern in the Hdh-Q111 model (Fig. S1 B and D, arrowheads). These findings reveal impairments in the acquisition of the cytoarchitecture of striatal subcompartments, therefore suggesting abnormalities in Hdh-Q111 MSN specification.
Fig. 1.
Comparative immunofluorescence micrographs of the E17.5 striatum revealed impaired acquisition of the Hdh-Q111 striatal cytoarchitecture. Compared to Hdh-Q18 embryos, the expression of Islet1 (A and F) and β-tubulin (C and H) were reduced in the Hdh-Q111 striatal germinative region (arrowheads). DARPP32 expression in the Hdh-Q111 striatum was also poorly organized in patch clusters (C and H, arrows) and the subcallosal streak appeared to be immature (arrowheads). Hdh-Q111 embryos failed to exhibit the patchy distribution of mGluR1 (D and I, arrowheads) and NeuN expression was visibly reduced (E and J). (Scale bar, 200 μm.)
Defects of Striatal MSN Lineage Elaboration and Cell Cycle Progression in Hdh-Q111 Mice.
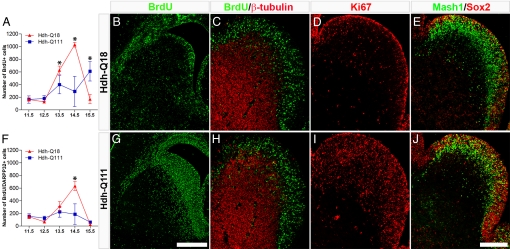
To further examine striatal lineage specification, we next defined the temporal profiles of cell cycle exit of neural progenitors by BrdU birth-dating paradigms (Fig. 2). Compared to the Hdh-Q18 model, Hdh-Q111 embryos displayed significantly lower numbers of striatal BrdU+ cells between E13.5 and E14.5 (Fig. 2A). Conversely, by E15.5 a higher number of Hdh-Q111 striatal neural species had exited the cell cycle. These cells were abnormally retained in the striatal subventricular and intermediate zones (SVZ and IZ) (Fig. 2 B and G) and few of them coexpressed DARPP32 (Fig. 2F).
Fig. 2.
Hdh-Q111 embryos display abnormal profiles of striatal neurogenesis and cell cycle progression. BrdU birth-dating analysis was performed using BrdU pulse administration at different developmental intervals (from E11.5 through E15.5), and brains were then harvested at E17.5 (A and F). Striatal BrdU+ cells (A) and BrdU+/DARPP32+ (F) cells were quantified and the mean ± SEM. of at least three independent biological replicas represented. *, all P values correspond to <0.05. The number of Hdh-Q111 cells born between E13.5 and E14.5 was significantly reduced, whereas the number of Hdh-Q111 neurons born at E15.5 was significantly increased when compared with Hdh-Q18 profiles. (B and G) Illustrate representative samples of E15.5 BrdU birth-dating. The presence of E14.5 striatal cells in S-phase were assessed by BrdU pulse labeling followed by embryo harvesting 30 min later (C and H). Equivalent profiles of BrdU+ cells, both negative and positive for β-tubulin were observed between Hdh-Q18 and Hdh-Q111 embryos. Profiles of expression of striatal Ki67 (D and I) and Sox2/Mash1 (E and J) were assessed by immunostaining at E14.5. Both Ki67 and Sox2 expression were enhanced in Hdh-Q111 embryos. (Scale bar, 500 μm in G and 200 μm in J.)
To define the nature of the changes in striatal birth-dating, we assessed the profiles of S-phase cells by BrdU pulse administration at E14.5. There were comparable patterns of BrdU+ cells, both positive and negative for β-tubulin, in Hdh-Q18 and Hdh-Q111 embryos, thus indicating that the complement of cells in S-phase was unchanged between models (Fig. 2 C and H). However, Hdh-Q111 embryos displayed higher numbers of Ki67+ cells (Fig. 2 D and I), suggesting that a greater fraction of these cells are in an active cell cycle state. Interestingly, at E15.5 the number of RC2+ radial glial intermediate progenitors was significantly higher in Hdh-Q111 than in Hdh-Q18 embryos (3.5 vs. 6.8% for Hdh-Q18 and Hdh-Q111 respectively; P = 0.0069) (Fig. S2A), in the absence of differences in the number of nestin+/RC2- cells. Despite the overall increase in RC2 immunoreactivity in Hdh-Q111 embryos, the expression of this marker in the medial aspect of the ventricular zone (VZ) of the ventral telencephalon was noticeably reduced (Fig. S2 B and C, arrowheads). Moreover, at this embryonic stage, electron microscopic analysis revealed that the number of cells in the Hdh-Q111 VZ exhibiting the typical radial glial morphology (Fig. S2D, asterisk) was substantially reduced and replaced by an abnormal population of rounded cells some with irregular and deeply invaginated nuclei (Fig. S2E, asterisk). These findings suggest that changes in cell cycle parameters previously observed are associated with abnormalities in the deployment of Hdh-Q111 radial glial intermediate progenitors.
To further define the abnormalities of Hdh-Q111 striatal intermediate progenitor cell cycle regulation and neuronal specification, we studied the profiles of expression of Sox2, a key regulator of neural stem and progenitor cell cycle progression and neurogenesis and Mash1, a seminal proneural factor of the ventral telencephalon at E14.5. We observed that the number of Sox2+ cells within the VZ/SVZ and IZ regions was higher in the Hdh-Q111 model, and many Sox2+ cells coexpressed Mash1 (Fig. 2 E and J). These results suggest that deregulation of Sox2 and Mash1 may be implicated in the abnormal program of Hdh-Q111 striatal neuronal lineage specification.
Complementary embryonic birth-dating experiments were used to further examine the profiles of striatal neurogenesis defined at PND2 (S3). In contrast to the more dorsal distribution of Hdh-Q18 striatal neurogenesis at E15.5, Hdh-Q111 cellular species were distributed throughout the striatum (Fig. S3 E arrowheads and F, respectively). Moreover, compared to Hdh-Q18 embryos, a substantially lower number of cells were elaborated between E16.5 and E17.5 in Hdh-Q111 embryos (Fig. S3 G–J). Paradoxically, at E18.5 Hdh-Q111 embryos displayed enhanced elaboration of striatal cells (Fig. S3 K and L). These data provide additional evidence for alterations in the temporospatial profiles of Hdh-Q111 striatal neurogenesis.
Striatal Hdh-Q111 Neural Stem Cell Species Exhibited Reduced Proliferation Potential, Enhanced Late-Stage Self-Renewal, Impaired Neural Lineage Restriction, and Abnormal MSN Subtype Specification.
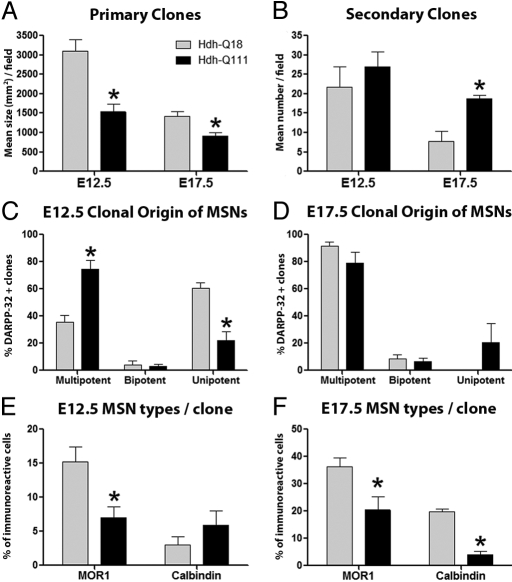
We next used in vitro clonal expansion and differentiation assays to determine the identity and functional properties of early (E12.5) and late (E17.5) embryonic striatal stem and progenitor species that give rise to MSN subtypes (Fig. 3). At both embryonic stages, Hdh-Q111 primary clones were significantly smaller than -Q18 clones (Fig. 3A) and their smaller size was not a consequence of enhanced cell death. Indeed E12.5 Hdh-Q111 clones showed fewer TUNEL positive cells, suggesting that the reduction in Hdh-Q111 clonal size was a consequence of changes in clonal proliferation (Fig. S4). The number of E17.5 Hdh-Q111 secondary clones was increased, compared to Hdh-Q18 clones (Fig. 3B), indicating enhanced late-stage Hdh-Q111 NSC self-renewal. Clonal differentiation assays at E12.5 demonstrated that twice the number of Hdh-Q111 MSNs originated from multipotent clones, whereas Hdh-Q18 MSNs were predominantly derived from unipotent clones (Fig. 3 C and D). Conversely, at E17.5 no differences were observed in the fraction of MSNs originating from multipotent clones (Fig. 3D). However, in contrast to Hdh-Q18 clones, a substantial number of Hdh-Q111 MSNs also originated from unipotent clones. These findings suggest the presence of stage-specific developmental impairments in Hdh-Q111 NSC maintenance and lineage restriction. The observation that MOR1+ and CB+ MSN subtypes are elaborated from individual clones at both E12.5 and E17.5 strongly suggest that striosome and matrix MSNs are generated from a common NSC (Fig. 3 E and F). The clonal analysis also revealed that the overall fraction of MOR1+ cells generated from early clones and both MOR1+ and CB+ cells generated from late clones were significantly reduced in the Hdh-Q111 model (Fig. 3 E and F), further supporting the existence of defects in Hdh-Q111 MSN subtype specification.
Fig. 3.
Hdh-Q111 neural stem and progenitor cell species exhibit reduced proliferation potential, enhanced late-stage self-renewal and impaired generation of MSN subtypes. Clonal expansion assays revealed reduced mean size of E12.5 and E17.5 Hdh-Q111 primary clones (A), while stem cell self-renewal was enhanced in Hdh-Q111 secondary clones at E17.5 (B). The lineage composition of clones that give rise to MSNs (DARPP32+ cells) (C and D) was examined by quantifying the proportion of clones that expressed three (multipotent), two (bipotent) or one (unipotent) neural lineage markers: β-tubulin, O4, and GFAP. Hdh-Q111 MSNs originated predominantly from multipotent clones at E12.5 (C), whereas a substantial fraction of these cells originated from unipotent clones at E17.5 (D). Compared to the Hdh-Q18 model, the fractions of MOR1+ cells generated from early clones (E) and of both MOR1+ and CB+ cells generated from late clones (F) were significantly reduced in the Hdh-Q111 model. Bars in A–F represent the mean ± SEM. of at least four independent biological replicas; *, all P values correspond to <0.05.
Deregulation of Key Members of the PP Factor Network During Hdh-Q111 Embryonic Striatal Development.
Alterations of Sox2 at E14.5 prompted us to also examine Sox2 expression at embryonic stages associated with striosome and matrix MSN specification (E12.5, E17.5, respectively). At E12.5, Sox2 expression was mostly localized to germinative zones of both models. However, scattered cells expressing low levels of Sox2 were also observed in the Hdh-Q18 mantle (Fig. 4 D and H). Immunoblot analysis of nuclear fractions of E12.5 LGE and CGE specimens demonstrated that Sox2 expression was significantly lower in the Hdh-Q111 than the Hdh-Q18 model (0.083 ± 0.01 vs. 0.25 ± 0.06, respectively; P = 0.0317). Conversely, at E17.5 Hdh-Q111 embryos exhibited higher Sox2 protein levels (0.27 ± 0.02 vs. 0.59 ± 0.09 for Hdh-Q18 and Hdh-Q111, respectively; P = 0.0149).
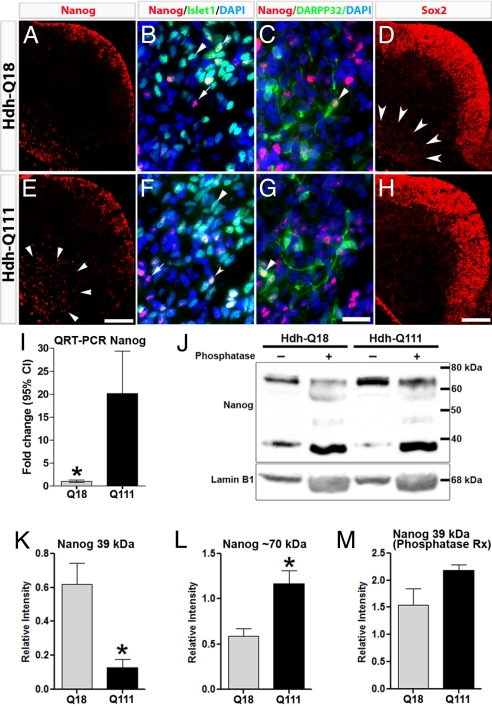
Fig. 4.
Expression profiles of Nanog and Sox2 are deregulated in Hdh-Q111 embryos at E12.5. Nanog displayed comparable expression profiles in LGE germinative zones of both models (A and E), whereas in the Hdh-Q111 mantle Nanog expression was substantially increased (arrowheads). Within the mantle region, Nanog colocalizes with the MSN markers, Islet1 (B and F, arrowheads) and DARPP32 (C and G, arrowheads). Sox2 immunoreactivity in the E12.5 LGE generative zone was equivalent in both models (D and H), whereas Sox2 expression (arrowheads) was reduced in the Hdh-Q111 mantle. (Scale bar, 100 μm (E and H) and 20 μm (G).) QRT-PCR analysis revealed that Nanog transcript expression was significantly increased in Hdh-Q111 relative to Hdh-Q18 specimens at E12.5 (I). Bars in I represent the mean ± 95%CI of four independent biological replicas. Immunoblot analysis of Nanog in E12.5 specimens (J) demonstrated a lower relative intensity of the approximately 39-kDa band (K) and a higher relative intensity of the approximately 70-kDa band (L) in the Hdh-Q111 compared to Hdh-Q18 specimens, with normalization of the levels of expression of the 39-kDa band following phosphatase treatment (M). Bars in K–M represent the mean ± SEM. of four independent biological replicas. *, all P values correspond to <0.05.
Alterations in Sox2 in the Hdh-Q111 model may suggest impairments of additional members of the core PP network (e.g., Nanog, Oct4). We found that Nanog was expressed at E12.5 within the germinative zones of the striatal anlagen at comparable levels in both models (Fig. 4 A and E). Conversely, at this time immunoreactivity of Nanog in the Hdh-Q111 mantle was substantially higher than in the Hdh-Q18 model (arrowheads). Consistently at E12.5, Nanog transcript expression was significantly higher in Hdh-Q111 relative to Hdh-Q18 specimens (20.14-fold change for Hdh-Q111 relative to -Q18, P < 0.001) (Fig. 4I). E12.5 immunoblot analysis revealed two discrete bands, one at approximately 39 kDa, the expected size for Nanog, and one at approximately 70 kDa (Fig. 4J). The relative intensity of the band at 39 kDa was significantly lower in Hdh-Q111 as compared to Hdh-Q18 specimens (Fig. 4K) (P < 0.01), whereas the relative intensity of the band at 70 kDa was significantly higher in Hdh-Q111, as compared to Hdh-Q18 models (Fig. 4L) (P < 0.008). Nanog is known to undergo posttranslational phosphorylation (13). As a corollary, phosphatase treatment revealed substantial and preferential increase in the intensity of the 39-kDa Nanog band in the Hdh-Q111, as compared to the Hdh-Q18 model (16.8-fold vs. 2.5-fold, respectively), resulting in comparable levels of the 39-kDa bands (Fig. 4M), thereby suggesting that in the Hdh-Q111 striatal anlagen Nanog is differentially phosphorylated. Interestingly, at E17.5 in both Hdh-Q18 and -Q111 models Nanog was preferentially phosphorylated, suggesting that Nanog may display developmental stage-dependent posttranslational modifications. Finally, immunolocalization studies between E12.5 and E14.5 revealed that Nanog is expressed in both models in many Islet1+ MSN precursors (Fig. 4 B and F) and in some DARPP32+ MSNs (Fig. 4 C and G, arrowheads and Fig. S5), whereas by E17.5 Nanog immunostaining was not detected (Fig. S6).
We next quantified protein levels of phosphorylated (active) forms of Stat3 and Smad1, two essential and interrelated factors regulating PP gene networks. Consistently, the E12.5 and E17.5 Hdh-Q111 striatal primordia displayed significant higher levels of nuclear phosphorylated Stat3 than those of Hdh-Q18 embryos (E12.5: 7.75 ± 1.3 vs. 14.03 ± 0.7 for Hdh-Q111 and Hdh-Q18, respectively; P = 0.005; E17.5: 0.29 ± 0.05 vs. 0.67 ± 0.03 for Hdh-Q111 and Hdh-Q18, respectively; P = 0.0002) (Fig. S7 A and C). Conversely, no differences between Hdh-Q18 and -Q111 specimens were detected for nuclear phosphorylated Smad-1 at E12.5 (0.73 ± 0.14 vs. 0.35 ± 01 for Hdh-Q18 and Hdh-Q111 respectively; P = 0.063) (Fig. S7B) and at E17.5 (0.38 ± 0.07 vs. 0.30 ± 0.01 for Hdh-Q18 and Hdh-Q111, respectively; P = 0.289) (Fig. S7D), although a trend toward lower levels of this protein were observed in Hdh-Q111 embryos. These findings suggest that deregulation of core PP factors in Hdh-Q111 developmental species is associated with corresponding and selective impairments in coregulatory PP-associated signaling pathways.
Discussion
We have identified molecular and cellular alterations in the developmental program of specification and maturation of MSNs and in the acquisition of the mature striatal cytoarchitecture in a knock-in model of HD. Our in vivo observations suggest the presence of defects in cell cycle progression consistent with abnormal phase lengthening or transient phase arrest in Hdh-Q111 striatal intermediate progenitors. This is supported by the finding of an overall increase in the number of Ki67+ cells in the E14.5 generative zone without a corresponding increase in the number of cycling cells in S-phase. Moreover, at this time, there is a reduction in the number of striatal cells exiting cell cycle with a corresponding increase in the number of intermediate progenitor species. Complementary in vitro studies support and extend the in vivo findings by demonstrating clonal size reductions in association with impairments of striatal NSC lineage restriction and specification of MSN subtypes, further illustrating the broad developmental consequences of Hdh-Q111-mediated deregulation of cell cycle parameters. The abnormalities of striatal neurogenesis are in contrast to the absence of consistent neurogenic abnormalities in other areas of the embryonic forebrain and diencephalon, thereby suggesting a developmental basis for the enhanced vulnerability of MSNs.
We also observed enhanced Hdh-Q111 NSC self-renewal at late embryonic stages together with high levels of Sox2, a factor that mediates NSC self-renewal, multilineage potential, and neurogenesis (14, 15). Our findings of deregulation of Sox2 in a HD knock-in model may not only explain the developmental abnormalities we have uncovered, but may also explain the impairments in cellular proliferation and neurogenesis reported during the postnatal period in HD animal models and in postmortem specimens from HD patients (16–18). Deficiency of Sox2 has previously been associated with impaired neurogenesis and neurodegeneration (19). These observations coupled with our present findings further reinforce the importance of precise stoichiometric relationships for mediating the developmental functions of the core PP factors (20).
The spectrum of neural stem and progenitor cell abnormalities identified in concert with deregulation of Sox2 in the Hdh-Q111 model prompted us to examine additional core PP factors. Although isolated reports have suggested that these PP factors (e.g., Oct4) play roles in adult stem cells and in somatic cancers, it is widely accepted that they display unique roles before organogenesis (21, 22). However, our observations suggest that not only Sox2 but also Nanog but not Oct4 (Fig. S8) exhibit dynamic and combinatorial profiles of expression in neural stem and progenitor cells within embryonic telencephalic generative zones, where they may be involved in orchestrating stem cell maintenance and multilineage potential. Surprisingly, Nanog differentially reappears within immature MSN differentiation zones, frequently colocalizing with stage-specific MSN markers, suggesting roles in MSN subtype specification and progressive maturation. Indeed, genes with recognized functions in MSN neurogenesis represent downstream targets of these PP factors, including Epha4, RARs, Foxp1, and Notch1 (23). Thus, Sox2 and Nanog may regulate multilineage potential and the fidelity of NSC-mediated differentiation programs in ways that parallel the regulation of pluripotency and organogenesis by embryonic stem cells. Because the range of mature neuronal subtypes is almost as broad as the spectrum of somatic lineages elaborated following ESC differentiation, the functions of these core PP genes (e.g., Nanog) may have been exapted during vertebrate and mammalian nervous system phylogeny to provide a flexible and dynamic regulatory apparatus to ensure the fidelity of evolving neuronal subtype identity and connectivity. In this context, it is conceivable that disruption of components of the PP network in the HD knock-in model may underlie the observed alterations in specification and progressive maturation of early born striosome MSN precursors and, in turn, disrupt the overall developmental program of striatal functional compartmentalization.
The complex compartmentalization of the striatum and the establishment of intrastriatal functional circuits depend on the sequential elaboration of MSNs in two asynchronous waves of striosome/matrix neurogenesis (24). Just as the array of gradient morphogens, growth factors, and other trophic influences present in the extracellular matrix exhibit spatiotemporally distinctive expression profiles, the segregation of MSN neurogenesis into two waves implies that the molecular pathways that sculpt neuronal precursors to their final mature phenotype are unique for each neurogenic phase (25–27). The common theme underlying the developmental abnormalities we observed in the HD knock-in model is deregulation of the temporal and spatial profiles of striatal neurogenesis. As a consequence, Hdh-Q111 striatal species may be exposed to inappropriate molecular cues with the potential to drive them into aberrant programs of neuronal differentiation and ultimately into dysfunctional mature MSNs. These considerations presuppose that before the occurrence of HD-mediated striatal cell death, functional abnormalities are present that reflect these putative pathological alterations in striatal function. Indeed, cumulative clinical and neuroimaging evidence is consistent with this notion (7, 9, 10), and complementary studies in presymptomatic animal models of HD have also reported alterations in synaptic plasticity (11, 28). Therefore, despite the apparent normalization observed after the initial striatal developmental delay, the early HD-associated aberrant developmental program likely results in irrevocable changes in cellular homeostasis and stress responses, including processes known to lead to neurodegeneration (29). Definitive examination of the role of these developmental abnormalities in HD pathogenesis will require substitution of the wild type for the mutant Htt alleles at sequential stages of MSN neurogenesis to assess their effects in differentially modifying the progression of the neurodegenerative phenotype.
Materials and Methods
Animal Model.
We used knock-in control (Hdh-Q18) and HD knock-in (Hdh-Q111) mouse models in which murine Htt gene exon 1 was replaced by the corresponding human normal and mutant forms, respectively (30). All studies were conducted with the approval of the Animal Institute of the Albert Einstein College of Medicine.
Tissue Processing and Immunostaining.
Cellular preparations were fixed and processed for immunocytochemistry, as described in ref. 31. Tissue sections for immunohistochemistry and samples for electron microscopy were processed using standard techniques (SI Materials and Methods).
In Vivo BrdU Birth-Dating and Labeling of Cells in S-Phase.
IP BrdU pulses were administered at different embryonic times and brains were subsequently harvested either at E17.5 or at PND2, as described in ref. 32. For BrdU labeling of proliferating cells, single IP doses of BrdU (50 mg/kg total) were administered to E14.5 pregnant mice followed by embryo harvesting 1 h later.
Neural Cell Clonal Assays.
The clonal assay was performed with specific modifications (SI Materials and Methods) of the procedure described elsewhere (33).
Subcellular Fractionation and Protein Immunoblot Analysis.
Nuclear and cytoplasmic extracts from embryonic day (E), 12.5 lateral (LGE), and caudal (CGE) ganglionic eminences and E17.5 striatal primordia (four biological replicas/strain/embryonic stage) were isolated and further processed for immunoblotting, as described in ref. 34. Primary antibodies were used at specific dilutions (SI Materials and Methods). Protein dephosphorylation was performed by incubating 25 μg of nuclear fractions with 20 U of calf intestinal alkaline phosphatase (Invitrogen) for 15 min. For quantification, band intensity was measured using Kodak 1D 3.6 software and normalized to the corresponding levels of Lamin B1.
Quantitative Real-Time PCR Analysis.
RNA from the striatal anlagen was extracted from four separate litters, each composed of 10–14 embryos and further processed for cDNA synthesis using standard techniques. QRT-PCR was performed with TaqMan chemistry (Applied Biosystems) (SI Materials and Methods).
Statistical Analysis.
Comparison of proportions was evaluated by Fisher exact test while mean values by Mann–Whitney U test or the t test depending on whether the data described or did not represent a Gaussian distribution. Unless otherwise indicated, the statistical differences between the Hdh-Q18 and Hdh-Q111 samples were assigned using a probability of at least <0.05.
Supplementary Material
Acknowledgments.
We thank Dr. Marcy MacDonald for supplying invaluable mouse models. This work was supported by stipends from Fundadesarrollo and the University of Zulia, Maracaibo, Venezuela (A.E.M.), National Institutes of Health Grants NS39802 and MH66290, and the Roslyn and Leslie Goldstein, Mildred and Bernard H. Kayden and Alpern Family Foundations (M.F.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912171106/DCSupplemental.
References
- 1.U.S.-Venezuela Collaborative Research Project. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's disease collaborative research group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Strong TV, et al. Widespread expression of the human and rat Huntington's disease gene in brain and nonneural tissues. Nat Genet. 1993;5:259–265. doi: 10.1038/ng1193-259. [DOI] [PubMed] [Google Scholar]
- 3.Vonsattel JP, et al. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Woda JM, et al. Inactivation of the Huntington's disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo. BMC Develop Biol. 2005;5:17. doi: 10.1186/1471-213X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiner A, et al. Neurons lacking huntingtin differentially colonize brain and survive in chimeric mice. J Neurosci. 2001;21:7608–7619. doi: 10.1523/JNEUROSCI.21-19-07608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robins-Wahlin TB, Lundin A, Dear K. Early cognitive deficits in Swedish gene carriers of Huntington's disease. Neuropsychology. 2007;21:31–44. doi: 10.1037/0894-4105.21.1.31. [DOI] [PubMed] [Google Scholar]
- 8.Paulsen JS, et al. Brain structure in preclinical Huntington's disease. Biological Psychiatry. 2006;59:57–63. doi: 10.1016/j.biopsych.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Blekher T, et al. Saccades in presymptomatic and early stages of Huntington disease. [see comment] Neurology. 2006;67:394–399. doi: 10.1212/01.wnl.0000227890.87398.c1. [DOI] [PubMed] [Google Scholar]
- 10.Reading SA, et al. Functional brain changes in presymptomatic Huntington's disease. Annal Neurol. 2004;55:879–883. doi: 10.1002/ana.20121. [DOI] [PubMed] [Google Scholar]
- 11.Cybulska-Klosowicz A, et al. Impaired learning-dependent cortical plasticity in Huntington's disease transgenic mice. Neurobiol Disease. 2004;17:427–434. doi: 10.1016/j.nbd.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Penney JB, Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington's disease. Annal Neurol. 1997;41:689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- 13.Yates A, Chambers I. The homeodomain protein Nanog and pluripotency in mouse embryonic stem cells. Biochem Soc Trans. 2005;33:1518–1521. doi: 10.1042/BST0331518. [DOI] [PubMed] [Google Scholar]
- 14.Suh H, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Episkopou V. SOX2 functions in adult neural stem cells. Trends Neurosci. 2005;28:219–221. doi: 10.1016/j.tins.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Batista CM, et al. A progressive and cell non-autonomous increase in striatal neural stem cells in the Huntington's disease R6/2 mouse. J Neurosci. 2006;26:10452–10460. doi: 10.1523/JNEUROSCI.2850-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis MA, et al. The distribution of progenitor cells in the subependymal layer of the lateral ventricle in the normal and Huntington's disease human brain. Neuroscience. 2005;132:777–788. doi: 10.1016/j.neuroscience.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 18.Lazic SE, et al. Neurogenesis in the R6/1 transgenic mouse model of Huntington's disease: Effects of environmental enrichment. Eur J Neurosci. 2006;23:1829–1838. doi: 10.1111/j.1460-9568.2006.04715.x. [DOI] [PubMed] [Google Scholar]
- 19.Ferri AL, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 20.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136:2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengner CJ, Welstead GG, Jaenisch R. The pluripotency regulator Oct4: A role in somatic stem cells? Cell Cycle. 2008;7:725–728. doi: 10.4161/cc.7.6.5573. [DOI] [PubMed] [Google Scholar]
- 23.Sharov AA, et al. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Kooy D, Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain Res. 1987;401:155–161. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- 25.Skogh C, Campbell K. Homotopic glial regulation of striatal projection neuron differentiation. Neuroreport. 2003;14:1037–1040. doi: 10.1097/01.wnr.0000073424.02536.22. [DOI] [PubMed] [Google Scholar]
- 26.Jensen JB, Bjorklund A, Parmar M. Striatal neuron differentiation from neurosphere-expanded progenitors depends on Gsh2 expression. J Neurosci. 2004;24:6958–6967. doi: 10.1523/JNEUROSCI.1331-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janis LS, Cassidy RM, Kromer LF. Ephrin-A binding and EphA receptor expression delineate the matrix compartment of the striatum. J Neurosci. 1999;19:4962–4971. doi: 10.1523/JNEUROSCI.19-12-04962.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy KP, et al. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington's disease mutation. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington's disease. Eur J Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 31.Mehler MF, Mabie PC, Zhu G, Gokhan S, Kessler JA. Developmental changes in progenitor cell responsiveness to bone morphogenetic proteins differentially modulate progressive CNS lineage fate. Dev Neurosci. 2000;22:74–85. doi: 10.1159/000017429. [DOI] [PubMed] [Google Scholar]
- 32.Mason HA, et al. Notch signaling coordinates the patterning of striatal compartments. Development. 2005;132:4247–4258. doi: 10.1242/dev.02008. [DOI] [PubMed] [Google Scholar]
- 33.Yung SY, et al. Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral forebrain progenitor species. Proc Natl Acad Sci USA. 2002;99:16273–16278. doi: 10.1073/pnas.232586699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao D, Pang Z, Morgan JI. The structure and proteolytic processing of Cbln1 complexes. J Neurochem. 2005;95:618–629. doi: 10.1111/j.1471-4159.2005.03385.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.