Abstract
We propose an integrated computational model for the network of cyclin-dependent kinases (Cdks) that controls the dynamics of the mammalian cell cycle. The model contains four Cdk modules regulated by reversible phosphorylation, Cdk inhibitors, and protein synthesis or degradation. Growth factors (GFs) trigger the transition from a quiescent, stable steady state to self-sustained oscillations in the Cdk network. These oscillations correspond to the repetitive, transient activation of cyclin D/Cdk4–6 in G1, cyclin E/Cdk2 at the G1/S transition, cyclin A/Cdk2 in S and at the S/G2 transition, and cyclin B/Cdk1 at the G2/M transition. The model accounts for the following major properties of the mammalian cell cycle: (i) repetitive cell cycling in the presence of suprathreshold amounts of GF; (ii) control of cell-cycle progression by the balance between antagonistic effects of the tumor suppressor retinoblastoma protein (pRB) and the transcription factor E2F; and (iii) existence of a restriction point in G1, beyond which completion of the cell cycle becomes independent of GF. The model also accounts for endoreplication. Incorporating the DNA replication checkpoint mediated by kinases ATR and Chk1 slows down the dynamics of the cell cycle without altering its oscillatory nature and leads to better separation of the S and M phases. The model for the mammalian cell cycle shows how the regulatory structure of the Cdk network results in its temporal self-organization, leading to the repetitive, sequential activation of the four Cdk modules that brings about the orderly progression along cell-cycle phases.
Keywords: cellular rhythms, oscillations, mitotic oscillator, model, systems biology
In the presence of sufficient amounts of growth factor (GF), mammalian cells quit a quiescent state, denoted G0, and start their progression in the cell cycle (1–3). During the G1 phase, cells pass the restriction point, which is a point of no return beyond which they are irreversibly engaged in the cell cycle and do not require the presence of GF to complete mitosis (1, 2). Progression in the cell cycle is controlled by the sequential, transient activation of a family of cyclin-dependent kinases (Cdks), which allow an ordered succession of the cell-cycle phases G1, S, G2, and M (4, 5), even though there appears to be a certain overlapping of the different cyclins and Cdks (6). The Cdk proteins are active only when forming a complex with their corresponding cyclin. The cyclin D/Cdk4–6, cyclin E/Cdk2, cyclin A/Cdk2, and cyclin B/Cdk1 complexes promote, respectively, progression in G1, the transition to DNA replication in S, progression in S and transition to G2, and finally the G2/M transition allowing entry into mitosis (3–7). Cdk regulation is achieved through a variety of mechanisms that include association with cyclins and protein inhibitors, phosphorylation–dephosphorylation (8), and cyclin synthesis or degradation (9).
A number of theoretical models for the cell cycle have been proposed. Initially, these models pertained to the early cell cycles in amphibian embryos (10–14), which are relatively fast and consist of only two phases, interphase and mitosis (15). Chen et al. (16) later proposed a detailed computational model for the yeast cell cycle, which accounts for the behavior of a large number of mutants. Theoretical models were subsequently proposed for portions of the mammalian cell cycle, particularly the G1/S transition and the restriction point (17–21). A generic model for the eukaryotic cell cycle has also been presented (22). We still lack a detailed, integrative model coupling the different cyclin/Cdk complexes that control the successive phases of the mammalian cell cycle, which would be capable of describing their repetitive, sequential activation. Models of this sort were proposed for the yeast cell cycle in which a key role is played by cell growth; in those models mitosis is controlled by cell mass, which is treated as a bifurcation parameter (16).
Here, we propose a detailed integrative model for the cyclin/Cdk network that drives the mammalian cell cycle and explore the conditions for its temporal self-organization. Building on previous work that showed the occurrence of oscillations in models for the cell cycle in embryos (10–14) and yeast (16), and in less detailed or partial models for the mammalian cell cycle (17–22), we focus on the conditions in which the cyclin/Cdk network may function as a self-sustained biochemical oscillator solely as a result of its regulatory structure. To this end we disregard the control by cell mass, which appears less stringent in mammalian cells than in yeast (23). The model for the Cdk network contains four coupled modules centered on cyclin D/Cdk4–6, cyclin E/Cdk2, cyclin A/Cdk2, and cyclin B/Cdk1. The activity of the cyclin/Cdk complexes is regulated both through phosphorylation–dephosphorylation and reversible association with the protein inhibitors p21 or p27 (8, 24). The model includes the retinoblastoma protein (pRB) and the transcription factor E2F, which, respectively, inhibit and promote progression in the cell cycle. The Cdk network itself controls the balance between pRB and E2F through phosphorylation. Additional regulations of cyclin/Cdk complexes occur in the form of negative feedback, which arises from Cdk-induced cyclin degradation (9), and positive feedback, which originates from the fact that Cdks indirectly promote their own activation (25).
The model predicts that in the presence of suprathreshold amounts of GF the regulatory interactions within the Cdk network can spontaneously give rise to sustained oscillations corresponding to the repetitive, ordered activation of the various cyclin/Cdk complexes along the cell-cycle phases. Considered in turn are the existence of a restriction point in G1 and the need for a fine-tuned balance between pRB and E2F for oscillations to occur. By incorporating the kinases ATR (26) and Chk1 (27) we show that a DNA replication checkpoint slows down the dynamics of the network without modifying its oscillatory nature. Finally, the model accounts for truncated cell cycles corresponding to endoreplication, in which multiple rounds of DNA replication occur in the absence of mitosis (28).
Model for the Repetitive Activation of Cdks in the Cell Cycle
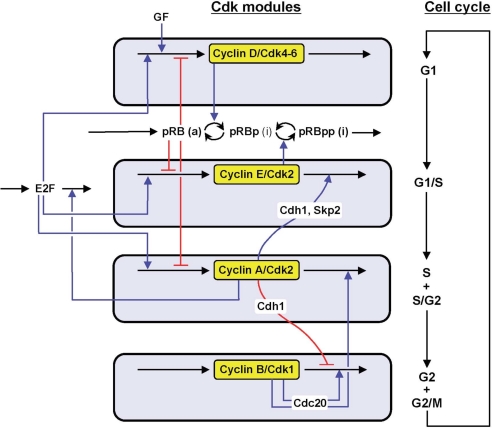
The model, schematized in Fig. 1, contains four modules corresponding to the sequential activation of the various cyclin/Cdk complexes. Modules 1–3 are centered on cyclin D/Cdk4–6, cyclin E/Cdk2, and cyclin A/Cdk2, respectively, whereas cyclin B/Cdk1 is at the core of module 4. The modules are coupled through multiple regulatory interactions, which are depicted in a more detailed manner in section 1 of SI Appendix, and more comprehensive schemes for modules 1–4 are presented in Figs. S1 and S2, together with a detailed description of each module. Variables are defined in Table S1, and a definition of parameters and a list of their numerical values are given in Table S2. The temporal evolution of the model is governed by a set of 39 kinetic equations, which are listed in section 2 of SI Appendix. These equations are based on mass action or Michaelian kinetics. To limit the complexity of the model we only consider the variation of protein levels without incorporating explicitly changes in the mRNAs.
Fig. 1.
Scheme of the model for the mammalian cell cycle. The model incorporates the four main cyclin/Cdk complexes centered on cyclin D/Cdk4–6, cyclin E/Cdk2, cyclin A/Cdk2, and cyclin B/Cdk1. Also considered are the effect of the GF and the role of the pRB/E2F pathway, which controls cell-cycle progression. Cyclin D/Cdk4–6 and cyclin E/Cdk2 elicit progression in G1 and the G1/S transition by phosphorylating and inhibiting pRB. The active, unphosphorylated form of pRB inhibits the transcription factor E2F, which promotes cell-cycle progression by inducing the synthesis of cyclins D, E, and A. Additional regulatory interactions are described in section 1 of SI Appendix where more detailed schemes for the whole network and each of its four modules are shown in Figs. S1 and S2. The combined effect of regulatory interactions between the four modules allows the cell to progress in a repetitive, oscillatory manner along the successive phases of the cell cycle, as depicted to the right.
Rather than attempting to attribute precise values to the parameters, many of which remain to be determined experimentally and vary in different cell types, we focus on the dynamic properties that emanate from the regulatory structure of the model. These properties will be explored over a large range of parameter values. The main goal of this study is to bring to light the modes of dynamic behavior that emerge from the intertwined regulations of the different modules forming the cyclin/Cdk network that drives the mammalian cell cycle.
Oscillatory Dynamics in the Presence of GF
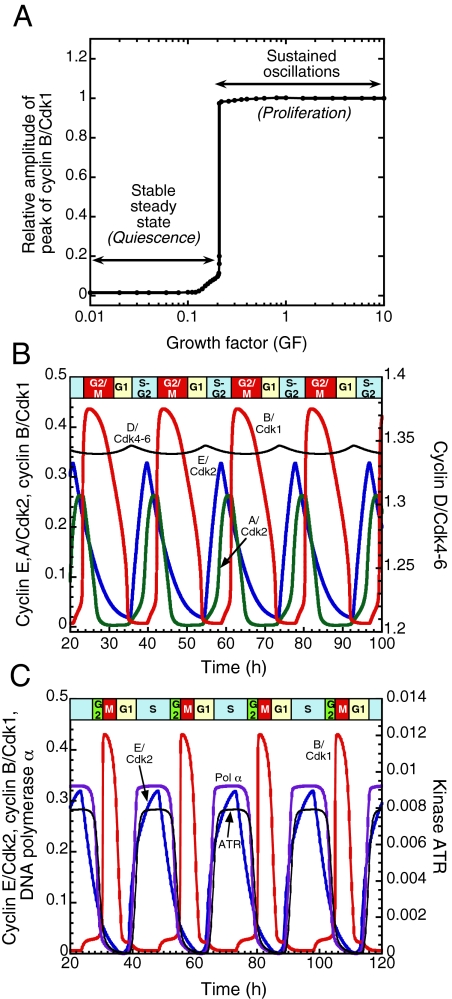
Healthy mammalian cells enter and progress in the cell cycle only in the presence of a sufficient amount of GF (1–3). Fig. 2A shows the dynamic behavior of the Cdk network as a function of the concentration of GF (see section 1 of SI Appendix for a detailed description of the role of GF). To characterize this behavior we plot the steady-state or maximum concentration in active cyclin B/Cdk1 complex as a function of GF. Before the addition of GF, cells reach a low, steady-state level of active cyclin/Cdk complexes. The model indicates that when GF exceeds a critical value (Fig. 2A) repetitive activation of the cyclin/Cdk complexes occurs in the form of self-sustained oscillations (Fig. 2B).
Fig. 2.
GF-induced oscillations in the Cdk network. (A) Below a sharp threshold in the concentration of GF, the Cdk network evolves to a stable steady state, whereas sustained oscillations occur above the threshold that corresponds to a bifurcation beyond which the steady state becomes unstable (see also Fig. 4). (B) Sustained oscillations correspond to the repetitive, ordered activation of the four cyclin/Cdk complexes. Cyclin D/Cdk4–6 and cyclin E/Cdk2 control progression in G1 and elicit the G1/S transition, whereas cyclin A/Cdk2 allows progression into S and G2. Finally, the peak in cyclin B/Cdk1 brings about the G2/M transition. The curves were generated by numerical integration of kinetic Eqs. 1–39 listed in section 2 of SI Appendix, for the parameter values listed in Table S2. Shown are the oscillations in the active forms of the cyclin/Cdk complexes. For cyclin D/Cdk4–6 the curve shows the evolution of the sum of the free form of the complex and its form bound to p21/p27. The oscillations are of the limit cycle type, i.e., they correspond in the phase plane to a unique closed trajectory (see Fig. 4B) that can be reached regardless of initial conditions. (C) Effect of the ATR/Chk1 checkpoint. The inclusion of the checkpoint lengthens the period, reduces the width of the peak in Cdk1, and results in a better separation of the peaks in cyclin E/Cdk2 and cyclin B/Cdk1. The curves showing the time evolution of cyclin E/Cdk2, cyclin B/Cdk1, DNA polymerase α, and kinase ATR were obtained by numerical integration of Eqs. 1–44 from SI Appendix for the parameter values listed in Table S2, with kce = 0.24 h−1 instead of 0.29 h−1 to further reduce the width in the peak of Cdk1. Concentrations are tentatively expressed in units of μM (see Table S2).
The ordered activation of the cyclin/Cdk complexes corresponds to the passage through the successive phases of the cell cycle (Fig. 2B). The peaks in the activity of cyclin D/Cdk4–6 and cyclin E/Cdk2 allow progression from G1 to S. The activity of cyclin A/Cdk2 rises in S and G2, while at the end of the cycle, the peak in cyclin B/Cdk1 brings about the G2/M transition. The level of cyclin D/Cdk4–6 remains elevated throughout the cycle, in agreement with experimental observations (29) and falls only when GF is removed. Below the GF threshold (Fig. 2A) cells are in a stable steady state, which can be associated with the quiescent phase G0. Above the threshold the repetitive activation of the Cdks can be associated with cell proliferation. Oscillations only occur in precise conditions, in a range bounded by critical parameter values. For an illustration see Fig. S4 A and B as a function of cyclins D and E and Fig. S4C as a function of phosphatase Cdc25 and the protein Cdh1 that controls cyclin B degradation.
Restriction Point in G1
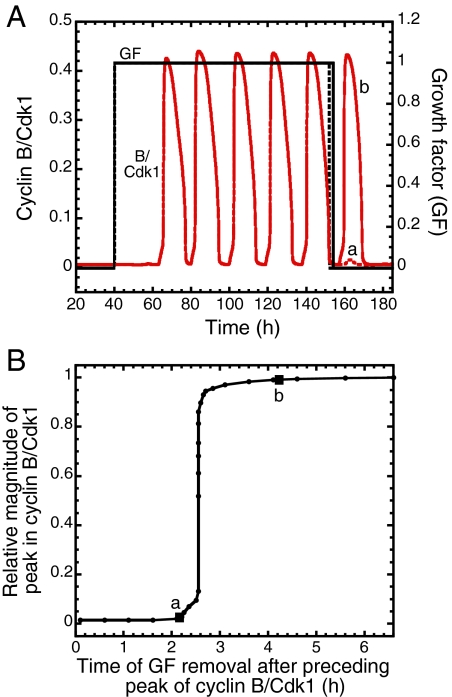
When GF is removed after a peak in Cdk1, oscillations disappear but a last, additional peak may or may not occur, depending on the time at which GF is withdrawn. If removal occurs before a critical time interval after the previous peak, oscillations vanish without production of an additional peak in Cdk1 (Fig. 3A, dashed line for GF and curve a for Cdk1). If, in contrast, GF is removed after the critical time interval, Cdk1 goes through an additional peak (Fig. 3A, solid line for GF and curve b for Cdk1). When plotting the relative magnitude of the additional peak in Cdk1 as a function of the time of removal of GF after the preceding peak in Cdk1, a sharp threshold is observed (Fig. 3B). The duration of the total cell cycle in the conditions of Figs. 2B and 3A is close to 19 h. In these conditions, the threshold interval predicted in Fig. 3B is of the order of 2.6 h. Given that the interval is calculated from the midpoint of the decrease in the preceding peak in Cdk1, it corresponds to a point in G1 because it occurs before the peak in cyclin E/Cdk2, which is associated with the G1/S transition.
Fig. 3.
Restriction point in G1. (A) When GF is removed early in G1 (vertical dashed line), the Cdk network returns directly to a stable steady state without producing the peak in cyclin B/Cdk1 associated with the G2/M transition (curve a). In contrast, when GF is removed slightly later in G1, after a critical time that defines the restriction point, cells complete the cell cycle, triggered by a peak in cyclin B/Cdk1 (curve b), before returning to G0. (B) The existence of a restriction point in G1 is reflected by the sharp threshold in the curve showing the magnitude of the peak in cyclin B/Cdk1 relative to the magnitude of the preceding peak in the presence of a suprathreshold level of GF, as a function of the time at which GF is removed after the preceding peak. In the case considered, the restriction point occurs ≈2.6 h after the end of the preceding cell cycle. The curves were generated by numerical integration as described in Fig. 2B for the same set of parameter values.
The restriction point is a point of no return located in the G1 phase of the cell cycle (1, 2, 30). The model suggests that the threshold in Fig. 3B corresponds to the restriction point in G1. Indeed, if GF is removed early in G1, cells miss the significant rise in Cdk1 required for the G2/M transition and return directly to a state characterized by a low level of cyclin B/Cdk1 (point a in Fig. 3B). If GF is removed later in G1, after the threshold, cells complete their cell cycle characterized by one additional peak of cyclin B/Cdk1 before returning to G0 (point b in Fig. 3B and curve b in Fig. 3A). A threshold in the activity of cyclin A/Cdk2 also exists as a function of the GF removal time. For the parameter values considered, this threshold occurs somewhat earlier in G1 and is less abrupt than the threshold observed for Cdk1. The cyclin A/Cdk2 threshold must be passed to initiate DNA replication, which is often taken experimentally as marker for passage through the restriction point (1, 30).
Control of Cell-Cycle Progression by the Balance Between E2F and pRB
pRB and the transcription factor E2F are two main regulators of the cell cycle (31, 32). pRB inhibits progression in the cell cycle by preventing the transcription of G1 cyclins D, E, and A and forming with E2F a dimeric, inactive complex. Formation of this complex counteracts the effect of E2F, which promotes the transcription of G1 cyclins (33). pRB is inactivated through phosphorylation by cyclin D/Cdk4–6 and cyclin E/Cdk2 (34) (see SI Appendix and Figs. S1 and S2).
The results indicate that progression in the cell cycle is controlled by the balance between E2F and pRB rather than by the absolute levels of these antagonistic factors: if the level of active pRB is too high, the cell stops in G1, whereas high levels of E2F promote cell-cycle progression (see SI Appendix and Fig. S3). In the model this balance is robust, because oscillations occur over several orders of magnitude of the rates of synthesis of pRB and E2F.
Mechanism of Oscillations in the Cyclin/Cdk Network
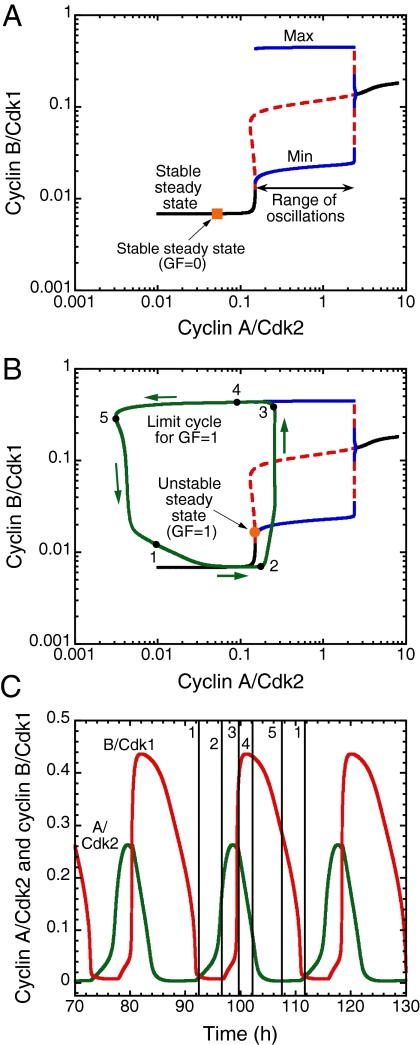
To clarify the nature of the dynamical transitions in the cell cycle it is useful to resort to bifurcation diagrams established for the various modules of the Cdk network, an approach pioneered by Novak, Tyson, and coworkers (12, 16, 21) in their models for the embryonic, yeast, and somatic cell cycles. The bifurcation diagram in Fig. 4A is obtained by determining the dynamic behavior of the Cdk1 module when the concentration of its input, cyclin A/Cdk2, is taken as a parameter governing the evolution of cyclin B/Cdk1. The diagram shows that as cyclin A/Cdk2 increases the steady-state level of cyclin B/Cdk1 is at first stable (the Cdk network cannot oscillate) before becoming unstable above a critical value of cyclin A/Cdk2, and then oscillations emerge. Beyond a second, higher critical value of cyclin A/Cdk2 the steady state recovers its stability and oscillations disappear.
Fig. 4.
Origin and mechanism of oscillatory behavior in the Cdk network driving the mammalian cell cycle. (A) Bifurcation diagram for the cyclin B/Cdk1 module. As a function of cyclin A/Cdk2 considered as a control parameter, black lines represent stable steady states, blue lines represent the minimum and maximum of the oscillations in cyclin B/Cdk1, and the red dashed line represents the unstable steady state. The stable steady state in the absence of GF is shown by an orange square. The bifurcation diagram indicates that sustained oscillations occur in the cyclin B/Cdk1 module when the level of cyclin A/Cdk2 exceeds a critical value. This situation prevails only in the presence of sufficient amounts of GF (see Fig. 2A). (B) Superimposed on the bifurcation diagram of A, the green curve shows the trajectory followed by the full system of 39 variables in the course of sustained oscillations, in conditions where the steady state (orange dot) is unstable, for GF = 1. This limit cycle trajectory represents a projection onto the cyclin B/Cdk1 versus cyclin A/Cdk2 phase plane, where cyclin A/Cdk2 now behaves as a variable, according to Eq. 22 in SI Appendix. Arrows indicate the direction of movement along the periodic trajectory. (C) Sustained oscillations. The vertical lines drawn at successive phases over one period of the oscillations correspond to points 1–5 on the limit cycle in B. The limit cycle in B and the curves in C were generated as in Fig. 2B, for the same set of parameter values, in the absence of the ATR/Chk1 checkpoint. As indicated in Fig. 2C, oscillations also occur in the presence of this checkpoint, with a narrower peak in Cdk1. Using cyclin A/Cdk2 as a parameter, the bifurcation diagram in A was established by means of the program AUTO (49) applied to the kinetic equations of the cyclin B/Cdk1 module, i.e., Eqs. 26–32 and 34–39 in SI Appendix from which the terms related to p27 were removed, so as to keep this module isolated from the other modules. The unstable steady state for GF = 1 in B was located by means of AUTO applied to Eqs. 1–39 in SI Appendix.
Rather than being a fixed parameter, cyclin A/Cdk2 behaves as a variable that evolves in the course of time, as prescribed by the first three modules of the Cdk network, according to Eq. 22 in SI Appendix. In response to such variation the Cdk1 module will evolve along the steady-state curve drawn on the bifurcation diagram of Fig. 4A. In Fig. 4B, superimposed on this bifurcation diagram is the projection onto the phase plane formed by cyclin B/Cdk1 and cyclin A/Cdk2 of the trajectory (green curve) followed by the full system of equations in the course of sustained oscillations, when GF is raised from zero up to a suprathreshold value. To further clarify the oscillatory dynamics we decompose one period of the oscillations in successive phases marked by points 1–5 on the closed curve in Fig. 4B and in the corresponding time series in Fig. 4C. The results of Fig. 4B suggest that the oscillations in Cdk1 can be viewed as a repetitive, transient excursion of Cdk1 into a domain of sustained oscillations. The first three modules of the Cdk network periodically push the Cdk1 module into this domain, from which it exits after one peak in Cdk1. The peak in Cdk1 indeed controls the dynamics of the other modules of the network by inducing a decrease in cyclin A/Cdk2 and cyclin B/Cdk1, through activation of Cdc20, which promotes degradation of cyclins A and B. The system thus resets and the first two modules cooperate to produce a new round of increase in cyclin A/Cdk2 that pushes again cyclin B/Cdk1 transiently into the oscillatory range.
Dynamics in the Presence of Checkpoints: Incorporation of the ATR/Chk1 Pathway
Checkpoints ensure that DNA replication and mitosis are correctly completed before the cell proceeds to the next phase of the cycle. Checkpoints are particularly crucial in the presence of cellular damage; if damage is too severe, the p53 pathway can induce apoptosis (35). Even during normal cell-cycle progression, the proper sequence of events must be highly regulated. To investigate how checkpoints affect the oscillatory dynamics of the Cdk network, we focus on one such endogenous checkpoint, mediated by the ATR/Chk1 pathway, which inhibits the phosphatases Cdc25 that activate Cdk2 and Cdk1. The checkpoint (see scheme in Fig. S5 and section 3 in SI Appendix, where the additional kinetic equations are given) is activated by cyclin E/Cdk2 at the initiation of DNA replication and ensures that mitosis will occur only when DNA replication is completed (26, 27, 36). Activation of this checkpoint can also be triggered after cellular damage, to delay cell cycle progression and allow for DNA repair (37).
Incorporating DNA polymerase, RNA primers synthesized by DNA polymerase, Cdc45, ATR, and Chk1 into the model allows us to characterize the dynamics of the mammalian cell cycle in the presence of the endogenous DNA replication checkpoint. The model indicates that the mere effect of the checkpoint is to slow down cell-cycle progression without altering the repetitive, oscillatory nature of cell-cycle dynamics (Fig. 2C). In the absence of the kinase ATR (Fig. 2B), the checkpoint is not activated and a partial overlap between the peaks in DNA polymerase and cyclin E/Cdk2 (corresponding to S phase) and cyclin B/Cdk1 occurs. Upon activating the checkpoint via cyclin E/Cdk2 in the presence of ATR (Fig. 2C), cell-cycle progression remains periodic but slows down : the period of the cell cycle now increases by several hours (the magnitude of the increase depends on parameters such as the rate of activation of ATR and the rate of cyclin E synthesis). The checkpoint thus acts as a braking mechanism (3) and creates a delay between the peaks in DNA polymerase and cyclin B/Cdk1, thereby improving the separation between S and G2/M. The duration of the peak in Cdk1, which appears to be large in Fig. 2B, is significantly reduced in Fig. 2C. As a result, the relative durations of the cell-cycle phases in the presence of the ATR/Chk1 checkpoint (Fig. 2C) are closer to those measured experimentally.
Discussion
To investigate the dynamics of the cyclin/Cdk network that controls the transitions between the successive phases of the mammalian cell cycle, we built a computational model incorporating the key regulatory interactions that couple four modules centered around cyclin D/Cdk4–6, cyclin E/Cdk2, cyclin A/Cdk2, and cyclin B/Cdk1. Of intermediate complexity, the model contains 39 variables, or 44 when including the ATR/Chk1 checkpoint. It is more comprehensive than partial or global models previously proposed for the mammalian cell cycle (17–22). It includes additional variables and more biochemical details about the regulation of the various cyclin/Cdk complexes, incorporates the role of a key checkpoint, and does not rely on control by cell mass.
The model shows that owing to the tight regulatory coupling of the four Cdk modules the cyclin/Cdk network is able to self-organize in time so as to operate in a sustained oscillatory manner corresponding to the repetitive, sequential activation of the cyclin/Cdk complexes. This periodic behavior occurs in a range that often extends over several orders of magnitude for many parameters (see, e.g., Figs. S3A and S4) so that sustained oscillations represent a robust mode of dynamic behavior of the Cdk network. Outside the domain of oscillatory behavior, which corresponds to cell proliferation, the cell cycle stops as the network reaches a stable steady state that can be associated with cellular quiescence.
The model accounts for a number of key properties of the mammalian cell cycle. First, sustained oscillations only occur above a critical level of GF (Fig. 2A). Without sufficient amount of GF cells remain in the quiescent phase. In the presence of suprathreshold levels of GF, cells enter the proliferative mode characterized by the repetitive, sequential activation of the cyclin/Cdk complexes that control the successive phases of the cell cycle. Second, the presence of GF is not required for generating an additional peak in Cdk1 when the time at which GF is removed in G1, after the preceding peak in Cdk1, exceeds a threshold value (Fig. 3). This result provides a dynamical basis for the existence of a restriction point in G1. Third, oscillations depend on a balance between the antagonistic effects of E2F and pRB, which promote or impede progression in the cell cycle, respectively (Fig. S3).
The model further matches experimental observations in a number of particular conditions. As shown in Fig. S6A (see section 4 in SI Appendix), the network can still oscillate in the presence of only Cdk1, which accounts for the observation that Cdk1 is sufficient for driving the mammalian cell cycle (38). Cell cycling can also occur spontaneously in the absence of pRB and GF (see Figs. S3A and S6B). This result holds with the observation that cells lacking the three forms of pRB (39) can proliferate despite serum starvation. The model also accounts for circadian entrainment of the cell cycle (40) upon coupling the latter to the circadian clock (see section 5 in SI Appendix and Fig. S7). Further comparison with experiments is presented in section 7 of SI Appendix.
Experimental observations (41) and theoretical studies (20, 41) indicate that bistability, i.e., the coexistence between two stable steady states in a given set of conditions may originate from the regulatory interactions between E2F and pRB. Such bistability can readily occur in the E2F/pRB module in the present model (see section 8 of SI Appendix). The phenomenon does not take place, however, for the parameter values listed in Table S2, which correspond to the conditions of Fig. 3. This finding implies that the sharp threshold observed in Fig. 3B for the restriction point does not necessarily rest on bistability in the E2F/pRB module, in contrast to conclusions based on the isolated module (41). The model suggests that the steep rise in the amplitude of the Cdk1 peak beyond the restriction point is associated with bistability in the Cdk1 module, which results from the positive feedback exerted by cyclin B/Cdk1 via its activation of Cdc25 and inhibition of Wee1 (see sections 1 and 8 of SI Appendix). Indeed, the rise in the amplitude of the peak in Cdk1 beyond the threshold in Fig. 3B becomes more progressive when the positive feedback loops leading to bistability are suppressed in the Cdk1 module. The occurrence of bistability in Cdk1 activation was established experimentally in amphibian egg extracts (13, 14), and recent observations in HeLa cells indicate that it plays a key role in the somatic cell cycle as well (42). One role of bistability in the various modules is to reinforce the all-or-none, relaxation nature of self-sustained oscillations in the Cdk network. Bistability thereby contributes to unify the two views of the cell cycle as dominoes versus clock (see ref. 43 and section 9 of SI Appendix).
We incorporated the intrinsic checkpoint based on the ATR/Chk1 pathway that inhibits Cdk2 and Cdk1 as long as DNA replication is not complete. The model shows (Fig. 2C) that the effect of this checkpoint is merely to act as a braking mechanism that allows for better separation between the S phase and the G2/M transition, without affecting qualitatively the oscillatory nature of the cell-cycle dynamics. Extrinsic checkpoints such as that involving the tumor suppressor p53 (35) activated upon DNA damage are expected to behave in a similar manner. In physiological conditions the checkpoint mediated by Chk1 plays a key role in cell-cycle progression (44). Its inclusion into the model brings the relative durations of the cell-cycle phases closer to experimental observations (Fig. 2 B and C).
Cancer is often linked to deregulation of the cell cycle. The present results suggest that a cancer cell behaves as a cell in which the mitotic clock fails to stop, in conditions where normal cells would settle in a stable steady state corresponding to quiescence or to a differentiated state. There are multiple entry points in which a modification of a biochemical parameter may induce the Cdk network to switch from a stable steady state to self-sustained oscillations. Many factors acting directly or indirectly on the Cdk network can trigger this transition and may therefore be viewed as oncogenic. For example, as shown in Fig. S4C, mutations reducing the activity of Cdh1, or overexpression of the phosphatases Cdc25 that activate Cdk2 or Cdk1, may tilt the balance toward proliferation (45, 46) by triggering the repetitive activation of Cdks when the cell is initially in a quiescent state.
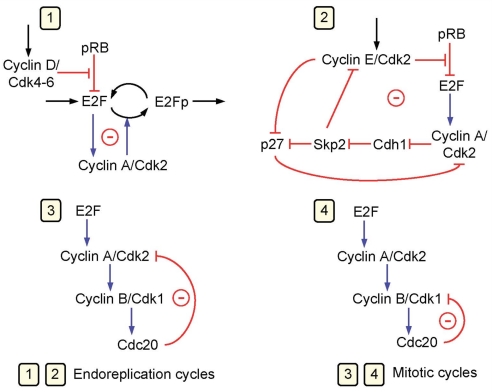
If the full Cdk network can globally operate in a periodic manner, the model predicts that it contains at least four oscillatory circuits, each of which can produce sustained oscillations on its own. When coupled, as occurs in physiological conditions, these circuits generally cooperate to produce the periodic, ordered activation of the cyclin/Cdk complexes that drive the successive phases of the cell cycle. The four oscillatory circuits in the Cdk network are schematized in Fig. 5. The oscillators all are based on negative feedback and were identified by numerical simulations. All contain cyclin A/Cdk2, but only two circuits also contain cyclin B/Cdk1 and can thus be viewed as mitotic oscillators producing a peak in cyclin B/Cdk1.
Fig. 5.
The Cdk network controlling the mammalian cell cycle contains multiple oscillatory circuits. Schematized are four circuits containing negative feedback loops that are capable of generating sustained oscillations in the model for the mammalian cell cycle. Cyclin A/Cdk2 is present in the four circuits, each of which can generate on its own sustained oscillations. In circuits 1 and 2, oscillations can occur in the absence of cyclin B/Cdk1, which is responsible for the entry of cells into mitosis. Oscillations in Cdk2, which controls DNA replication, occur in these circuits without any peak in Cdk1, a phenomenon known as endoreplication (28). In oscillatory circuits 3 and 4, mitotic oscillations involving repetitive activation of cyclin B/Cdk1 can occur, based on a negative feedback exerted via the protein Cdc20, which allows the degradation of either cyclin A or cyclin B in these circuits. In physiological conditions, all four oscillatory circuits synchronize to produce the ordered, repetitive activation of the different modules forming the Cdk network that drives the mammalian cell cycle.
In contrast, two subnetworks predict oscillations in cyclin A/Cdk2 in the absence of coupling to Cdk1. Even when the four oscillatory circuits are coupled, and thus include Cdk1, simulations indicate that these subnetworks may sometimes produce oscillations in Cdk2 without accompanying oscillations in Cdk1, or several peaks in Cdk2 may be produced for each peak in Cdk1. The former phenomenon corresponds to endoreplication, i.e., multiple rounds of DNA replication in the absence of mitosis (28, 47). Oscillatory circuits 1 and 2 produce Cdk1-independent Cdk2 oscillations and are therefore associated with endoreplication, whereas circuits 3 and 4 involve Cdk1 oscillations and are associated with periodic cell division. The possibility of endoreplication was previously reported in a model for the yeast cell cycle (48) and in a generic model for the eukaryotic cell cycle (22). Oscillatory circuit 4 is in fact closely related to the mitotic oscillator driving the early cell cycles in amphibian embryos (10–15). Rapid cycling likely associated with an oscillatory subnetwork involving cyclin B/Cdk1 was revealed by treatments perturbing the normal operation of the Cdk network (42).
The present results suggest that the sequential activation of the Cdk modules in the Cdk network is brought about by temporal self-organization corresponding to the global, periodic operation of the mammalian cell cycle. The first three modules of the network (see Fig. 1) centered on cyclin D/Cdk4–6, cyclin E/Cdk2, and cyclin A/Cdk2 cooperate to induce the transient firing of the last, embryonic-like, oscillatory module centered on cyclin B/Cdk1. The two modules at the top of the network elicit the increase in cyclin A/Cdk2 in module 3 that transiently drives module 4 into the domain of sustained oscillations (Fig. 4). The resulting pulse in cyclin B/Cdk1 triggers successively the decrease in cyclin A/Cdk2, the associated exit of circuit 4 from the oscillatory domain, and the return to conditions leading to the resumption of a new cell cycle.
Supplementary Material
Acknowledgments.
We thank the referees for fruitful suggestions. This work was supported by Fonds de la Recherche Scientifique Médicale Grant 3.4607.99, Belgian Federal Science Policy Office Grant IAP P6/25 (“BioMaGNet: Bioinformatics and Modeling - From Genomes to Networks”), and the European Union through Network of Excellence BioSim Contract LSHB-CT-2004-005137. C.G. is supported by a research fellowship from the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.J.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903827106/DCSupplemental.
References
- 1.Pardee AB. A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci USA. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle. 2002;1:103–110. [PubMed] [Google Scholar]
- 3.Morgan DO. The Cell Cycle: Principles of Control. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 4.Nurse P. Cyclin-dependent kinases and cell cycle control. ChemBioChem. 2002;3:596–603. doi: 10.1002/1439-7633(20020703)3:7<596::AID-CBIC596>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: Does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 7.Pines J. Four-dimensional control of the cell cycle. Nat Cell Biol. 1999;1:73–79. doi: 10.1038/11041. [DOI] [PubMed] [Google Scholar]
- 8.Morgan DO. Principles of Cdk regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 9.Félix MA, Labbé JC, Dorée M, Hunt T, Karsenti E. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 1990;346:379–382. doi: 10.1038/346379a0. [DOI] [PubMed] [Google Scholar]
- 10.Tyson JJ. Modeling the cell division cycle: cdc2 and cyclin interactions. Proc Natl Acad Sci USA. 1991;88:7328–7332. doi: 10.1073/pnas.88.16.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldbeter A. A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase. Proc Natl Acad Sci USA. 1991;88:9107–9111. doi: 10.1073/pnas.88.20.9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak B, Tyson JJ. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J Cell Sci. 1993;106:1153–1168. doi: 10.1242/jcs.106.4.1153. [DOI] [PubMed] [Google Scholar]
- 13.Sha W, et al. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci USA. 2003;100:975–980. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomerening JR, Sontag ED, Ferrel JE., Jr Building a cell cycle oscillator: Hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 15.Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 16.Chen KC, et al. Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguda BD, Tang Y. The kinetic origins of the restriction point in the mammalian cell cycle. Cell Prolif. 1999;32:321–335. doi: 10.1046/j.1365-2184.1999.3250321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu Z, MacLellan WR, Weiss JN. Dynamics of the cell cycle: Checkpoints, sizers, and timers. Biophys J. 2003;85:3600–3611. doi: 10.1016/S0006-3495(03)74778-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu Z, Weiss JN, MacLellan WR. Regulation of the mammalian cell cycle: A model of the G1-to-S transition. Am J Physiol. 2003;284:C349–C364. doi: 10.1152/ajpcell.00066.2002. [DOI] [PubMed] [Google Scholar]
- 20.Swat M, Kel A, Herzel H. Bifurcation analysis of the regulatory modules of the mammalian G1/S transition. Bioinformatics. 2004;20:1506–1511. doi: 10.1093/bioinformatics/bth110. [DOI] [PubMed] [Google Scholar]
- 21.Novak B, Tyson JJ. A model for restriction point control of the mammalian cell cycle. J Theor Biol. 2004;230:563–579. doi: 10.1016/j.jtbi.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 22.Csikasz-Nagy A, Battogtokh D, Chen KC, Novak B, Tyson JJ. Analysis of a generic model of eukaryotic cell-cycle regulation. Biophys J. 2006;90:4361–4379. doi: 10.1529/biophysj.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlon I, Raff M. Differences in the way a mammalian cell and yeast cells coordinate cell growth and cell-cycle progression. J Biol. 2003;2:7. doi: 10.1186/1475-4924-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitomi M, et al. p27Kip1 and cyclin–dependent kinase 2 regulate passage through the restriction point. Cell Cycle. 2006;5:2281–2289. doi: 10.4161/cc.5.19.3318. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- 27.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 28.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: More for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 29.Matsushime H, et al. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zetterberg A, Larsson O, Wiman KG. What is the restriction point? Curr Opin Cell Biol. 1995;7:835–842. doi: 10.1016/0955-0674(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 31.Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 32.Chau BN, Wang JY. Coordinated regulation of life and death by RB. Nat Rev Cancer. 2003;3:130–138. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- 33.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 35.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 36.Dart DA, Adams KE, Akerman I, Lakin ND. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S phase. J Biol Chem. 2004;279:16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez Y, et al. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 38.Santamaria D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 39.Sage J, et al. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 41.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 42.Pomerening JR, Ubersax JA, Ferrell JE., Jr Rapid cycling and precocious termination of G1 phase in cells expressing CDK1AF. Mol Biol Cell. 2008;19:3426–3441. doi: 10.1091/mbc.E08-02-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray AW, Kirschner MW. Dominoes and clocks: The union of two views of the cell cycle. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- 44.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 45.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: Key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 46.Fujita T, Liu W, Doihara H, Wan Y. Regulation of Skp2–p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am J Pathol. 2008;173:217–228. doi: 10.2353/ajpath.2008.070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacAuley A, Cross JC, Werb Z. Reprogramming the cell cycle for endoreduplication in rodent trophoblast cells. Mol Biol Cell. 1998;9:795–807. doi: 10.1091/mbc.9.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novak B, Tyson JJ. Modeling the control of DNA replication in fission yeast. Proc Natl Acad Sci USA. 1997;94:9147–9152. doi: 10.1073/pnas.94.17.9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doedel EJ. AUTO: A program for the automatic bifurcation analysis of autonomous systems. Congr Numer. 1981;30:265–284. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.