Abstract
The traditional site-directed spin labeling (SDSL) method, which utilizes cysteine residues and sulfhydryl-reactive nitroxide reagents, can be challenging for proteins that contain functionally important native cysteine residues or disulfide bonds. To make SDSL amenable to any protein, we introduce an orthogonal labeling strategy, i.e., one that does not rely on any of the functional groups found in the common 20 amino acids. In this method, the genetically encoded unnatural amino acid p-acetyl-L-phenylalanine (p-AcPhe) is reacted with a hydroxylamine reagent to generate a nitroxide side chain (K1). The utility of this scheme was demonstrated with seven mutants of T4 lysozyme, each containing a single p-AcPhe at a solvent-exposed helix site; the mutants were expressed in amounts qualitatively similar to the wild-type protein. In general, the EPR spectra of the resulting K1 mutants reflect higher nitroxide mobilities than the spectra of analogous mutants containing the more constrained disulfide-linked side chain (R1) commonly used in SDSL. Despite this increased flexibility, site dependence of the EPR spectra suggests that K1 will be a useful sensor of local structure and of conformational changes in solution. Distance measurements between pairs of K1 residues using double electron electron resonance (DEER) spectroscopy indicate that K1 will also be useful for distance mapping.
Keywords: EPR, nitroxides, T4 lysozyme
Site-directed spin labeling (SDSL) has become a powerful method for in vitro studies of both soluble and membrane proteins of arbitrary molecular weight (1–3). In traditional SDSL, a unique cysteine residue is introduced into a recombinant protein via site-directed mutagenesis, and then reacted with a sulfhydryl-specific nitroxide reagent to generate a covalently linked nitroxide side chain. A number of different nitroxide side chains have been used in SDSL, but that designated R1 is the most widely used (Fig. 1A). To study proteins containing reactive native cysteines by SDSL, these residues are replaced by a nonreactive analog (e.g., serine). However, this approach is not always practical because the sulfhydryl group plays a number of important biological roles. In these cases, a labeling strategy that does not rely on the sulfhydryl or any of the other reactive functional groups found in the 20 standard amino acids is needed.
Fig. 1.
SDSL strategies based on cysteine and p-acetylphenylalanine. (A) Reaction of a thiol-containing protein with a sulfhydryl-specific nitroxide reagent generates a disulfide-linked nitroxide side chain (R1). (B) Reaction of a protein containing p-acetylphenylalanine with a ketone-specific reagent (HO-4120) results in a new, ketoxime-linked spin label side chain (K1). (C) Ribbon model of wild-type T4L (PDB ID code 1L63) (17), highlighting the solvent-exposed sites used in this study with spheres at α-carbons. (D) Deconvoluted ESI mass spectra of a reaction mixture of T4L 72p-AcPhe with HO-4120 after 0, 12, or 24 h. (see Materials and Methods for conditions). (E and F) Area-normalized EPR spectra of T4L mutants bearing either the K1 or R1 side chain recorded in a 30% (wt/vol) sucrose solution at 293 K (solid black trace). Overlaid on each spectrum is a nonlinear least-squares fit to the MOMD model (gray dashed trace). Arrows mark the position of a second dynamic component in the spectra of 68K1 and 72K1.
One elegant approach is to genetically incorporate an unnatural amino acid that has unique (“orthogonal”) chemical functionality in the context of the native protein, which allows a spectroscopic probe to be introduced via reaction with a reagent specific for the new functional group (4). In this strategy, a nonnative suppressor tRNA and aminoacyl-tRNA synthetase pair is used to incorporate the unnatural amino acid into the recombinant protein in response to a unique amber (TAG) stop codon (5). To date, >40 unnatural amino acids have been genetically encoded (6–10), with a subset of these amino acids containing chemically reactive functional groups for selective incorporation of biophysical probes.
For this study, the unnatural amino acid p-acetyl-L-phenylalanine (p-AcPhe) (11) was selected for introducing a nitroxide spin label, because it contains the chemically versatile keto functional group that is not present in any of the common amino acids and readily reacts with hydroxylamines in aqueous solution (4). This particular unnatural amino acid can be site-specifically incorporated in response to the nonsense amber codon into recombinant proteins expressed in Escherichia coli (11), Saccharomyces cerevisiae (12), and mammalian cells (13).
Here, we report the preparation of p-AcPhe mutants of T4 lysozyme (T4L), the crystal structure of one of them, and the reaction of each with a hydroxylamine reagent (HO-4120) that yields a ketoxime-linked nitroxide side chain designated K1 (Fig. 1B). EPR spectra of K1 at the solvent-exposed helical sites examined reveal a higher mobility of the nitroxide compared with R1 at the same site, as anticipated from the longer K1 side chain relative to R1. Despite the high mobility of K1, site dependence of the EPR spectra suggest that K1 will be a useful reporter of local structure, and distance measurements between pairs of residues (K1/K1, K1/R1, R1/R1) in T4L by using double electron electron resonance (DEER) show that K1 can be used for distance mapping, particularly for resolving conformational changes.
Results
Preparation and Spin Labeling of T4L p-AcPhe Mutants.
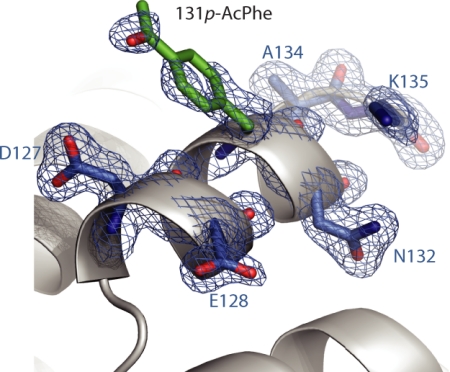
Seven mutants of T4L with a single p-AcPhe residue at each of the sites shown in Fig. 1C were constructed in a his-tagged, cysteine-free background (4), designated WT**. Using an improved system for p-AcPhe incorporation in E. coli (10), high yields of the mutants were obtained in amounts qualitatively similar to the cysteine-free wild-type protein (WT*) (14). The selected sites reside on the solvent-exposed surface of helices where the mutation should produce minimal perturbation to the protein and where the ketone function is accessible for reaction with nucleophiles. One of these sites, 131, has served as a “reference” site for understanding the internal motion of the R1 side chain on solvent-exposed helices (15, 16). To compare with the structure of 131R1, the 1.8-Å crystal structure of a T4L mutant with p-AcPhe substituted at site 131 was solved and refined to an R-factor of 18.9% (Fig. 2); refinement statistics are provided in supporting information (SI) Table S1. At the level of the backbone fold, the structure is essentially identical to WT* (PDB ID code 1L63) (17), as superposition of the two structures yields an average rmsd of 0.27 Å for backbone atoms. As shown in Fig. 2, the p-AcPhe side chain adopts a single {X1,X2} rotamer (X1 = −87°, X2 = −37°; dihedral angles defined in Fig. 1B) that is similar to one of the three preferred rotamers of the parent phenylalanine (see Discussion). In contrast, the 131R1 side chain adopts two equally populated {X1, X2} rotamers (16). Assuming that the K1 derivative of p-AcPhe adopts the same conformation, the single {X1,X2} rotamer will substantially limit the spatial distribution of the nitroxide, an important consideration for use of K1 in distance measurement.
Fig. 2.
Crystal structure of T4L 131p-AcPhe. Electron density map for T4L 131p-AcPhe calculated as an unweighted 2Fo-Fc composite omit map (marine mesh) and contoured at 1.0 σ. For clarity, only a stick model and electron density for 131p-AcPhe and selected nearby residues are shown (as indicated) with the protein backbone displayed as a ribbon model.
The acid-catalyzed reaction of p-AcPhe with the hydroxylamine nitroxide (HO-4120) to generate the K1 side chain was followed by electrospray ionization (ESI) liquid chromatography MS (example in Fig. 1D). Following 12 h of reaction with a tenfold molar excess of HO-4120 at pH 4 and 37°C, a product with a mass increase of 168 ± 1 Da was identified for each mutant (Table S2), which corresponds to the theoretical mass of the predicted product shown in Fig. 1B within the uncertainty of measurement. After 24 h, only the mass of the spin-labeled protein could be detected for all of the mutants tested (e.g., 72p-AcPhe, Fig. 1D), showing that the labeling reaction is either at or very near completion under these conditions. At pH 7 and 37 °C, no product was detected after 24 h of reaction. Although all reactions for the present report were carried out at pH 4, preliminary data indicate that the reaction can be carried out at pH 7 in the presence of p-methoxyaniline as a catalyst (see Fig. S1).
Thermostability of the p-AcPhe and K1 Mutants.
To ascertain the effect of p-AcPhe and K1 substitutions on the stability of T4L, thermal unfolding was monitored by CD spectropolarimetry at 223 nm. For comparison, thermal unfolding of the analogous R1 mutants was also studied. A reversible, two-state transition was observed in all cases investigated (Fig. S2); the thermodynamic parameters obtained from fitting the melting curves to a two-state van 't Hoff unfolding model (18) are provided in Table S3. Destabilizations due to p-AcPhe are small, ranging from 0.2 to 0.6 kcal/mol. A K1 substitution is destabilized by an additional 0–0.9 kcal/mol relative to the analogous p-AcPhe mutant (Table S3), but the overall change in thermostability relative to WT** is generally within the range associated with a natural amino acid substitution at a solvent-exposed helical site (19), as previously observed for R1 (20). The analogous R1 mutants are destabilized by 0.1–0.5 kcal/mol (Table S3).
Internal Motion of the K1 Side Chain Compared with R1.
The room temperature EPR spectrum of each K1 mutant in 30% (wt/vol) sucrose is shown in Fig. 1E. For comparison, the spectra of the analogous R1 mutants are shown in Fig. 1F; with the exception of 86R1, the spectra of the R1 mutants were previously reported (20, 21). To evaluate the relative motions of the nitroxides, the spectra were fit to the macroscopic order microscopic disorder (MOMD) model (22); the fits are shown as dotted traces in Figs. 1 E and F, and parameters determined from the fits are listed in Table S4. As discussed in detail in refs. 15 and 16, the intrinsic internal motion of the R1 side chain at many solvent exposed helical sites is strongly constrained by an intraresidue interaction of the disulfide group with the backbone, which gives rise to a weakly ordered nitroxide motion and a characteristic EPR spectrum illustrated here by 68R1, 72R1, 86R1, and 131R1. At other sites, multiple dynamic states of the nitroxide are observed in the spectrum (e.g., 65R1 and 69R1), which can arise from interactions of the nitroxide with nearby groups in the different substates (23–25). Qualitative interpretation of spectra in terms of local structure and backbone dynamics has been discussed (26).
In contrast to the R1 spectra that reflect an ordered nitroxide motion, the majority of K1 spectra (i.e., 65K1, 69K1, 86K1, 109K1, 131K1) reflect a rapid isotropic motion with effective correlation times (τ) less than 2 ns; site-to-site variation in τ for these mutants (Table S4) may result from differences in local backbone motions. The spectra of 68K1 and 72K1 each have two components, one that corresponds to a rapid isotropic motion that is similar to the aforementioned mutants and a second that reflects an ordered anisotropic motion (denoted by arrows in Fig. 1E), amounting to ≈90% and 50% of the population of 68K1 and 72K1, respectively (Table S4). Because this more restricted motion is site specific, it likely arises from an interaction of the spin-label side chain with nearby groups on the protein surface (see Discussion).
Interspin Distance Measurements with Pulsed Dipolar Spectroscopy.
Measurement of R1 interspin distances based on magnetic dipole–dipole interactions in spin-labeled proteins is an important tool for mapping structure in solution, particularly for cases in which NMR methods are not well suited. DEER spectroscopy (27) has revolutionized interspin distance measurement by extending the distance of detectable dipole–dipole interactions to ≈70 Å, making it a suitable technique for structure mapping in large proteins and complexes (28, 29). Equally important, the distribution of interspin distances can be accurately determined. The primary data obtained in DEER spectroscopy is the amplitude of an electron spin echo of an observed spin as a function of time, corresponding to the application of a pulse that inverts the magnetization of an interacting spin. The dipolar evolution function, obtained by correcting for an exponential background that results from random intermolecular dipolar interactions, is a sum of oscillatory functions whose frequencies correspond to the dipolar interaction strengths for the distances represented. The Fourier transform of the dipolar evolution function is the dipolar spectrum, and either may be fit to obtain the interspin distance probability distribution.
To explore the utility of K1 for distance mapping, three T4L double mutants, each containing a pair of K1 spin labels selected from the set 68K1, 109K1, and 131K1, were studied by DEER spectroscopy. For comparison, analogous R1/R1 and K1/R1 mutants were also studied. The dipolar evolution function and the interspin distance probability distribution for each mutant investigated are shown in Fig. 3 A and B, respectively; the most probable interspin distance and the width of the distribution are provided in Table S5. The interspin distance distribution arises from spatial distribution of the nitroxide and not from an uncertainty in measurement (30). The substitutions were made at sites in well-ordered regions of the protein (17), so it is likely that the distance distributions primarily reflect spatial delocalization of the nitroxide resulting from multiple rotamers of the spin label side chain.
Fig. 3.
Pulsed EPR studies of T4L mutants bearing either two K1 side chains (black), a K1 and an R1 side chain (red), or two R1 side chains (green). (A) Background subtracted dipolar evolution. (B) Area-normalized distance probability distribution from Tikhonov regularization of the data in A. (C and D) Projection contours of the nitroxide spin locations of R1 and K1 calculated from the nine measured distance distributions in B. Probable locations of the nitroxide spins are shown as a gradient from red to yellow and overlaid on a ribbon model of T4L from the crystal structure (PDB ID code 150L, chain C) (46).
In all cases, the most probable distance is longer and the overall distance distribution broader for the K1/K1 mutants pairs compared with the analogous R1/R1 pair; the latter finding was anticipated from the substantially higher mobility (“flexibility”) of the K1 side chain compared with R1 (Fig. 1 E and F).
To evaluate the extent to which the nitroxide is delocalized in the K1 side chain compared with R1, a previously described strategy (31) was used to determine the spatial probability distribution of each nitroxide within the geometric plane defined by the three residues of the various pairs. This analysis utilizes the distance probability distributions of all nine double mutants given in Fig. 3B, and the result is presented in Fig. 3C for R1 and Fig. 3D for K1. The spatial probability is color coded with a gradient from red (low probability) to yellow (high probability). The tangential elongations of the probability distributions result from the limited dataset used, so the distributions are likely to be narrower in this dimension (30). Even at this relatively low resolution, the map reveals that the probability distributions of R1 and K1 are very similar at 131, where the p-AcPhe side chain adopts a single {X1,X2} rotamer (Fig. 2); the small satellite density of 131K1 in Fig. 3D (arrow) is likely an artifact resulting from the limited data set. On the other hand, the distributions of K1 at 109 and 68 are considerably broader than those for R1. For 68K1, and perhaps for 109K1, the distribution is bimodal, probably reflecting multiple rotamers of K1. In this regard, it is significant that the spectrum of 68K1 reveals two dynamic states (Fig. 1E).
Discussion
The purpose of this study was to develop an SDSL strategy that does not rely on any of the functional groups found in the 20 common amino acids. The genetically encoded unnatural amino acid p-AcPhe together with the hydroxylamine nitroxide introduced here serves this purpose. This approach should be applicable to a wide variety of proteins because this particular unnatural amino acid can be incorporated into recombinant proteins expressed from bacterial (11), yeast (12), and mammalian expression systems (13); the only mutation necessary is at the site (or at sites) where introduction of the spin label is desired. Moreover, the p-AcPhe amino acid and the K1 derivative produce tolerably small changes in protein thermal stability (Table S3), and, at least for the 131p-AcPhe mutant, essentially no change in backbone fold. It should be noted that the reduction in free energy of unfolding due to an R1 or K1 substitution could be due to stabilization of the unfolded form rather than destabilization of the native fold.
The reaction of ketones with hydroxylamine compounds has been found to proceed fastest under mildly acidic conditions because of an acid-catalyzed dehydration step (32), and pH 4 was used for forming K1 in the present study. Although the reaction conditions are more extreme than those for forming R1 (and the reaction rate many orders of magnitude slower), the K1 labeling scheme should be applicable to all but relatively unstable proteins or those with acidic isoelectric points where aggregation might be a problem. In such instances, it appears that the HO-4120 labeling reaction can be carried out at pH 7 in the presence of p-methoxyaniline (under an N2 atmosphere; Fig. S1), which has been shown to catalyze the reaction between hydroxylamines and ketones (33).
Model for the Internal Motion of K1.
The fast nanosecond isotropic motion of K1 observed at many sites (Fig. 1E), including the 131 reference site, clearly results from internal motions within the K1 side chain rather than from large-amplitude backbone fluctuations on that time scale. This conclusion follows from the fact that the sites selected all lie within well-ordered helical regions of T4L (17) and that the R1 spectra at the same sites (Fig. 1F) reveal an ordered anisotropic motion characteristic of stable helices with little backbone motion on the nanosecond time scale (20). The K1 side chain has six bonds about which rotations could contribute to the motion of the distal nitroxide ring (in the form of torsional oscillations, rotameric transitions, or both); the C N bond (dihedral X4) is assumed to be rigid with respect to rotation and to adopt a trans configuration, which was observed in structural studies of an analogous ketoxime compound (34). The crystal structure of 131p-AcPhe reveals a single rotameric state of the phenyl group (Fig. 2), suggesting that rotations about the first two bonds might be restricted at this site and others. Torsional oscillations of X3 are assumed to be small on the nanosecond time scale due to conjugation between the aromatic ring and the ketoxime linkage. In contrast, rotations about the three bonds proximal to the nitroxide ring (corresponding to torsions of X5, X6, and X7) are probably only restricted by steric clashes within the side chain. Hence, it is tentatively assumed that rotations about these three bonds are primarily responsible for the large-amplitude isotropic motion reflected in the EPR spectra (Fig. 1E). In contrast, the ordered anisotropic motion of R1 at these same sites (Fig. 1F) arises from motions about just two bonds proximal to the nitroxide ring (15, 16), one fewer than for K1.
N bond (dihedral X4) is assumed to be rigid with respect to rotation and to adopt a trans configuration, which was observed in structural studies of an analogous ketoxime compound (34). The crystal structure of 131p-AcPhe reveals a single rotameric state of the phenyl group (Fig. 2), suggesting that rotations about the first two bonds might be restricted at this site and others. Torsional oscillations of X3 are assumed to be small on the nanosecond time scale due to conjugation between the aromatic ring and the ketoxime linkage. In contrast, rotations about the three bonds proximal to the nitroxide ring (corresponding to torsions of X5, X6, and X7) are probably only restricted by steric clashes within the side chain. Hence, it is tentatively assumed that rotations about these three bonds are primarily responsible for the large-amplitude isotropic motion reflected in the EPR spectra (Fig. 1E). In contrast, the ordered anisotropic motion of R1 at these same sites (Fig. 1F) arises from motions about just two bonds proximal to the nitroxide ring (15, 16), one fewer than for K1.
It is intriguing that a second component reflecting hindered motion is observed in the spectra of 68K1 and 72K1 (arrows, Fig. 1E), but not 65K1 or 69K1. All four sites reside in helix C (Fig. 1C), but 68 and 72 lay on the face that is in close proximity to other elements in the tertiary structure (i.e., helices A and E). Energy minimization of the K1 side chain shows that X3 adopts either 0° or 180° due to the conjugation between C N and the aromatic ring. Based on the {X1,X2} rotamer observed in the crystal structure of 131p-AcPhe, molecular modeling of 68K1 and 72K1 suggests that the two possible X3 isomers have chemically distinct environments, one in which the nitroxide is free in solution and a second that can contact nearby elements of the tertiary structure, thus accounting for the two components observed in the EPR spectra. Because such interactions depend strongly on the tertiary fold, this result suggests that K1 will likely be a useful sensor for local structure and conformational changes.
N and the aromatic ring. Based on the {X1,X2} rotamer observed in the crystal structure of 131p-AcPhe, molecular modeling of 68K1 and 72K1 suggests that the two possible X3 isomers have chemically distinct environments, one in which the nitroxide is free in solution and a second that can contact nearby elements of the tertiary structure, thus accounting for the two components observed in the EPR spectra. Because such interactions depend strongly on the tertiary fold, this result suggests that K1 will likely be a useful sensor for local structure and conformational changes.
Measuring Interspin Distances with K1.
Although a number of spin labels have been developed for SDSL (35), the R1 side chain is almost exclusively used for distance mapping studies by pulsed EPR methods (31, 36–39). The results presented here suggest that K1 will also be useful for such measurements, although the distance probability distribution will likely be broader than for R1 (Fig. 3). The broader distribution, which can accurately be determined by experiment, leads to an increased uncertainty of the relationship between the nitroxide position and Cα, which is important for static structure mapping. However, changes in the probability distribution due to a conformational change in the protein can accurately reflect corresponding changes in the Cα position provided that the rotameric distribution of the side chain itself remains the same. Judicious selection of sites for the introduction of the spin label, based on some structural information, can generally yield this situation as previously discussed (30). Briefly, sites are selected to lay on solvent-exposed surfaces where the nitroxide does not make tertiary interactions, and where the EPR spectrum does not change between conformational states. The first condition can be verified by the EPR spectrum (20); the second condition indicates that only the interspin distance is altered during the transition, not the local environment. The sensitivity to conformational changes is optimized when the two labels point away from one another (i.e., those in which the projection of one Cα–Cβ vector onto the other gives an angle of 180°).
In summary, we have shown site-specific labeling of a genetically incorporated unnatural amino acid containing the ketone group. Proteins not amenable to traditional spin labeling methods based on sulfhydryl reactive reagents can now be studied by SDSL. We envision that this new ketoxime linked spin label will be useful for detecting conformational changes via continuous wave EPR spectroscopy, and for quantitatively characterizing the changes via distance measurement using pulsed EPR methods (e.g., DEER). Also, it should not be overlooked that this technique allows for the introduction of two different spectroscopic probes into a protein via sulfhydryl and keto groups, or for attaching a protein to a solid support at a specific site while at the same time providing chemistry for site-specific attachment of a sulfhydryl spectroscopic probe. Finally, it is likely that this methodology for spin labeling can be extended to other orthogonal chemistries (e.g., “click” chemistry with alkyne- or azide-containing amino acids), or perhaps directly to amino acids containing nitroxide side chains. Exploring these possibilities provides an avenue for optimizing the properties of the spin label side chain for specific purposes.
Materials and Methods
Synthesis of Hydroxylamine Reagent HO-4120.
The reagent HO-4120 (Fig. 1B) was prepared according to the general procedure of Scheme 1. Materials used in the synthesis were obtained from Sigma–Aldrich. Melting points given are uncorrected.
Scheme 1.
Reagents and conditions. (A) N-hydroxyphthalimide (1.05 equiv.), Et3N (2.0 equiv.), DMF, r.t. 3h, 82%. (B) N2H4· H2O (5.0 equiv.), EtOH, 12h, r.t. then PbO2 (1.0 equiv.), O2, 30 min. 43%.
Synthesis of N-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-ylmethoxy)phthal-imide radical [2].
To a stirred solution of N-hydroxyphthalimide (1.71 g, 10.5 mmol) and allylic bromide (40) [1] (10.0 mmol) in dry DMF (10 mL), Et3N (2.02 g, 20.0 mmol) was added at 0 °C, and the reddish-brown mixture was stirred at room temperature (r.t.) for 3 h and then the mixture was poured onto water (200 mL) and the precipitate was filtered, air-dried, and used without further purification in the next step. Yield: 2.58 g (82%), yellow solid, mp 158–160 °C, Rf: 0.55 (CHCl3/Et2O, 2:1). MS EI m/z (%): 315 (M+, 22), 153 (68), 137 (91), 123 (100). IR (nujol): 1,785, 1,730 (C O), 1650 (C
O), 1650 (C C) cm−1. Composition calculated for C17H19N2O4: C, 64.75; H, 6.07; N, 8.88. Found: C, 64.59; H, 6.04; N, 8.84.
C) cm−1. Composition calculated for C17H19N2O4: C, 64.75; H, 6.07; N, 8.88. Found: C, 64.59; H, 6.04; N, 8.84.
Synthesis of 3-aminooxymethyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl radical [3] (i.e., HO-4120).
To a stirred solution of compound 2 (4.0 mmol) in EtOH (30 mL), N2H4–H2O (1.0g, 20.0 mmol) was added dropwise, and the mixture was stirred overnight at r.t. The white precipitate was removed by filtration, the EtOH was evaporated, and the residue was redissolved in CHCl3 (20 mL), and the nonsoluble white precipitate was again removed by filtration. The organic phase was washed with water (10 mL), dried (MgSO4), and PbO2 (956 mg, 4.0 mmol) was added, and O2 was bubbled through the solution for 30 min. The mixture was filtered, and the solvent was removed from the filtrate in vacuo to provide a crude oil that was further purified on silica gel column (elution with CHCl3–MeOH, 99:1 then 80:20) to provide hydroxylamine 3 as orange oil. During purification acetone-free solvents were used. Yield: 320 mg (43%), orange oil, Rf: 0.56 (CHCl3/MeOH, 9:1). MS EI m/z (%): 185 (M+, 15), 153 (47), 138 (85), 123 (48), 81 (100). IR (neat): 3320, 3260 (NH2), 1590 (C C) cm−1. Composition calculated for C9H17N2O2: C, 58.38; H, 9.25; N, 15.12. Found: C, 58.33; H, 9.13; N, 15.01.
C) cm−1. Composition calculated for C9H17N2O2: C, 58.38; H, 9.25; N, 15.12. Found: C, 58.33; H, 9.13; N, 15.01.
Preparation and Spin Labeling of p-AcPhe T4L Mutants.
Amber (TAG) mutants of T4L were generated by the QuikChange Site-Directed mutagenesis method (Stratagene) in a histidine-tagged genetic background (4), designated WT** in this text. Additional information regarding the site-directed mutagenesis method and the pET101/D-TOPO-T4L genetic construct is found in the SI Text. T4L mutants containing p-AcPhe were expressed similar to the method described by Brustad et al. (4); see SI Text for details of the procedure used in this study. Briefly, unnatural amino acid incorporation in E. coli requires two plasmids, one for expression of the gene of interest (containing the TAG codon at the site of interest) and a second that contains the orthogonal tRNA and aminoacyl-tRNA synthetase specific for the unnatural amino acid; in this study, the recently developed pSUPAR plasmid (10) specific for p-AcPhe was used. The unnatural amino acid p-AcPhe (SynChem) was added to the LB medium at inoculation to a final concentration of 2 mM. Mutants were purified by the method described by Fleissner et al. (16), and found to be >95% pure by SDS/PAGE.
Purified mutants were passed over a desalting column (HiPrep 26/10; GE Healthcare) equilibrated with labeling buffer A [50 mM monobasic sodium phosphate, 25 mM NaCl (pH 4.0)], concentrated to a final concentration of 0.1–1 mM and then reacted with a 10-fold molar excess of HO-4120 for 12–48 h at 37 °C. Excess spin reagent was removed by using the above-mentioned desalting column, eluting with a buffer of 50 mM Mops, 25 mM NaCl (pH 6.8) (buffer B). For the three cysteine containing mutants, a 10-fold molar excess of 2,2,5,5-tetramethyl-pyrroline-1-oxyl methanethiosulfonate (40) was added to the elution and allowed to react at ambient temperature overnight; excess spin reagent was removed as just described. Spin labeled T4L was concentrated to ≈1 mM in an Amicon Ultra-5 concentrator (Millipore).
Preparation of R1 and R1/R1 T4L Mutants.
Single- and double-cysteine mutants of T4L were generated by site-directed mutagenesis in the pseudo wild-type (WT*) genetic background (14), which contains the mutations C54T and C97A. Additional information regarding the mutagenic primers and the pET11a-T4L genetic construct is found in the SI Text. The mutants were expressed, purified, and spin labeled with 2,2,5,5-tetramethyl-pyrroline-1-oxyl methanethiosulfonate (40) according to Fleissner et al. (16).
EPR Spectroscopy.
For CW EPR experiments, samples consisted of 400 μM protein in labeling buffer B containing 30% wt/vol sucrose. Spectra were collected at X-band over 100 G on a Bruker Elexys 580 spectrometer fitted with a high-sensitivity resonator using 20 mW incident microwave power and 1 G field modulation amplitude at 100 kHz. Fits of the data to the MOMD model (22) were performed according to the procedure described in Fleissner et al. (16). See additional information in SI Text.
The four-pulse DEER experiment was conducted on a Bruker Elexys 580 spectrometer fitted with an MS-2 split ring resonator according to Altenbach et al. (31). Samples of 200 μM spin-labeled T4L containing 20% glycerol (vol/vol) were loaded into quartz capillaries and then flash-frozen in liquid nitrogen; data were collected at 80 K. A custom program written in LabVIEW was used to subtract the exponential background from the primary data and to determine distance distributions by fitting the resulting dipolar evolution function with Tikhonov regularization (41). For the 2D projection maps shown in Fig. 3 C and D, the geometric positions of the nitroxide spins were optimized by using a Levenberg–Marquardt algorithm according to the procedure described by Altenbach et al. (31). See SI Text for more detail.
CD Spectropolarimetry.
Thermal dentaturation studies were conducted on a Jasco-810 spectropolarimeter equipped with a Peltier temperature controller. Samples consisted of 0.5 mg/mL protein in 20 mM potassium phosphate, 25 mM KCl (pH 3.0.) Ellipticity was monitored at 223 nm, and data were collected at 10-s intervals as the sample temperature was increased from 20 to 80 °C at a rate of 2 °C per minute. Denaturation curves were fit to the equation for a two-state transition of a monomer between a folded and an unfolded state assuming a 2.5 kcal/mol·deg (42) change in the heat capacity of the folded and unfolded forms (18) by a nonlinear least-squares procedure.
X-Ray Crystallography.
The 131p-AcPhe mutant used for crystallization contains the mutations N68C/A93C in the WT* background; these mutations generate a disulfide-linked T4L dimer that facilitates crystallization and yields crystals isomorphous to WT* (43). Similar to the crystallization of other T4L mutants, the hanging-drop method was used to obtain protein crystals using phosphate as the precipitant; see SI Text for more detail. Diffraction experiments were conducted on a single crystal (cryoprotected in mineral oil) maintained in a nitrogen gas cryostream (100 K). Diffraction data were collected and processed according to the method described by Fleissner et al. (16). A molecular replacement solution was found by using WT* T4L (PDB ID code 1C6T) as a starting model with glycine substituted at positions 68, 93, and 131. The model was iteratively built with COOT (44) and refined with PHENIX (45).
Mass Spectrometry.
Electrospray ionization (ESI) mass spectrometry was performed on a single-Quad Agilent 1100 series LCMS with tandem 1100 series ESI mass spectrometer equipped with an Agilent Zorbax 5 μM 300SB-C8 column. Data were collected between 500 and 2000 m/z to obtain raw ESI spectra. Protein masses were calculated by using Agilent Chemstations software with the ESI deconvolution package.
Supplementary Material
Acknowledgments.
We thank Evan K. Brooks (University of California, Los Angeles) for excellent technical assistance, Dr. Joe Horwitz for assistance with the CD spectropolarimetry experiments, Sebastian Peuker for assistance with the p-methoxyaniline catalyzed reaction, and the UCLA–DOE X-Ray Crystallography Core Facility, which was supported by U.S. Department of Energy Grant DE-FC02-02ER63421. This work was supported by National Institutes of Health Grants R01EY05216 (to W.L.H), RT32EY007026 (to M.R.F.), R01GM062159 (to P.G.S.), and the Jules Stein Professor Endowment (W.L.H.). Synthesis of new spin label reagents was supported by Hungarian National Research Funds OTKA T048334 and OTKA-NKTH K67597 (to K.H. and T.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The structure factors and atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3HWL).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912009106/DCSupplemental.
References
- 1.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 3.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 4.Brustad EM, Lemke EA, Schultz PG, Deniz AA. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J Am Chem Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 6.Xie J, Schultz PG. A chemical toolkit for proteins—An expanded genetic code. Nat Rev Mol Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 8.Brustad E, Bushey ML, Brock A, Chittuluru J, Schultz PG. A promiscuous aminoacyl-tRNA synthetase that incorporates cysteine, methionine, and alanine homologs into proteins. Bioorg Med Chem Lett. 2008;18:6004–6006. doi: 10.1016/j.bmcl.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 9.Brustad E, et al. A genetically encoded boronate-containing amino acid. Angew Chem Int Ed Engl. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cellitti SE, et al. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9268–9281. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Brock A, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 14.Matsumura M, Matthews BW. Control of enzyme activity by an engineered disulfide bond. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 15.Columbus L, Kálai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: Analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 16.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson H, Anderson DE, Dao-pin S, Matthews BW. Analysis of the interaction between charged side chains and the alpha-helix dipole using designed thermostable mutants of phage T4 lysozyme. Biochemistry. 1991;30:9816–9828. doi: 10.1021/bi00105a002. [DOI] [PubMed] [Google Scholar]
- 18.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews BW. Studies on protein stability with T4 lysozyme. Adv Protein Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]
- 20.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 21.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 22.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg–Marquardt algorithm. J Magn Reson Series A. 1996;120:155–189. [Google Scholar]
- 23.Guo Z, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17:228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z, Cascio D, Hideg K, Kálai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16:1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langen R, Oh KJ, Cascio D, Hubbell WL. Crystal structures of spin labeled T4 lysozyme mutants: Implications for the interpretation of EPR spectra in terms of structure. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 26.Kusnetzow AK, Altenbach C, Hubbell WL. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pannier M, Vei S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole–dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 28.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 29.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 30.Eaton SS, Eaton GR. In: Biological Magentic Resonance Vol 19: Distance Measurements in Biological Systems by EPR. Berliner LJ, Eaton SS, Eaton GR, editors. New York: Springer; 2000. p. 21. [Google Scholar]
- 31.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jencks WP. Studies on the mechanism of oxime and semicarbazone formation. J Am Chem Soc. 1959;81:475–481. [Google Scholar]
- 33.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic catalysis of oxime ligation. Angew Chem-Int Edit. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 34.Glidewell C, Low JN, Skakle JMS, Wardell JL. Hydrogen-bonded R22 (8) dimers in (E)-{[2-(phenyldiazenyl)phenyl]ethylideneaminooxy}acetic acid. Acta Crystallogr E. 2004;60:o1560–o1562. [Google Scholar]
- 35.Mchaourab HS, Kálai T, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme: Effect of side chain structure. Biochemistry. 1999;38:2947–2955. doi: 10.1021/bi9826310. [DOI] [PubMed] [Google Scholar]
- 36.Alexander N, Bortolus M, Al-Mestarihi A, Mchaourab H, Meiler J. De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure. 2008;16:181–195. doi: 10.1016/j.str.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatnagar J, Freed JH, Crane BR. Rigid body refinement of protein complexes with long-range distance restraints from pulsed dipolar ESR. Methods Enzymol. 2007;423:117–133. doi: 10.1016/S0076-6879(07)23004-6. [DOI] [PubMed] [Google Scholar]
- 38.Hanson SM, et al. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci USA. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hankovszky HO, Hideg K, Lex L. Nitroxyls. VII. Synthesis and reactions of highly reactive 1-oxyl-2,2,5,5-tetramethyl-2,5-dihydropyrrole-3-ylmethyl sulfonates. Synthesis. 1980:914–916. [Google Scholar]
- 41.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Eriksson AE, Baase WA, Matthews BW. Similar hydrophobic replacements of Leu99 and Phe153 within the core of T4 lysozyme have different structural and thermodynamic consequences. J Mol Biol. 1993;229:747–769. doi: 10.1006/jmbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- 43.Heinz DW, Matthews BW. Rapid crystallization of T4 lysozyme by intermolecular disulfide cross-linking. Protein Eng. 1994;7:301–307. doi: 10.1093/protein/7.3.301. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 46.Faber HR, Matthews BW. A mutant T4 lysozyme displays five different crystal conformations. Nature. 1990;348:263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.